Abstract
Spinal muscular atrophy (SMA) is an autosomal recessive motor neuron disease with a high incidence and is the most common genetic cause of infant mortality. SMA is primarily characterized by degeneration of the spinal motor neurons that leads to skeletal muscle atrophy followed by symmetric limb paralysis, respiratory failure, and death. In humans, mutation of the Survival Motor Neuron 1 (SMN1) gene shifts the load of expression of SMN protein to the SMN2 gene that produces low levels of full-length SMN protein because of alternative splicing, which are sufficient for embryonic development and survival but result in SMA. The molecular mechanisms of the (a) regulation of SMN gene expression and (b) degeneration of motor neurons caused by low levels of SMN are unclear. However, some progress has been made in recent years that have provided new insights into understanding of the cellular and molecular basis of SMA pathogenesis. In this review, we have briefly summarized recent advances toward understanding of the molecular mechanisms of regulation of SMN levels and signaling mechanisms that mediate neurodegeneration in SMA.
Keywords: SMA, SMN, JNK, ROCK, ZPR1, MND
Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disorder of early childhood caused by the deletion or mutation of Survival Motor Neuron 1 (SMN1) gene. SMA affects 1 in 6,000 to 1 in 10,000 individuals worldwide.1 Humans have two copies of SMN gene located on chromosome 5q13 that are identified as SMN1 (telomeric) and SMN2 (centromeric).2 The SMN2 gene is almost identical to the SMN1 gene but is unable to produce sufficient amount of full-length transcripts because of a C to T transition in the coding exon 7 that causes alternative splicing and skipping of exon 7, resulting in a truncated protein lacking exon 7 (SMNΔ7) that is not fully functional and degrades rapidly.2–4 However, SMN2 produces low levels (5%–10%) of the full-length SMN protein that are sufficient for survival but result in SMA. The severity of SMA disease inversely correlates with the SMN2 copy number.5–7 Low levels of SMN protein result in the degeneration of spinal motor neurons and cause muscle weakness that is followed by symmetric limb paralysis, respiratory failure, and death.8,9
Currently, there is no treatment for SMA. The development of therapeutic treatments requires understanding of the molecular mechanisms involved in the regulation of gene expression and neurodegeneration. The molecular mechanisms of regulation of SMN2 gene expression and the mechanisms of motor neuron degeneration caused by low levels of SMN in SMA are unclear. However, recent studies have provided insights into the regulation of SMN2 gene expression that may help develop suitable therapeutic strategies. In addition, recent advances toward understanding the signaling pathways activated by low levels of SMN that might mediate neurodegeneration in SMA have provided insights into non-SMN targets as potential therapeutic targets to prevent neurodegeneration.
This review focuses on the role of cellular signaling pathways, extracellular regulated kinase (ERK)/ELK-1, JAK2/signal transducer and activator of transcription 5 (STAT5), and AKT/cAMP response element-binding protein (CREB), in the regulation of transcription of SMN2 gene. In addition, this review discusses the role of Rho kinase (ROCK) and the recently identified c-Jun NH2-terminal kinase (JNK) signaling pathways in mediating neurodegeneration associated with the pathogenesis of SMA.
Regulation of SMN2 Gene Expression
All forms of SMA are caused by insufficient levels of full-length SMN protein, ranging from the most severe type 0 (onset in utero), severe type I (onset 0–6 months), intermediate type II (onset ~6–18 months), mild type III (onset >18 months), and mildest type IV (onset >30 years).8–10 The onset and severity of SMA disease inversely correlate with the amount of full-length SMN protein produced by varying SMN2 copy numbers present in patients with severity ranging from type I to type IV.5–7 Restoration of SMN levels within the central nervous system (CNS), including spinal motor neurons, using transgenic expression of SMN results in the rescue of phenotype, alleviation of SMA pathologies, and increase in lifespan of mice with SMA-like disease.11–14 These findings suggest that restoration of SMN levels in the CNS is sufficient to reduce the severity of disease and improve the SMA phenotype. The SMN2 gene represents a positive modifier and an attractive therapeutic target for producing higher amounts of SMN protein by manipulating the transcription of SMN2 gene.7,15 Understanding the mechanisms of control of the transcriptional regulation of SMN2 gene is one of the important areas of investigation that may lead to identification of viable cellular therapeutic targets to generate sufficient amounts of SMN for the treatment of SMA.
Both SMN1 and SMN2 genes are regulated transcriptionally during cell growth and differentiation.16 Analysis of promoter regions of SMN1 and SMN2 genes shows identical sequences consisting of common cis-regulatory elements required for the initiation and regulation of transcription.17,18 However, both SMN genes show differential expression in neurons and nonneuronal cell types.17 The differential expression of SMN genes in different cell types might be because of the presence of two transcription initiation sites: the first transcription site is located 163 base pairs upstream of the translation start site and the second site is located 246 base pairs upstream of the translation start site.16 A regulatory region of approximately 5 kb upstream of the transcription start site might be involved in the transcriptional regulation of the SMN genes. The upstream regulatory regions (5′-UTR) of the SMN genes contain binding sites for known trans-acting factors, such as ELK-1 (E26 transformation specific [ETS] like or ETS domain containing), CREB, and STAT5 (signal transducers and activator of transcription) that could regulate transcription.17–20
Recent studies have indicated the role of modulation of ELK-1 and CREB activities by mitogen-activated protein kinase (MAPK) signaling pathways in the regulation of SMN2 gene expression. The intracellular Calcium/calmodulin-dependent kinase II (CaMKII)/phosphatidylinositol-3 kinase (PI3K)/AKT/CREB cascade that is known to be a downstream mediator of N-methyl-D-aspartate (NMDA) receptor signaling was found to be activated in the spinal cord explant cultures from mouse models with SMA-like disease, the Taiwanese SMA type II mouse model21 and the severe SMA type I mouse model,22 upon treatment with NMDA.23 Treatments of the spinal cord cocultures with inhibitors for kinases, CaMKII (KN-93) and PI3K (LY294002), abolished NMDA-mediated increase in the levels of SMN. However, treatment with NMDA and U0126, inhibitor of MEK/ERK/ETS like (ELK) pathway that is known to be a target for CaMKII, did not change the levels of SMN expression induced by NMDA. In vivo studies show that the treatment of mice with NMDA improved phenotype, including lifespan of the SMA type II mice.23
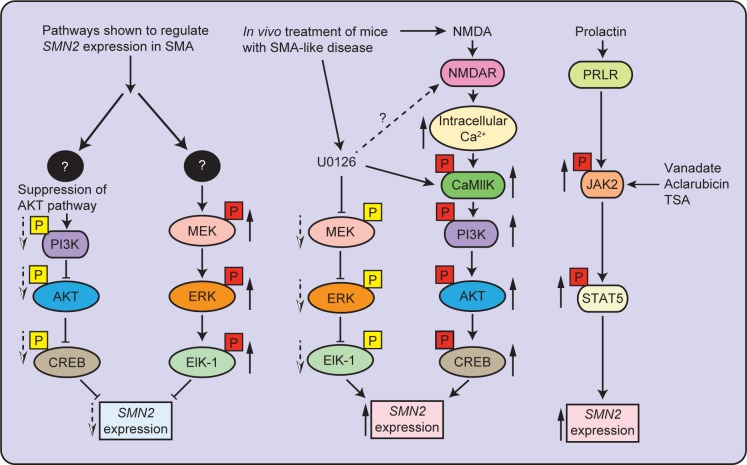
In vitro studies indicated that the presence of intracellular crosstalk between ERK and AKT pathways and shifting of balance of activation from ERK to AKT pathway by inhibition of MEK/ERK/ELK pathway result in increased SMN2 gene expression.24 In vivo inhibition of ERK pathway using the MEK inhibitor (U0126) resulted in the activation of CaMKII/AKT/CREB cascade and an increase in SMN levels in the spinal cords from severe SMA-like mice. Treatment of severe SMA mice with U0126 resulted in improvement of disease phenotype with reduced loss of motor neurons and increased lifespan.24 A recent study showed that the reduced expression of the insulin-like growth factor-1 receptor (Igf-1r) gene results in neuroprotection and improvement in the phenotype of SMA mice. Reduction in IGF-1R levels causes activation of the AKT/CREB pathway and inhibition of the ERK/ELK-1 pathway, which results in higher levels of SMN.25 Together, findings from these studies suggest that the activation of ERK/ELK-1 pathway negatively regulates SMN2 expression and the activation of AKT/CREB pathway stimulates SMN2 expression to increase the levels of full-length SMN. Simultaneous inhibition of ERK pathway and stimulation of AKT pathway results in the upregulation of SMN2 expression in SMA. A graphical summary of signaling pathways regulating the SMN2 gene expression in SMA is presented in Figure 1.
Figure 1.
Mechanisms of regulation of SMN2 gene expression in SMA. SMA is caused by low levels of SMN protein translated from full-length transcripts (5%–10%) generated from the SMN2 gene. Increase in the transcription of SMN2 gene generates higher levels of full-length SMN protein. Signaling pathways identified in SMA that may regulate expression of the SMN2 gene are presented. The MEK/ERK/ELK-1 pathway is activated in SMA and negatively regulates SMN2 expression. Inhibition of ERK pathway in vivo in SMA mice using MEK inhibitor (U0126) results in the upregulation of SMN2 expression by the activation of the PI3K/AKT/CREB pathway. Treatment with NMDA also results in the activation of the AKT/CREB pathway that results in the upregulation of SMN2 expression. The activation of JAK2/STAT5 pathway in vivo by treatment with peptide hormone PRL results in an increase in SMN levels in SMA mice. Solid up arrows (red box) show increase in phosphorylation and dotted down arrows (yellow box) show decrease in phosphorylation.
The effect of different classes of small cell permeable compounds has been examined on increasing levels of SMN protein by enhancing transcription that improve disease phenotype in mice with SMA. These compounds include quinazoline compounds (eg, RG3039) that function as inhibitors of RNA decapping enzyme (DcpS)26–28 and have been shown to improve the disease phenotype, including the lifespan of mice with SMA in different SMA mouse models, severe SMAΔ7 model29–31 and intermediate Smn2B/− model.32 Benefits of RG3039 treatment were observed in the improvement of SMA phenotype, such as increase in the number of spinal motor neurons and increase in the number of SMN-containing gems.31,32 However, in vivo increase in SMN levels was not significant in mice with SMA.32 Another set of compounds known as histone deacetylase (HDAC) inhibitors are valproic acid (VPA), trichostatin A (TSA), LBH589, M344, suberoylanilide hydroxamic acid (SAHA), sodium butyrate, and phenylbutyrate that are shown to increase the levels of SMN.33–41 Other small compounds that have been shown to increase SMN levels include hydroxyurea,42,43 resveratrol,44 and a new class of compound, LDN-76070, whose precise mode of action remains to be examined, which improved the phenotype of SMA mice.45 However, the detailed mechanism of action of these compounds on the regulation of SMN2 expression remains to be studied.
The role of Janus kinase (JAK)/STAT signaling pathway is also shown in the regulation of SMN2 expression. The JAK tyrosine kinase interacts with cytokine and prolactin (PRL) receptors and relays signal downstream by phosphorylation of STAT group of transcription factors; which regulates transcription and are essential for mammalian developmental process, including cell survival, proliferation and differentiation, migration, apoptosis, neuroprotection, and immune cell and mammary gland development.46–50 Cell permeable compounds, sodium valproate, TSA, and aclarubicin, have been shown to activate STAT5 in SMA-like mouse embryonic fibroblasts and motor-neuron-like (NSC34) cells transfected with human SMN2 and induce SMN2 expression.20,51 In addition, a peptide hormone PRL that is known to activate JAK2/STAT5 pathway52 is shown to increase SMN2 expression in neuronal (NT2) cells.19 In vivo activation of JAK2/STAT5 pathway by administration of PRL in mice with severe SMA (SMAΔ7 mice) causes an increase in SMN levels that improves disease phenotype and increases the lifespan of SMA mice.19 The role of JAK2/STAT5 pathway in the regulation of SMN2 expression is presented in graphical form (Fig. 1).
An alternative method has been developed to generate full-length SMN from the SMN2 gene by modifying the processes involved in RNA biogenesis, such as transcription and pre-mRNA processing and splicing using transcriptional activators, small nuclear U RNA, small compounds, and antisense oligonucleotides (ASO) to correct splicing.53–59 The small compounds, pseudocantharidins, a phosphatase (PP2A) activator, which dephosphorylates Tra2-β1, a splicing factor,60 and VPA, a drug approved by the U.S. Food and Drug Administration, which upregulates the levels of Tra2-β1, result in increased incorporation of exon 7 and enhances the levels of full-length transcripts by partially correcting splicing.35 A new class of cell permeable compounds (SMN-C1, SMN-C2, and SMN-C3, developed by PTC Therapeutics) has also been shown to correct SMN2 splicing and improve the phenotype of SMA mice.61 The molecular mechanisms of splicing involved in the exclusion/inclusion of exon 7 in the transcripts generated by the SMN2 gene are recently reviewed elsewhere in detail along with the use of ASO in the correction of SMN2 gene splicing as a potential therapeutic strategy for the treatment of SMA.62–64 The ASO-based approach to correct splicing and increase the levels of SMN is one of the promising therapeutic approaches currently under different phases of clinical trials.54,65
Intracellular Signaling Pathways that Mediate Motor Neuron Degeneration in SMA
In SMA, muscular atrophy is a result of degeneration of spinal motor neurons caused by low levels of SMN protein. SMN is a ubiquitously expressed protein, but why selectively lower spinal cord motor neurons degenerate remains unclear.66,67 The degeneration of motor neurons suggests that the low levels of SMN are unable to support the essential cellular functions required for the survival and maintenance of neurons. The defects in cellular functions, including mRNA biogenesis caused by reduced levels of SMN, might result in the activation of intracellular stress signaling pathways that mediate neurodegeneration in SMA. The intracellular mechanisms that are triggered by the low levels of SMN and mediate neurodegeneration remain unclear. However, a noticeable progress has been made recently to understand the intracellular signaling cascades activated by the low levels of SMN that might mediate neurodegeneration in SMA. The ROCK and the JNK signaling pathways have been shown to be activated by the low levels of SMN in in vitro and in vivo SMA models.
The RhoA/ROCK signaling pathway in SMA
The role of RhoA (a small GTPase) and the immediate downstream target ROCK, RhoA/ROCK signaling, is established in the regulation of cytoskeleton dynamics essential for neuronal growth, differentiation, pathfinding, retraction, and degeneration.68–70 Alterations in the activity of ROCK and its downstream targets, including profilin IIa, cofilin, lim kinases (LIMK), myosin regulatory light chain, and myosin light chain phosphatase (MYPT), are associated with human diseases.71,72 SMN has been shown to interact with ROCK and profilin.73,74 It is suggested that the low levels of SMN result in a free pool of profilin IIa and cause an increase in ROCK/profilin complexes that leads to hyperphosphorylation of profilin IIa in SMA. In vitro studies with knockdown of SMN in neuron-like cells (PC12 and NSC34) indicated the activation of RhoA/ROCK and the phosphorylation of downstream targets, such as profilin IIa, cofilin, LIMK, and MYPT, and suggested that the ROCK pathway might be associated with the pathogenesis of SMA.75,76 It is clear that the low levels of SMN result in the activation of ROCK; however, there is some inconsistency in the literature on the modulation of downstream targets of ROCK that might be because of the use of different cellular and animal SMA models.77,78
The activation of both ROCK and ERK pathways in SMA indicates a possibility of crosstalk because ERK and ROCK can inhibit each other. However, in in vitro SMA cell model, activated ERK was unable to affect the levels of activated ROCK.79 It is possible that in SMN-depleted neuronal cells, ERK activation contributes toward promoting neuronal outgrowth and negatively regulates SMN2 expression with phosphorylation of ELK-1, whereas hyperactivation of ROCK may inhibit neurite outgrowth. A possibility of crosstalk between neurotrophic growth factor signaling and ROCK pathway to regulate neurite outgrowth is also indicated in SMA.80,81 Another possibility of crosstalk exists between ROCK and phosphatase and tensin homolog (PTEN deleted on chromosome 10) pathways because ROCK interacts and phosphorylates PTEN.82 PTEN hydrolyzes phosphatidylinositol (3,4,5)-triphosphate, a second messenger that activates PI3K, and inhibits the activation of AKT mediated by PI3K.83 The downregulation of phospho-AKT is shown in the spinal cords of SMAΔ7 mice and human SMA patients.84
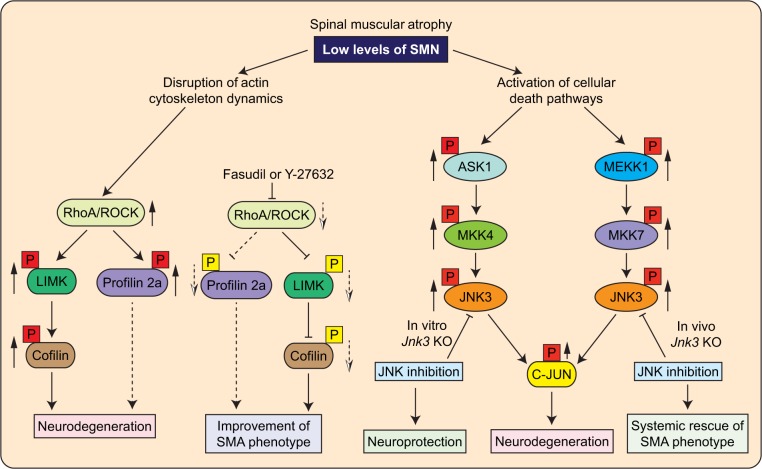
Therefore, ROCK activation in SMA might be involved in the activation of PTEN that leads to inactivation of PI3K/AKT cascade. However, in vivo modulation of PTEN activity under SMA conditions remains to be examined. Nevertheless, studies with the knockdown of PTEN in cultured SMN-deficient motor neurons85 and in mice with SMA have shown beneficial effects on the growth of motor neurons, reduction in the severity of disease, and increase in the lifespan of SMAΔ7 mice.86 A graphical model representing the activation of RhoA/ROCK pathway in SMA is shown in Figure 2.
Figure 2.
The molecular mechanisms that mediate neurodegeneration in SMA. SMA is characterized by degeneration of spinal motor neurons caused by low levels of SMN. SMN deficiency results in the activation of intracellular signaling pathways that mediate the degeneration of neurons in SMA. Rho/ROCK pathway is activated in mice with intermediate SMA and mediates neurodegeneration by disruption of cytoskeleton stability. Inhibition of Rho/ROCK pathway with inhibitors Y-27632 and Fasudil results in the improvement of NMJ pathology and SMA phenotype. The JNK signaling pathway is activated in the spinal cords of SMA patients and SMAΔ7 mice. Two JNK signaling modules, ASK1/MKK4/JNK3 and MEKK1/MKK7/JNK3, mediate in vivo phosphorylation of c-Jun that causes the degeneration and apoptosis of neurons. Genetic inhibition of the JNK pathway by Jnk3 knockout results in the neuroprotection and systemic amelioration of SMA in mice. Black boxes with question marks represent upstream targets, which mediate the effects of changes stemming from the low levels of SMN that remain to be identified. Solid up arrows (red box) show increase in phosphorylation and dotted down arrows (yellow box) show decrease in phosphorylation. Dotted line connectors represent possibilities that need to be confirmed with further studies.
Interestingly, pharmacological inhibition of ROCK using inhibitors (Y-27632 or Fasudil) resulted in a marked increase in the lifespan of an intermediate SMA mouse model (Smn2B/−) without any change in the SMN transcription and protein levels.87–89 However, ROCK inhibition did not result in an increase in the numbers of spinal motor neurons and did not prevent SMN-dependent neuromuscular junction (NMJ) denervation. The improvement in the SMA phenotype might be because of the improvement in the functionality of motor neurons and NMJs and the increase in the skeletal muscle (tibialis anterior) fiber size due to reduction in the levels of phospho-LIMK and phospho-cofilin in SMA mice treated with ROCK inhibitors. The reduction in the levels of ROCK downstream targets, phospho-LIMK and phospho-cofilin, may help stabilize the actin cytoskeleton and improve the functionality of SMN-deficient neuronal and nonneuronal cells.77
The JNK signaling pathway in SMA
The role of JNK group of kinases has been established in neuronal cell growth, differentiation and apoptosis, CNS morphogenesis, memory, and synaptic plasticity.90,91 The JNK group of MAPK is encoded by three genes, Jnk1, Jnk2, and Jnk3, that generate a total of 10 transcripts for multiple isoforms. The Jnk1 and Jnk2 genes show ubiquitous expression, but the Jnk3 gene is mainly expressed in neurons, with some expression in the heart and testis.91,92 The role of JNK has been implicated in neurodegeneration caused by alteration of microtubule stability induced by JNK-mediated phosphorylation of microtubule-associated proteins, including MAP1B, MAP2, Tau, and stathmin (microtubule-destabilizing family of proteins); JNK pathway has been indicated as a potential therapeutic target for the treatment of neurodegenerative diseases, such as Parkinson’s and Alzheimer’s diseases.93–95
The low levels of SMN in neurons cause neurodegeneration in SMA. The stress-activated protein kinases are known to be activated by a variety of extracellular stress signals, such as growth factors, cytokines, and ultraviolet light.91 SMN deficiency may result in intracellular stress that might activate intracellular signaling cascade and lead to neurodegeneration in SMA. We have recently shown the activation of the JNK signaling pathway in the spinal cords of SMA patient and SMAΔ7 mice.84 JNK was activated in cultured spinal cord motor neurons from SMA mice. Knockdown of SMN using RNAi also resulted in JNK activation in cultured neurons. Reduced AKT phosphorylation in the SMA spinal cords is consistent with suppression of AKT pathway during JNK activation.96
Two MAPK signaling modules were identified that lead to the activation of JNK in SMA. These two MAPK signaling modules consist of three-tier of kinases, MAP kinase kinase kinase (MAP3K), MAP kinase kinase (MAP2K), and MAPK. Two MAP3Ks (ASK1 and MEKK1) and two MAP2Ks (MKK4 and MKK7) were activated to phosphorylate JNK (MAPK) in the spinal cords from SMA. Activation of both MKK4 and MKK7 is shown to be required for full in vivo activation of JNK.97 Activation of ASK1 and MEKK1 suggested the possibility of two signaling modules may be involved in the JNK activation because both MKK4 and MKK7 are known to be activated by ASK1 and MEKK1.98 It has been shown that Gemin5, a part of the SMN complex, interacts with ASK1, MKK4, and JNK in 293T cells.99 Gemin5 might act as a scaffold for ASK1/MKK4/JNK signaling module to maintain the specificity of signaling. Low levels of SMN complexes would result in a free pool of Gemin5 that might increase the levels of ASK1/MKK4/JNK complex resulting in higher levels of activated JNK in SMA neurons.
Scaffolding of MEKK1/MKK7/JNK complex by neuron-specific JNK-interacting protein 3 may activate MKK7.100 The marked difference in the levels of activation of MKK4 compared to MKK7 suggests the tight regulation and specificity of the activation of signaling modules. Preferential activation of neuron-specific isoform, JNK3 (MAPK), was detected in SMN-deficient neurons.84 JNK3 deficiency resulted in the protection of cultured neurons with low levels of SMN, suggesting that JNK3 may be a potential target for SMA therapeutic interventions. This study identified two signaling modules, ASK1/MKK4/JNK3 and MEKK1/MKK7/JNK3, that may mediate JNK activation and neurodegeneration in SMA.84 A graphical model representing the activation of JNK by two signaling modules in SMA is shown in Figure 2.
Furthermore, in vivo studies by genetic inhibition of JNK3 in SMAΔ7 mice resulted in the systemic rescue of SMA phenotype, including reduction in the loss of spinal cord motor neurons and muscle degeneration, improvement in muscle fiber thickness, muscle growth, gross motor function and overall growth, and increase in lifespan.84 Interestingly, genetic inhibition of the JNK3 did not alter the levels of SMN in mice with SMA. The findings from this study suggest that the amelioration of SMA phenotype in SMA mice by JNK3 deficiency is SMN independent, and JNK3 represents a non-SMN target. Genetic elimination of the Jnk3 gene in SMA mice validated JNK3 as a potential (non-SMN) therapeutic target.84
Other potential signaling molecules and pathways in SMA
A few other proteins have been identified that may be a part of the intracellular signaling mechanisms contributing toward SMA pathogenesis, including modifier proteins that alter disease phenotype. Humans with homozygous SMN1 deletion and identical SMN2 copy numbers show discordant phenotypes compared to their siblings, suggesting the possibility of SMA modifier genes in addition to SMN2.101–103 Recent studies have identified genes located outside of the 5q SMA locus, such as plastin 3 (PLS3, Chr Xq23) and zinc finger protein 1 (ZPR1, Chr 11q23.3), that have been shown to modify the severity of SMA disease.103,104 PLS3 levels were upregulated in unaffected female SMA patients compared to affected SMA patients (siblings). PLS3 is a calcium-dependent actin-bundling protein and shown to regulate axonogenesis by increasing the levels of F-actin.103,105 Overexpression of PLS3 in cultured SMN-deficient neurons corrected axonal growth defects. PLS3 overexpression moderately improved the SMA phenotype by delaying axon pruning that resulted in improved NMJ functionality in Taiwanese SMA mouse model.106 In another study, PLS3 overexpression did not modify the severity of SMAΔ7 mouse model.107
The reasons for moderate to no improvement in different mouse models are unclear. It is possible that in addition to PLS3 overexpression in unaffected individuals, there would be other proteins/factors whose levels could be altered in a gender-specific manner that contribute to PLS3-dependent discordant phenotype in SMA type II/III patients. It is unclear whether PLS3 overexpression will also provide beneficial effects in severe SMA but warrants further studies. However, identification of the molecular mechanism that upregulates PLS3 levels in unaffected individuals with homozygous SMN1 deletion will provide insights into the alteration of levels of other potential targets that may be operating synergistically with PLS3 in SMA.
ZPR1 is an evolutionary-conserved essential protein108 that is a component of the receptor tyrosine kinase signaling pathways and interacts with the epidermal growth factor receptor and platelet-derived growth factor receptor in quiescent cells.109,110 Treatment of quiescent cells with mitogens or serum results in the formation of ZPR1 complexes with translation elongation factor EF-1a and SMN proteins and translocation to the nucleus.111,112 ZPR1 interacts with SMN and is required for accumulation of SMN in subnuclear bodies, including gems and Cajal bodies. Interaction of ZPR1 with SMN is disrupted in SMA patients, and both ZPR1 and SMN fail to accumulate into nuclear bodies. The defect in nuclear accumulation of SMN is the cellular defect in SMA that may affect the biochemical function of SMN associated with its localization to nuclear bodies. Notably, the severity of SMA disease correlates negatively with the number of SMN bodies.6
ZPR1 is downregulated in SMA patients.104,113 The reduced expression of ZPR1 causes progressive loss of spinal motor neurons in mice.114 The low levels of ZPR1 increase the severity of disease and decrease the lifespan of mice with SMA.104 Overexpression of ZPR1 in fibroblast derived from SMA type I patients restores the accumulation of SMN in subnuclear bodies and increases the levels of SMN. ZPR1 overexpression in spinal motor neurons from SMA mice rescues axonal growth defects.
The role of ZPR1–SMN complexes in the growth and maintenance of neurons is unclear. However, ZPR1 may contribute to the functions of SMN, including mRNA splicing because ZPR1 is a part of the SMN containing cytoplasmic spliceosomal small nuclear ribonucleoprotein (snRNP) complexes and interacts with snurportin 1.115 ZPR1 deficiency causes defects in cellular distribution of snRNPs and in pre-mRNA splicing similar to SMN deficiency.112,116,117 In addition, ZPR1 complexes may also contribute to overall RNA biogenesis, including splicing and transcription.109,118
A recent study showed that ubiquitin-like modifier activating enzyme 1 (UBA1) interacts with SMN and disruption of ubiquitination pathway contributes to the severity of SMA disease.119 The ubiquitination pathway is shown to regulate the stability of SMN protein120 and is involved in mediating synaptic and axonal degeneration.121 Mutations in the human UBE1 (UBA1) gene cause X-linked infantile SMA.122 The reduced levels of UBA1 and the increased levels of β-catenin in SMA mouse models [severe SMA (Smn−/−; SMN2+/+) and Taiwanese SMA (Smn−/−; SMN2tg/0)] indicate an increase in β-catenin signaling that may influence the transcriptional regulation of critical genes.119 However, the expression of specific genes altered by the increased levels of β-catenin that may contribute to SMA pathogenesis remains to be examined. Interestingly, pharmacological inhibition of β-catenin with quercetin, a cell permeable flavonoid,123 improves neuromuscular pathology in different animal models, Drosophila SMA model,124 Zebrafish SMA model,125 and Taiwanese SMA mouse model21 by Gillingwater’ s group.119 However, the inhibition of β-catenin did not improve systemic pathology in SMA mice. Nevertheless, the alteration of ubiquitin homeostasis and β-catenin signaling in SMA suggests that targeting of this pathway may have therapeutic potential to reduce the severity of SMA disease.119
Recent advances made to understand the molecular mechanisms that regulate the expression of SMN2 gene and the cellular mechanisms triggered by the low levels of SMN that mediate neurodegeneration in SMA have provided insights into SMN-dependent and SMN-independent mechanisms and the potential non-SMN therapeutic targets that laid a foundation to develop new strategies for therapeutic intervention in SMA. A summary of the signaling pathways regulating SMN2 expression and the molecular mechanisms mediating the neurodegeneration in SMA is presented in Table 1. In addition, the molecular targets that have been tested to examine the therapeutic potential in preclinical studies using SMA animal models are identified. Recent studies have also provided insights into the complexity of SMA disease as a multisystem disorder, in which the primary pathogenesis is the degeneration of the spinal cord motor neurons and muscle atrophy, accompanied by complications in the development and functioning of multiple nonneuronal organs, including the heart, liver, pancreas, vasculature, respiratory system (lungs, diaphragm, and phrenic nerve), and gastrointestinal system reviewed in recent publications.84,126 Collectively, these advances in the field of SMA point to the development of combinatorial treatments to simultaneously increase the levels of SMN and prevent neurodegeneration using non-SMN targets and SMN-independent mechanisms to restore the normal function of neuronal and nonneuronal tissues and organs.
Table 1.
Signaling pathways and the molecular targets of spinal muscular atrophy.
| MOLECULAR TARGETS/PATHWAYS | CELLULAR MODELa | COMPOUNDS/GENES | OUTCOMES | ANIMAL MODELb | COMPOUNDS/GENES | OUTCOMES | REFERENCES |
|---|---|---|---|---|---|---|---|
| HDAC (Histone deacetylase) | SMA patient fibroblast Organotypic hippocampal slice cultures from rat |
Valproic acid (VPA) (Inhibitor) | Increase in FL-SMN2 mRNA, splicing factors-Htra2-β1 and SR SF2/ASF and SRp20 protein levels. | SMAΔ7 mouse model127 Taiwanese SMA mouse model21 Severe SMA-like mouse model22 | Trichostatin A (TSA), Sodium butyrate SAHA (Inhibitors) | Increase in SMN levels. Improvement in SMA pheno-type and increase in lifespan. |
35a 41a 34b 37b 39b |
| ROCK (Rho kinase) | PC12 cells NSC34 (motor neuron-like cells) HEK293 cells |
Y-27632 (Inhibitor) | Enhanced neurite outgrowth in SMN-deprived NSC34 cells. | Smn2B/− SMA mouse model89 | Y-27632, Fasudil | Increase in skeletal muscle fiber and postsynaptic endplate size. Improvement in SMA phe-notype and increase in lifespan. No change in SMN levels (SMN-independent). |
75a 76a 79a 87b 88b |
| NMDA receptor | Co-cultures of spinal cord expiants and muscle cells | NMDA | Increase in SMN levels. | Severe SMA-like mouse model22 Taiwanese SMA mouse model21 |
NMDA | Increase in SMN levels. Improvement in SMA pheno-type and increase in lifespan. |
23a,b |
| MEK/ERK/ELK-1 | Co-cultures of mouse spinal cord expiants and muscle cells Myogenic precursor cells from SMA type I patients |
U0126 (MEK inhibitor) | Increase in SMN levels. | Severe SMA-like mouse model22 | U0126, AZD6244 (MEK inhibitors) | Increase in SMN levels. Improvement in SMA pheno-type and increase in lifespan. |
24a,b |
| JAK2/STAT5 | SMA-like MEFs SMN2-NSC34 cells SMA-patient lymphocytes MN-1 cells NT2 cells |
Sodium vanadate, TSA and aclarubicin Prolactin, Aurintri-carboxylic acid (ATA; STAT5 activator) |
Increase in SMN levels and nuclear gems, and enhanced axonal outgrowth. Increase in SMN levels. |
SMAΔ7 mouse model127 | Prolactin | Increase in SMN levels. Improvement in SMA pheno-type and increase in lifespan. |
51a 20a 19a,b |
| DcpS (RNA decapping enzyme) | NSC34 (motor neuron-like cells) | D156844 (Inhibitor) | Increase in SMN levels. | Taiwanese SMA model21 and Smn2B/− SMA mouse model89 SMAΔ7 mouse model127 SMAΔ7 mouse model127 |
RG3039 (Inhibitor), D156844, D156844+ follistatin | Improvement in SMA pheno-type and increase in lifespan. Improvement in SMA pheno-type and increase in lifespan with minimal change in SMN levels. Improvement in SMA phenotype and increase in lifespan. |
28a 32b 31b 29b 30b |
| UBA1/β-catenin | NSC34 (motor neuron-like cells) | UBEI-41 (UBA1 inhibitor) | Increase in β-catenin levels | Taiwanese SMA model21 Zebrafish SMA model125 Drosophila SMA model124 |
Quercetin (β-catenin inhibitor) | Improvement in neuromuscular, but not systemic pathology. No change in SMN levels (SMN-independent). |
119a,b |
| PLS3 | HEK293 PC12 cells Spinal cord neurons from SMA mice |
Overexpression of PLS3 | Increase in F-actin levels, stabilization of growth cones, improved axonogenesis and neurite growth. | Zebrafish SMA model125 Taiwanese SMA mouse model21 SMAΔ7 model127 |
Overexpression of PLS3 | Rescued axonal outgrowth defects in motor neurons from SMA mouse and in zebrafish. Improved NMJs, stabilization of axons and increased muscle fiber size. No improvement in SMA phenotype |
103a,b 106b 107b |
| ZPR1 | SMA patient fibroblast Spinal cord motor neurons from SMA mice |
Overexpression of ZPRI | Increase in SMN levels and number of gems. Neurite growth stimulation and rescue of axonal growth defects. | Generation of new mild SMA-like model (Smn−/+; Zpr1−/+)104 Generation of new SMAΔ7 model with Zpr1−/+ (SZ)104 |
Reduced Zpr1−/+ gene dosage. | Increased loss of motor neurons. Hyper-myelination of phrenic nerve. Decrease in lifespan of SMA mice. | 104a,b |
| IGF-1R | MN-1 cells Myogenic precursor cells from SMA type I patients |
Mouse lgf-1r siRNA, Human IGF-1R siRNA |
Increase in SMN levels. | Generation of Taiwanese SMA mouse model with (lgf-1r+/−)25 | Reduced lgf-1r+/− gene dosage. | Increase in SMN levels Improvement in SMA phenotype and increase in lifespan. |
25a,b |
| JNK3 | Neuron-based SMA model (Primary cerebellar granule neurons and SMN knockdown with siRNA) Primary neurons from Jnk3+/+ and Jnk3−/− mice |
JNK3-deficiency | Reduced degeneration of SMN-deficient neurons. | Generation of new SMAΔ7 model with Jnk3−/−-null background (SMA-J3)84 | Genetic inhibition of JNK3 by knockout of the Jnk3 gene. | Reduced spinal motor neuron degeneration, improved motor function and muscle growth. Systemic improvement in SMA phenotype with increase in lifespan. No change in SMN levels (SMN-independent). |
84a,b |
Notes:
In vitro studies using cellular models.
In vivo studies using animal models.
Footnotes
ACADEMIC EDITOR: Lora Watts, Editor in Chief
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 946 words, excluding any confidential comments to the academic editor.
FUNDING: This study was supported by the research grant to LG from the National Institutes of Health (R01 NS064224). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Provenance: the authors were invited to submit this paper.
Author Contributions
Conceived and designed the topic and structure of the review: LG. Prepared first draft of the manuscript: SA. Contributed to the writing of the manuscript: KB and AK. Made critical revisions and prepared final version: LG. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.D’Amico A, Mercuri E, Tiziano FD, Bertini E. Spinal muscular atrophy. Orphanet J Rare Dis. 2011;6:71. doi: 10.1186/1750-1172-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monani UR, Lorson CL, Parsons DW, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 5.Cusco I, Barcelo MJ, Rojas-Garcia R, et al. SMN2 copy number predicts acute or chronic spinal muscular atrophy but does not account for intrafamilial variability in siblings. J Neurol. 2006;253:21–25. doi: 10.1007/s00415-005-0912-y. [DOI] [PubMed] [Google Scholar]
- 6.Lefebvre S, Burlet P, Liu Q, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 7.Prior TW, Krainer AR, Hua Y, et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet. 2009;85:408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 9.Dubowitz V. Major Problems in Clinical Pediatrics. Vol. 16. London; Philadelphia: Saunders; 1978. Muscle disorders in childhood; pp. 1–282. [PubMed] [Google Scholar]
- 10.Dubowitz V. Very severe spinal muscular atrophy (SMA type 0): an expanding clinical phenotype. Eur J Paediatr Neurol. 1999;3:49–51. doi: 10.1053/ejpn.1999.0181. [DOI] [PubMed] [Google Scholar]
- 11.Foust KD, Wang X, McGovern VL, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Gogliotti RG, Quinlan KA, Barlow CB, Heier CR, Heckman CJ, Didonato CJ. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J Neurosci. 2012;32:3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hua Y, Sahashi K, Hung G, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Passini MA, Bu J, Roskelley EM, et al. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirth B, Brichta L, Schrank B, et al. Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet. 2006;119:422–428. doi: 10.1007/s00439-006-0156-7. [DOI] [PubMed] [Google Scholar]
- 16.Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene. 2001;279:109–117. doi: 10.1016/s0378-1119(01)00758-2. [DOI] [PubMed] [Google Scholar]
- 17.Boda B, Mas C, Giudicelli C, et al. Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur J Hum Genet. 2004;12:729–737. doi: 10.1038/sj.ejhg.5201217. [DOI] [PubMed] [Google Scholar]
- 18.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farooq F, Molina FA, Hadwen J, et al. Prolactin increases SMN expression and survival in a mouse model of severe spinal muscular atrophy via the STAT5 pathway. J Clin Invest. 2011;121:3042–3050. doi: 10.1172/JCI46276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ting CH, Lin CW, Wen SL, Hsieh-Li HM, Li H. STAT5 constitutive activation rescues defects in spinal muscular atrophy. Hum Mol Genet. 2007;16:499–514. doi: 10.1093/hmg/ddl482. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh-Li HM, Chang JG, Jong YJ, et al. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 22.Monani UR, Sendtner M, Coovert DD, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in SMN(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 23.Biondi O, Branchu J, Sanchez G, et al. In vivo NMDA receptor activation accelerates motor unit maturation, protects spinal motor neurons, and enhances SMN2 gene expression in severe spinal muscular atrophy mice. J Neurosci. 2010;30:11288–11299. doi: 10.1523/JNEUROSCI.1764-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branchu J, Biondi O, Chali F, et al. Shift from extracellular signal-regulated kinase to AKT/cAMP response element-binding protein pathway increases survival-motor-neuron expression in spinal-muscular-atrophy-like mice and patient cells. J Neurosci. 2013;33:4280–4294. doi: 10.1523/JNEUROSCI.2728-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biondi O, Branchu J, Ben Salah A, et al. IGF-1R reduction triggers neuroprotective signaling pathways in spinal muscular atrophy mice. J Neurosci. 2015;35:12063–12079. doi: 10.1523/JNEUROSCI.0608-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarecki J, Chen X, Bernardino A, et al. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh J, Salcius M, Liu SW, et al. DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem Biol. 2008;3:711–722. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butchbach ME, Singh J, Thorsteinsdottir M, et al. Effects of 2, 4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet. 2010;19:454–467. doi: 10.1093/hmg/ddp510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris AW, Butchbach ME. The effect of the DcpS inhibitor D156844 on the protective action of follistatin in mice with spinal muscular atrophy. Neuromuscul Disord. 2015;25:699–705. doi: 10.1016/j.nmd.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Meerbeke JP, Gibbs RM, Plasterer HL, et al. The DcpS inhibitor RG3039 improves motor function in SMA mice. Hum Mol Genet. 2013;22:40744083. doi: 10.1093/hmg/ddt257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gogliotti RG, Cardona H, Singh J, et al. The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum Mol Genet. 2013;22:4084–4101. doi: 10.1093/hmg/ddt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreassi C, Angelozzi C, Tiziano FD, et al. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet. 2004;12:59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 34.Avila AM, Burnett BG, Taye AA, et al. Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brichta L, Hofmann Y, Hahnen E, et al. Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet. 2003;12:2481–2489. doi: 10.1093/hmg/ddg256. [DOI] [PubMed] [Google Scholar]
- 36.Brichta L, Holker I, Haug K, Klockgether T, Wirth B. In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann Neurol. 2006;59:970–975. doi: 10.1002/ana.20836. [DOI] [PubMed] [Google Scholar]
- 37.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci U S A. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garbes L, Riessland M, Holker I, et al. LBH589 induces up to 10-fold SMN protein levels by several independent mechanisms and is effective even in cells from SMA patients non-responsive to valproate. Hum Mol Genet. 2009;18:3645–3658. doi: 10.1093/hmg/ddp313. [DOI] [PubMed] [Google Scholar]
- 39.Riessland M, Ackermann B, Forster A, et al. SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet. 2010;19:1492–1506. doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 40.Riessland M, Brichta L, Hahnen E, Wirth B. The benzamide M344, a novel histone deacetylase inhibitor, significantly increases SMN2 RNA/protein levels in spinal muscular atrophy cells. Hum Genet. 2006;120:101–110. doi: 10.1007/s00439-006-0186-1. [DOI] [PubMed] [Google Scholar]
- 41.Sumner CJ, Huynh TN, Markowitz JA, et al. Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 42.Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH. Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann Neurol. 2005;58:194–202. doi: 10.1002/ana.20548. [DOI] [PubMed] [Google Scholar]
- 43.Liang WC, Yuo CY, Chang JG, et al. The effect of hydroxyurea in spinal muscular atrophy cells and patients. J Neurol Sci. 2008;268:87–94. doi: 10.1016/j.jns.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Dayangac-Erden D, Bora G, Ayhan P, et al. Histone deacetylase inhibition activity and molecular docking of (e)-resveratrol: its therapeutic potential in spinal muscular atrophy. Chem Biol Drug Des. 2009;73:355–364. doi: 10.1111/j.1747-0285.2009.00781.x. [DOI] [PubMed] [Google Scholar]
- 45.Cherry JJ, Osman EY, Evans MC, et al. Enhancement of SMN protein levels in a mouse model of spinal muscular atrophy using novel drug-like compounds. EMBO Mol Med. 2013;5:1035–1050. doi: 10.1002/emmm.201202305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19:225–268. doi: 10.1210/edrv.19.3.0334. [DOI] [PubMed] [Google Scholar]
- 47.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 48.Digicaylioglu M, Lipton SA. Erythropoietin-mediated neuroprotection involves cross-talk between Jak2 and NF-kappaB signalling cascades. Nature. 2001;412:641–647. doi: 10.1038/35088074. [DOI] [PubMed] [Google Scholar]
- 49.Li WX. Canonical and non-canonical JAK-STAT signaling. Trends Cell Biol. 2008;18:545–551. doi: 10.1016/j.tcb.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murray PJ. The JAK-STAT signaling pathway: input and output integration. J Immunol. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- 51.Andreassi C, Jarecki J, Zhou J, et al. Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- 52.Goffin V, Binart N, Clement-Lacroix P, et al. From the molecular biology of prolactin and its receptor to the lessons learned from knockout mice models. Genet Anal. 1999;15:189–201. doi: 10.1016/s1050-3862(99)00025-x. [DOI] [PubMed] [Google Scholar]
- 53.Dal Mas A, Rogalska ME, Bussani E, Pagani F. Improvement of SMN2 pre-mRNA processing mediated by exon-specific U1 small nuclear RNA. Am J Hum Genet. 2015;96:93–103. doi: 10.1016/j.ajhg.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howell MD, Singh NN, Singh RN. Advances in therapeutic development for spinal muscular atrophy. Future Med Chem. 2014;6:1081–1099. doi: 10.4155/fmc.14.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nizzardo M, Simone C, Dametti S, et al. Spinal muscular atrophy phenotype is ameliorated in human motor neurons by SMN increase via different novel RNA therapeutic approaches. Sci Rep. 2015;5:11746. doi: 10.1038/srep11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palacino J, Swalley SE, Song C, et al. SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol. 2015;11:511–517. doi: 10.1038/nchembio.1837. [DOI] [PubMed] [Google Scholar]
- 58.Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Kelemen O, van Santen MA, et al. Synthesis and characterization of pseudocantharidins, novel phosphatase modulators that promote the inclusion of exon 7 into the SMN (survival of motoneuron) pre-mRNA. J Biol Chem. 2011;286:10126–10136. doi: 10.1074/jbc.M110.183970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naryshkin NA, Weetall M, Dakka A, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science. 2014;345:688–693. doi: 10.1126/science.1250127. [DOI] [PubMed] [Google Scholar]
- 62.Porensky PN, Burghes AH. Antisense oligonucleotides for the treatment of spinal muscular atrophy. Hum Gene Ther. 2013;24:489–498. doi: 10.1089/hum.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh NN, Lee BM, DiDonato CJ, Singh RN. Mechanistic principles of antisense targets for the treatment of spinal muscular atrophy. Future Med Chem. 2015;7:1793–1808. doi: 10.4155/fmc.15.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sivanesan S, Howell MD, Didonato CJ, Singh RN. Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Transl Neurosci. 2013;4:1–7. doi: 10.2478/s13380-013-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Faravelli I, Nizzardo M, Comi GP, Corti S. Spinal muscular atrophy—recent therapeutic advances for an old challenge. Nat Rev Neurol. 2015;11:351–359. doi: 10.1038/nrneurol.2015.77. [DOI] [PubMed] [Google Scholar]
- 66.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 68.Da Silva JS, Medina M, Zuliani C, Di Nardo A, Witke W, Dotti CG. RhoA/ROCK regulation of neuritogenesis via profilin IIa-mediated control of actin stability. J Cell Biol. 2003;162:1267–1279. doi: 10.1083/jcb.200304021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 70.Hall A, Nobes CD. Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos Trans R Soc Lond B Biol Sci. 2000;355:965–970. doi: 10.1098/rstb.2000.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 72.Schofield AV, Bernard O. Rho-associated coiled-coil kinase (ROCK) signaling and disease. Crit Rev Biochem Mol Biol. 2013;48:301–316. doi: 10.3109/10409238.2013.786671. [DOI] [PubMed] [Google Scholar]
- 73.Giesemann T, Rathke-Hartlieb S, Rothkegel M, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with SMN in nuclear gems. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 74.Sharma A, Lambrechts A, Hao le T, et al. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 75.Bowerman M, Shafey D, Kothary R. SMN depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J Mol Neurosci. 2007;32:120–131. doi: 10.1007/s12031-007-0024-5. [DOI] [PubMed] [Google Scholar]
- 76.Nolle A, Zeug A, van Bergeijk J, et al. The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum Mol Genet. 2011;20:4865–4878. doi: 10.1093/hmg/ddr425. [DOI] [PubMed] [Google Scholar]
- 77.Coque E, Raoul C, Bowerman M. ROCK inhibition as a therapy for spinal muscular atrophy: understanding the repercussions on multiple cellular targets. Front Neurosci. 2014;8:271. doi: 10.3389/fnins.2014.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hensel N, Rademacher S, Claus P. Chatting with the neighbors: crosstalk between Rho-kinase (ROCK) and other signaling pathways for treatment of neurological disorders. Front Neurosci. 2015;9:198. doi: 10.3389/fnins.2015.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hensel N, Stockbrugger I, Rademacher S, et al. Bilateral crosstalk of rho- and extracellular-signal-regulated-kinase (ERK) pathways is confined to an unidirectional mode in spinal muscular atrophy (SMA) Cell Signal. 2014;26:540–548. doi: 10.1016/j.cellsig.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 80.Bruns AF, van Bergeijk J, Lorbeer C, et al. Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc Natl Acad Sci U S A. 2009;106:12747–12752. doi: 10.1073/pnas.0900122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hensel N, Ratzka A, Brinkmann H, Klimaschewski L, Grothe C, Claus P. Analysis of the fibroblast growth factor system reveals alterations in a mouse model of spinal muscular atrophy. PLoS One. 2012;7:e31202. doi: 10.1371/journal.pone.0031202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li G, Liu L, Shan C, et al. RhoA/ROCK/PTEN signaling is involved in AT-101-mediated apoptosis in human leukemia cells in vitro and in vivo. Cell Death Dis. 2014;5:e998. doi: 10.1038/cddis.2013.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: the long and the short of it. Trends Biochem Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Genabai NK, Ahmad S, Zhang Z, Jiang X, Gabaldon CA, Gangwani L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum Mol Genet. 2015;24(24):6986–7004. doi: 10.1093/hmg/ddv401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ning K, Drepper C, Valori CF, et al. PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Hum Mol Genet. 2010;19:3159–3168. doi: 10.1093/hmg/ddq226. [DOI] [PubMed] [Google Scholar]
- 86.Little D, Valori CF, Mutsaers CA, et al. PTEN depletion decreases disease severity and modestly prolongs survival in a mouse model of spinal muscular atrophy. Mol Ther. 2015;23:270–277. doi: 10.1038/mt.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bowerman M, Beauvais A, Anderson CL, Kothary R. Rho-kinase inactivation prolongs survival of an intermediate SMA mouse model. Hum Mol Genet. 2010;19:1468–1478. doi: 10.1093/hmg/ddq021. [DOI] [PubMed] [Google Scholar]
- 88.Bowerman M, Murray LM, Boyer JG, Anderson CL, Kothary R. Fasudil improves survival and promotes skeletal muscle development in a mouse model of spinal muscular atrophy. BMC Med. 2012;10:24. doi: 10.1186/1741-7015-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hammond SM, Gogliotti RG, Rao V, Beauvais A, Kothary R, DiDonato CJ. Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late. PLoS One. 2010;5:e15887. doi: 10.1371/journal.pone.0015887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coffey ET. Nuclear and cytosolic JNK signalling in neurons. Nat Rev Neurosci. 2014;15:285–299. doi: 10.1038/nrn3729. [DOI] [PubMed] [Google Scholar]
- 91.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 92.Gupta S, Barrett T, Whitmarsh AJ, et al. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 93.Borsello T, Forloni G. JNK signalling: a possible target to prevent neurodegeneration. Curr Pharm Des. 2007;13:1875–1886. doi: 10.2174/138161207780858384. [DOI] [PubMed] [Google Scholar]
- 94.Junyent F, Verdaguer E, Folch J, et al. Role of JNK in neurodegenerative diseases. In: Torrero DM, Haro D, Valles J, editors. Recent Advances in Pharmaceutical Sciences II. Transworld Research Network; 2012. pp. 15–28. [Google Scholar]
- 95.Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sunayama J, Tsuruta F, Masuyama N, Gotoh Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J Cell Biol. 2005;170:295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weston CR, Davis RJ. The JNK signal transduction pathway. Curr Opin Genet Dev. 2002;12:1421. doi: 10.1016/s0959-437x(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 99.Kim EK, Noh KT, Yoon JH, et al. Positive regulation of ASK1-mediated c-Jun NH(2)-terminal kinase signaling pathway by the WD-repeat protein Gemin5. Cell Death Differ. 2007;14:1518–1528. doi: 10.1038/sj.cdd.4402157. [DOI] [PubMed] [Google Scholar]
- 100.Kelkar N, Gupta S, Dickens M, Davis RJ. Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol Cell Biol. 2000;20:1030–1043. doi: 10.1128/mcb.20.3.1030-1043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hahnen E, Forkert R, Marke C, et al. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet. 1995;4:1927–1933. doi: 10.1093/hmg/4.10.1927. [DOI] [PubMed] [Google Scholar]
- 102.McAndrew PE, Parsons DW, Simard LR, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oprea GE, Krober S, McWhorter ML, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet. 2012;21:2745–2758. doi: 10.1093/hmg/dds102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lyon AN, Pineda RH, Hao le T, Kudryashova E, Kudryashov DS, Beattie CE. Calcium binding is essential for plastin 3 function in SMN-deficient motoneurons. Hum Mol Genet. 2014;23:1990–2004. doi: 10.1093/hmg/ddt595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ackermann B, Krober S, Torres-Benito L, et al. Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum Mol Genet. 2013;22:1328–1347. doi: 10.1093/hmg/dds540. [DOI] [PubMed] [Google Scholar]
- 107.McGovern VL, Massoni-Laporte A, Wang X, et al. Plastin 3 expression does not modify spinal muscular atrophy severity in the 7 SMA mouse. PLoS One. 2015;10:e0132364. doi: 10.1371/journal.pone.0132364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mishra AK, Gangwani L, Davis RJ, Lambright DG. Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes. Proc Natl Acad Sci U S A. 2007;104:13930–13935. doi: 10.1073/pnas.0704915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Galcheva-Gargova Z, Gangwani L, Konstantinov KN, et al. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol Biol Cell. 1998;9:2963–2971. doi: 10.1091/mbc.9.10.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Galcheva-Gargova Z, Konstantinov KNI, Wu H, Klier FG, Barrett T, Davis RJ. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science. 1996;272:1797–1802. doi: 10.1126/science.272.5269.1797. [DOI] [PubMed] [Google Scholar]
- 111.Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol. 1998;143:1471–1484. doi: 10.1083/jcb.143.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- 113.Helmken C, Hofmann Y, Schoenen F, et al. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum Genet. 2003;114:11–21. doi: 10.1007/s00439-003-1025-2. [DOI] [PubMed] [Google Scholar]
- 114.Doran B, Gherbesi N, Hendricks G, Flavell RA, Davis RJ, Gangwani L. Deficiency of the zinc finger protein ZPR1 causes neurodegeneration. Proc Natl Acad Sci U S A. 2006;103:7471–7475. doi: 10.1073/pnas.0602057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum Mol Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gangwani L, Flavell RA, Davis RJ. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol Cell Biol. 2005;25:27442756. doi: 10.1128/MCB.25.7.2744-2756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 118.Gangwani L. Deficiency of the zinc finger protein ZPR1 causes defects in transcription and cell cycle progression. J Biol Chem. 2006;281:40330–40340. doi: 10.1074/jbc.M608165200. [DOI] [PubMed] [Google Scholar]
- 119.Wishart TM, Mutsaers CA, Riessland M, et al. Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J Clin Invest. 2014;124:1821–1834. doi: 10.1172/JCI71318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Korhonen L, Lindholm D. The ubiquitin proteasome system in synaptic and axonal degeneration: a new twist to an old cycle. J Cell Biol. 2004;165:27–30. doi: 10.1083/jcb.200311091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ramser J, Ahearn ME, Lenski C, et al. Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet. 2008;82:188–193. doi: 10.1016/j.ajhg.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park CH, Chang JY, Hahm ER, Park S, Kim HK, Yang CH. Quercetin, a potent inhibitor against beta-catenin/Tcf signaling in SW480 colon cancer cells. Biochem Biophys Res Commun. 2005;328:227–234. doi: 10.1016/j.bbrc.2004.12.151. [DOI] [PubMed] [Google Scholar]
- 124.Chan YB, Miguel-Aliaga I, Franks C, et al. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 125.McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (SMN) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hamilton G, Gillingwater TH. Spinal muscular atrophy: going beyond the motor neuron. Trends Mol Med. 2013;19:40–50. doi: 10.1016/j.molmed.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 127.Le TT, Pham LT, Butchbach ME, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]