Recent studies have identified mutations in the dynein heavy chain gene (DYNC1H1), which lead to 2 closely related human motor neuropathies: a dominant spinal muscular atrophy with lower extremity predominance (SMALED) and axonal Charcot-Marie-Tooth (CMT) disease.1,2 We describe the identification of a novel mutation (p.G807S) in DYNC1H1 as the cause of SMALED.
Case report.
This study received approval from the People's Hospital of Jiangsu Province Human Studies Committee Institutional Review Board for experiments using human subjects. We obtained written informed consent from all subjects (or their guardians) participating in the study. The affected boy was born after 41 weeks of gestation. Pregnancy was uneventful, and birth weight was 3,080 g. His initial development was normal, and head control was recognized at 3–4 months. Late infantile motor development was mildly delayed, and he could walk unassisted at 2 years. An unstable gait persisted thereafter, and he was referred to us at 3 years and 6 months of age for evaluation. On examination he was unable to run, climbed stairs on all fours, walked unsteadily, and fell frequently. In contrast, his arms showed age-appropriate strength and no sensory loss could be appreciated. No cognitive delay was present, and he required a normal educational plan in nursery school. The patient is currently 7 years old and attends a regular primary school.
EMG of the tibialis anterior and genioglossus muscles showed evidence of denervation and reinnervation, consistent with non–length-dependent motor neuron disease. Neither serum transaminase nor creatine kinase was elevated, and brain MRI revealed normal findings. Muscle CT demonstrated severe atrophy and lipid degeneration, predominantly in the bilateral quadriceps femoris muscle. However, the upper limbs and distal lower limbs were not affected. A muscle biopsy from the quadriceps femoris muscle demonstrated severe grouping atrophy of type 2 fibers, with a massive increase in the amount of fibrous tissue and sparse enlarged type 1 fibers. Genomic analysis data showed a heterozygous mutation c.2419G>A in exon 8 of the DYNC1H1 gene, causing a glycine to serine substitution (p.G807S).
Three members of the subject's family have been diagnosed with SMALED. In his mother and grandfather, disease development has steadily progressed and both show moderate proximal lower limb–dominant muscle weakness and atrophy. They can walk unassisted, and both show a waddling gait and positive Gower sign. No sensory disturbance or ataxia is noted, and they do not have any intellectual disability. The 3 patients showed no pathologic mutations in SMN1, SMN2, UBA1, or VAPB.
Discussion.
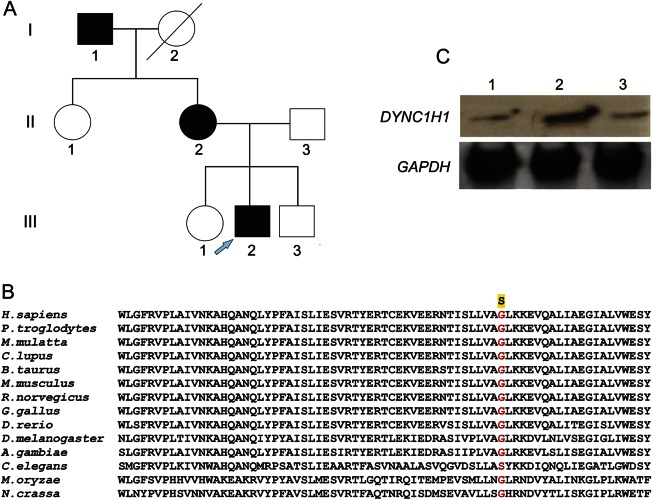
The proband in this study presented with a quadriceps femoris muscle that was almost exclusively involved in the early course of the disease, and the proximal lower limb was predominantly involved throughout disease progression. The subject eventually developed early childhood onset of proximal leg weakness with muscle atrophy, and clinical findings support non–length-dependent motor neuron disease without sensory involvement. His presentation and clinical course were characteristic of SMALED. We found a missense mutation [p.G807S (c.2419G>A)] on exon 8 in DYNC1H1 (figure, A), a mutation not previously been reported as causative for SMALED or CMT disease. The heterozygous missense mutation was confirmed in I-1, II-2, and III-2 (figure, A), and the position is highly conserved (figure, B). This missense mutation was not found in 187 control samples. Clinical features of affected individuals in this family are essentially consistent with those of SMALED, the hallmarks of which are early childhood onset of proximal leg weakness with muscle atrophy and non–length-dependent motor neuron disease without sensory involvement.
Figure. DYNC1H1 mutation in pedigree with dominant spinal muscular atrophy and effect on the dynein complex.
(A) The pedigree of a family with a dominant spinal muscular atrophy with lower extremity predominance. The proband is marked by an arrow. (B) Alignment of the DYNC1H1 protein showing high conservation of the mutated position in exon 8 of DYNC1H1. (C) Dynein from the proband and his parents' fibroblasts were incubated with microtubules in the presence of adenosine triphosphate. After centrifugation, binding of dynein was assessed by immunoblotting for intermediate chain using mouse monoclonal to dynein intermediate chain 1 antibody (ab6304, Abcam). Glyceraldehyde 3-phosphate dehydrogenase was used as internal control. Line 1: mother; line 2: father; line 3: proband.
Thirteen missense mutations identified in human cases of CMT or SMALED are located in the same tail domain of DYNC1H1, which is involved in protein dimerization.3–5 To assess the consequence of mutations in the DYNC1H1 tail domain, we examined the stability and function of dynein complexes purified from dermal fibroblasts derived from the proband and his parents. We observed that dynein complexes from cells containing the DYNC1H1 G807S mutation (the proband and his mother) showed markedly decreased binding to microtubules in the presence of adenosine triphosphate, compared to complexes from the father (figure, C). Our data suggest that the G807S mutation may disrupt dynein complex formation in a dominant negative manner. We further sequenced the DYNC1H1 whole genome and found a splicing variant in an intron involving the reciprocal third nucleotide before position 12685 (c.12685-3C>T) that coexisted with the G807S mutation in both the proband and his mother. This mutation could also affect the function of dynein complexes. Together, our clinical features illustrate a possible clinical-genetic phenotype of SMALED associated with newly identified DYNC1H1 mutations. However, further data will be required to establish possible genotype-phenotype correlations.
Acknowledgments
Acknowledgment: The authors thank patients and controls for having contributed to this study.
Footnotes
Author contributions: Study concept and design: Q.N. and Q.J.; acquisition of data: Q.N. and X.W.; analysis and interpretation of data: Q.N. and M.S.; drafting of the manuscript: Q.N. and Q.J.; critical revision of the manuscript for intellectual content: Q.N. and Q.J.; technical and material support: M.S.; study supervision: Q.J.
Study funding: Funding for this study was provided by grants from the National Natural Science Foundation of China (#30870125) and the Jiangsu Province Natural Science Foundation (#SBK201402229).
Disclosure: Dr. Niu, Dr. Wang, Dr. Shi, and Dr. Jin report no disclosures. Go to Neurology.org/ng for full disclosure forms. The Article Processing Charge was paid by the authors.
References
- 1.Weedon MN, Hastings R, Caswell R, et al. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2011;89:308–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harms MB, Ori-McKenney KM, Scoto M, et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology 2012;78:1714–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eschbach J, Sinniger J, Bouitbir J, et al. Dynein mutations associated with hereditary motor neuropathies impair mitochondrial morphology and function with age. Neurobiol Dis 2013;58:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Punetha J, Monges S, Franchi ME, et al. Exome sequencing identifies DYNC1H1 variant associated with vertebral abnormality and spinal muscular atrophy with lower extremity predominance. Pediatr Neurol 2015;52:239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scoto M, Rossor AM, Harms MB, et al. Novel mutations expand the clinical spectrum of DYNC1H1-associated spinal muscular atrophy. Neurology 2015;84:668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]