Abstract
Objective:
To assess the presence of DEPDC5 mutations in a cohort of patients with epileptic spasms.
Methods:
We performed DEPDC5 resequencing in 130 patients with spasms, segregation analysis of variants of interest, and detailed clinical assessment of patients with possibly and likely pathogenic variants.
Results:
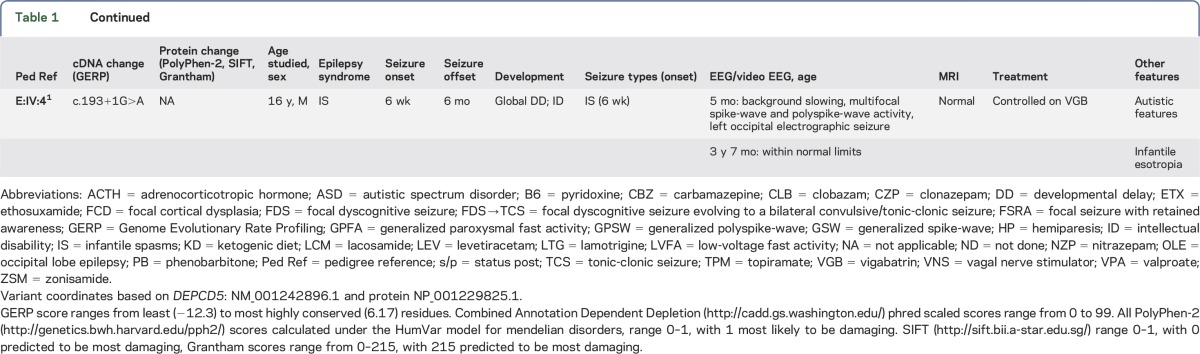
We identified 3 patients with variants in DEPDC5 in the cohort of 130 patients with spasms. We also describe 3 additional patients with DEPDC5 alterations and epileptic spasms: 2 from a previously described family and a third ascertained by clinical testing. Overall, we describe 6 patients from 5 families with spasms and DEPDC5 variants; 2 arose de novo and 3 were familial. Two individuals had focal cortical dysplasia. Clinical outcome was highly variable.
Conclusions:
While recent molecular findings in epileptic spasms emphasize the contribution of de novo mutations, we highlight the relevance of inherited mutations in the setting of a family history of focal epilepsies. We also illustrate the utility of clinical diagnostic testing and detailed phenotypic evaluation in characterizing the constellation of phenotypes associated with DEPDC5 alterations. We expand this phenotypic spectrum to include epileptic spasms, aligning DEPDC5 epilepsies more with the recognized features of other mTORopathies.
Autosomal dominant mutations in DEPDC5 cause familial focal epilepsy with variable foci (FFEVF).1,2 FFEVF is characterized by seizures arising from different cortical regions in different family members, and onset ranges from infancy to adulthood.1,3–7 Clinical presentation is highly variable and reduced penetrance of ∼66% is usual.1,2 Families may show patterns that are effectively subsets of FFEVF, such as a phenotypically homogeneous pattern of autosomal dominant nocturnal frontal lobe epilepsy; individuals with rolandic epilepsy have also been described.1,2,8–10 Recently, DEPDC5 mutations were reported in patients with various brain malformations, challenging the long-held distinction between lesional and nonlesional epilepsies.11–13
DEPDC5 forms part of the GATOR1 complex, a negative regulator of the mammalian target of rapamycin (mTOR) pathway.14 Mutations in other mTOR pathway proteins TSC1 and TSC2 lead to tuberous sclerosis complex (TSC). Infantile spasms are common in TSC, the prototypical disorder of the mTOR pathway.
Infantile spasms are part of the clinical triad of epileptic spasms, hypsarrhythmia, and developmental arrest or regression that forms the infantile spasms syndrome or West syndrome.15 While epileptic spasms typically begin in infancy at around 6 months, later onset may occur. The etiology of infantile spasms is an important determinant of developmental outcome but is unknown in one-third of cases.16,17 Given that spasms are a frequent feature in TSC and we observed spasms in 1 family in our initial report of DEPDC5 in FFEVF,1 we systematically searched for DEPDC5 variation in 130 patients with epileptic spasms.
METHODS
Standard protocol approvals, registrations, and patient consents.
Patients were recruited from the epilepsy clinic at Austin Health, from the practices of investigators, and by referral for epilepsy genetics research from around Australia and internationally. Electroclinical phenotyping was performed as previously outlined. In patients with DEPDC5 variants, brain MRI was systematically reviewed by a pediatric neuroradiologist. Informed consent was obtained from the patient or his or her parent or legal guardian. The study was approved by the Institutional Review Boards of Austin Health and The University of Washington.
Targeted resequencing of DEPDC5.
We used molecular inversion probes to capture all exons and 5 base pairs of flanking intronic DEPDC5 sequence; next-generation sequencing and data analysis were performed as described previously in 130 patients with epileptic spasms of unknown etiology.15,18 Known epileptic encephalopathy genes had been excluded in many cases (unpublished data, Carvill et al., January 2015).18
We considered only nonsynonymous, splice site, or frameshift variants that were present at an allele frequency <1% in ∼61,000 exomes of the Exome Aggregation Consortium (ExAC) data set (to exclude single nucleotide polymorphisms) (http://exac.broadinstitute.org/) for further analysis and performed segregation analysis for these rare variants in available family members. We considered truncating variants to be pathogenic and missense variants that were either inherited from an affected parent or arose de novo to be possibly pathogenic. Maternity/paternity was confirmed using the PowerPlex S5 system (Promega, Madison, WI). We included an additional novel DEPDC5 case, identified through commercial genetic testing (D:II:1), and additional phenotypic data on cousins from our earlier report (family E).1
Accession numbers.
DEPDC5: mRNA NM_001242896.1 and protein NP_001229825.1.
RESULTS
Molecular analysis.
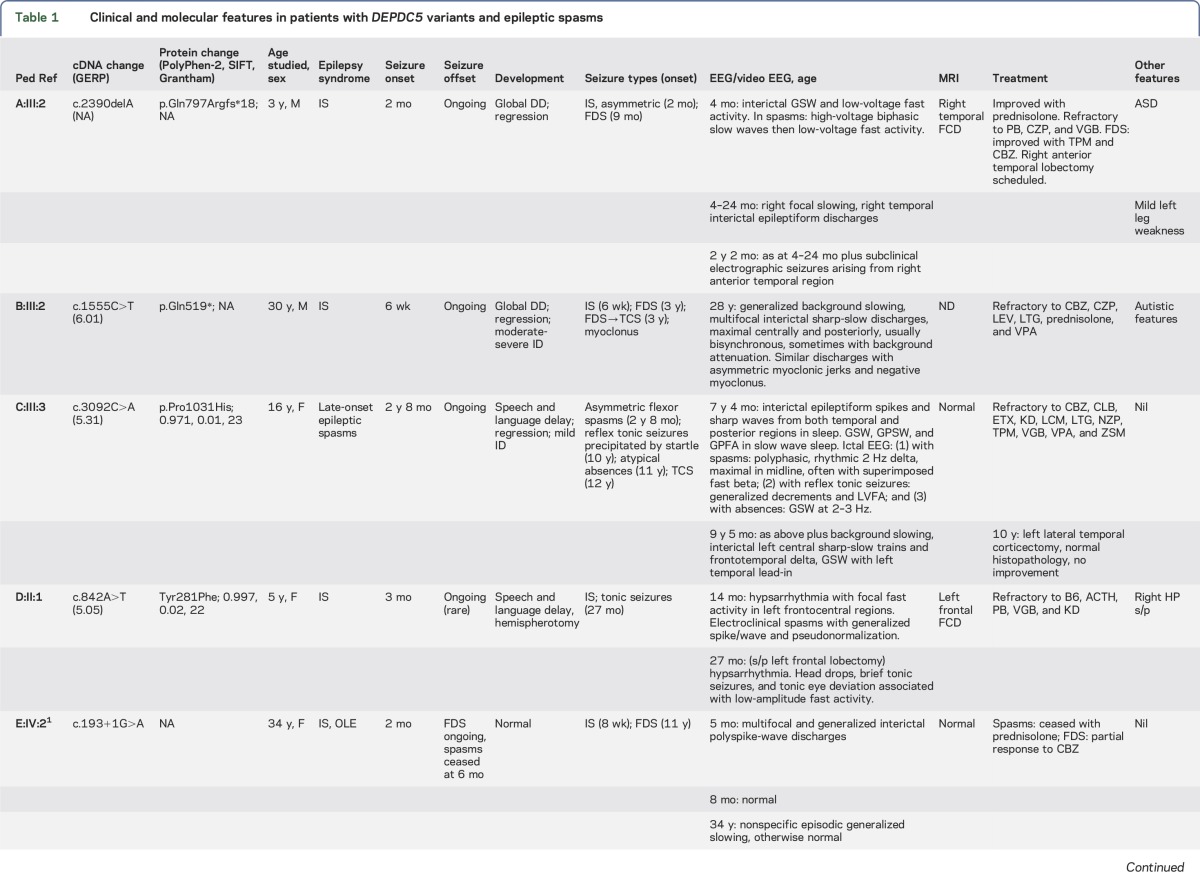
We sequenced all DEPDC5 target base pairs to a depth of 50× at a median of 90% across all samples. We identified likely pathogenic variants in 3 of 130 (2.3%) patients with epileptic spasms of unknown etiology (table 1, figures 1 and 2). A total of 92 of 130 patients had West syndrome, while 38 had epileptic spasms in association with other epileptic encephalopathies. In 2 cases (families A and C), the DEPDC5 variants occurred de novo. Patient A:III:2 had a truncating mutation not present in controls. Patient B:III:2 inherited a truncating mutation not seen in controls from a father with frontal lobe epilepsy. Patient C:III:3, who was of Chinese descent, had a de novo missense variant that disrupted a highly conserved nucleotide, which was predicted to be damaging by 2 of 3 in silico tools (table 1). This variant occurred in 71 individuals in ExAC, including 66 of 8,732 individuals of East Asian descent (minor allele frequency [MAF] 0.7%).
Table 1.
Clinical and molecular features in patients with DEPDC5 variants and epileptic spasms
Figure 1. Pedigrees of families with DEPDC5 variants and epileptic spasms.
We describe 6 patients in 5 families with DEPDC5 variants and spasms. Variants arose de novo in 2 cases (families A and C) and were inherited in 3 (families B, D, and E).
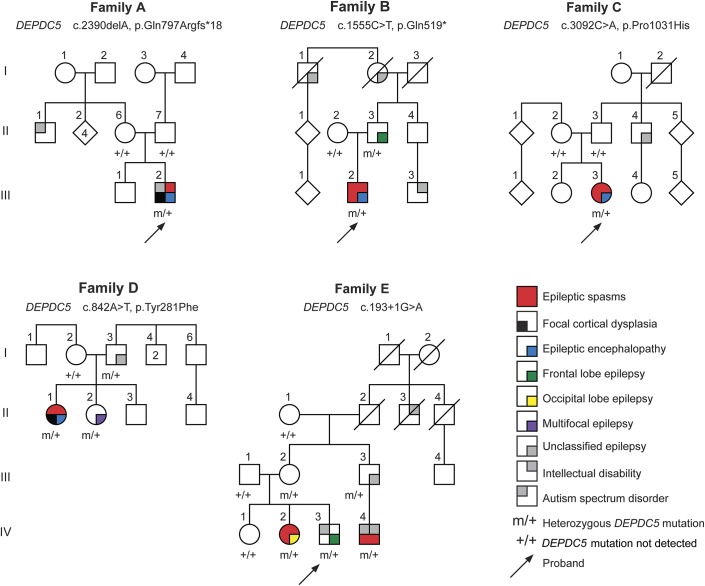
Figure 2. Distribution of DEPDC5 variants in patients and controls.
Upper panel of the schematic shows all previously reported truncating mutations (black)1,2,8–13 and those described in this study (red). Lower panel shows all missense mutations in previous studies (black) and those described in this study (red), numbers in parentheses show the highest population MAF from the ExAC data set, and black lines show the missense variants present in ExAC and variable frequencies. Many of the missense mutations described in patients are present at appreciable frequencies in controls, and there are many missense variants across the gene. It will be vital to perform functional experiments to test the functional effect of these variants, to understand whether and how they cause disease, and to understand the incomplete penetrance that is a common feature of this disorder. ExAC = Exome Aggregation Consortium; MAF = minor allele frequency.
We describe 2 additional families with inherited DEPDC5 variants in whom 1 or more affected family members presented with spasms (table 1, figure 1). Family D, ascertained through commercial testing, had a Tyr281Phe variant, which is highly conserved and predicted to be damaging by 2 of 3 tools (table 1). This variant was present in 5 of 67,552 (MAF 0.0007%) individuals of non-Finnish European descent. Family E carries a splice site mutation, and we describe 2 patients with infantile spasms from this previously reported family.1
Clinical characterization.
Spasms were the presenting seizure type in each case, with onset at 6–12 weeks in 5 cases and late onset at 2 years, 8 months in 1. Two had easily controlled spasms, both with offset at 6 months. In 4 cases, spasms had focal electroclinical features. EEG features included multifocal epileptiform abnormalities, generalized paroxysmal fast activity, and slow spike waves (figure e-1 at Neurology.org/ng). Hypsarrhythmia was seen in 1 patient. Three cases had later focal seizures with impaired awareness and onset from 9 months to 13 years; all were refractory. Atypical absence seizures beginning at age 11 occurred in C:III:3.
One patient, E:IV:2, had normal intellect and no developmental regression with spasm onset; at 34 years, she was a professional. Three patients showed regression with seizure onset and 1 was never normal. Patient C:III:3 showed an additional later decline at age 15 years. Three had autistic features.
Brain MRI revealed temporal focal cortical dysplasia (FCD) in A:III:2 and frontal FCD in D:II:1 (figure e-2). MRI was normal in 3 cases and not performed in 1. Epilepsy surgery was performed in 2 cases. D:II:1 had a histologically confirmed left frontal FCD type IIA (figure e-3). After anatomic left frontal lobectomy, the patient was seizure-free for 6 months and then had return of head drops and tonic seizures. Repeat surgery with a functional left hemispherectomy resulted in seizure freedom for 2.5 years. She now has monthly staring spells. Left lateral temporal corticectomy in C:III:3 was unsuccessful; pathology was normal.
DISCUSSION
We first identified DEPDC5 in familial focal epilepsy, and here we show its relevance to epileptic spasms, illustrating the convergence of phenotypes in genetic mutations of the mTOR pathway. Our findings suggest that greater significance should be attributed to a family history of focal seizures in patients with epileptic spasms. Affected family members had focal epilepsies emanating from different cortical regions, consistent with the pattern of FFEVF.
Infantile spasms have an identifiable etiology in ∼60% cases and may include hypoxic-ischemic or metabolic encephalopathies, malformations, infection, and chromosomal anomalies.16 A family history of spasms is rare but has been described in conditions such as TSC and specific genetic diseases such as ARX.19 The importance of de novo mutations has recently been emphasized, with a pathogenic mutation attributed to a single gene identified in 5%–16% of cases (n = 268 from 3 studies using next-generation sequencing technologies) (table e-1). The most frequently mutated genes were STXBP1 (n = 6), CDKL5 (n = 2), KCNQ2 (n = 2), and ALG13 (n = 2).18,20,21 Our finding of a DEPDC5 variant in up to 2.3% of patients in our cohort suggests that this gene may be one of the more frequent genes associated with epileptic spasms, taking into account that this cohort had been previously screened for many of the known genes. As only 1 patient showed classic hypsarrhythmia, the cohort may have some fundamental differences from other studies of infantile spasms in which hypsarrhythmia is essential for inclusion.
We identified 3 truncating mutations: 1 occurred de novo and the remaining 2 were inherited. This is in keeping with previous reports in which the overwhelming majority of pathogenic DEPDC5 mutations resulted in premature truncation of the protein (figure 2), suggesting that DEPDC5 mutations cause disease by haploinsufficiency of the protein. This is further supported by the presence of only 15 truncating variants in the ∼61,000 exomes in ExAC. However, we report 2 missense variants, Pro1031His (MAF 0.7%) and Tyr281Phe (MAF 0.007%), and there are more than 400 missense variants in ExAC. Tyr281Phe is exceedingly rare, and incomplete penetrance of DEPDC5 mutations may explain the presence of these variants in the population. However, the 0.7% MAF of Pro1031His (arose de novo in family C) seems too high in this population to be explained solely by incomplete penetrance, and this result needs to be interpreted with caution. This may also be true for several reported missense mutations also present in controls at low frequencies (figure 2).1,8 It will be important to develop robust functional experiments to assess the pathogenicity of these missense variants in the future.
We have combined a targeted resequencing approach in 130 patients and results from clinical diagnostics and extended phenotyping in a known mutation-positive family to determine several notable features that highlight the variability of onset and outcome of DEPDC5-associated spasms. Our findings expand the DEPDC5 phenotypic spectrum to include more severe epilepsies presenting with spasms. First, the outcome may be excellent with normal intellect, although most of our patients had intellectual disability with or without autistic features. Second, seizures were controlled with monotherapy in 2 patients. Third, 1 patient had later onset of spasms in the third year with further cognitive decline in adolescence. Fourth, 2 patients had malformations with FCD; in 1, surgery resulted in seizure improvement. Our findings show that DEPDC5 variants are associated with FCD type IIA. Surprisingly, only 1 patient showed hypsarrhythmia on EEG; however, all had highly abnormal EEGs with abundant epileptiform activity, which can be associated with epileptic spasms. The absence of classic hypsarrhythmia means that these patients differ from those with West syndrome, which has been the focus of many recent genomic studies.
Of note, spasms were present in patients with FCD (2 patients) and those without FCD (3 patients) after careful scrutiny of the MRI. Because many patients with focal epilepsies and DEPDC5 mutations have normal MRI,1,11,12 the presence of detectable lesions is not necessary for the development of seizures. Rather, loss-of-function mutations in DEPDC5, an inhibitor of the mTOR pathway, presumably lead to excessive signaling of this pathway, which has many functions that could conceivably contribute to hyperexcitability. Moreover, exactly how DEPDC5 mutations lead to a cortical malformation is not known. It has been hypothesized that the presence of a “second hit” is required for the development of these lesions.11 This scenario would be analogous to TSC, in which a second mutation in the mTOR regulators TSC1 and TSC2 is reported in some tumors of patients with TSC.22 In patients with DEPDC5 mutations, this “second hit” could occur either on the other allele or on another gene involved in the mTOR pathway. Alternatively, an acquired cause, such as a human papillomavirus, has been conjectured to be a “second hit” in FCD.23 In the future, deep targeted or even whole-exome sequencing should be performed on resected tissue to explore this hypothesis.
Given the incomplete penetrance of DEPDC5 mutations and the discovery of both inherited and de novo mutations, molecular approaches for epileptic spasms should interrogate both inheritance patterns. The detection of inherited mutations has important reproductive counseling implications for families of children with spasms, which needs to incorporate increased risk for comorbidities such as intellectual disability and autism spectrum disorders. The recognition of spasms in DEPDC5 epilepsies parallels other mTORopathies such as TSC and STE20-related kinase adaptor alpha syndrome.24 Although many DEPDC5 epilepsies are milder, the convergence of the phenotypic spectrum and molecular pathways suggests that targeted mTOR therapies may benefit patients with the more severe DEPDC5 disorders.24
Supplementary Material
GLOSSARY
- ExAC
Exome Aggregation Consortium
- FCD
focal cortical dysplasia
- FFEVF
familial focal epilepsy with variable foci
- MAF
minor allele frequency
- mTOR
mammalian target of rapamycin
- TSC
tuberous sclerosis complex
Footnotes
Supplemental data at Neurology.org/ng
AUTHOR CONTRIBUTIONS
G.L.C.: drafted/revised the manuscript, study design, acquisition of data, and data analysis. D.E.C.: drafted/revised the manuscript, acquisition of data, and data analysis. B.M.R.: data analysis and drafted/revised the manuscript. J.M.M.: data analysis. J. Saykally and M.Z.: acquisition of data and data analysis. A.L.S.: data analysis. L.D.: acquisition of data and revised the manuscript. K.B.H.: data analysis and drafted/revised the manuscript. S.M.: data analysis. R.J.L., A.S.H., S.A.M., S.F.B., and J. Sullivan: acquisition of data and data analysis. I.E.S. and H.C.M.: drafted/revised the manuscript, study design, acquisition of data, and data analysis.
STUDY FUNDING
This work was supported by funding from the NIH (NINDS 5R01NS069605) to H.C.M. H.C.M. is a recipient of the Burroughs Wellcome Fund Career Award for Medical Scientists. G.L.C. is supported by a postdoctoral fellowship from the Epilepsy Foundation and the Lennox and Lombroso Fund and the NIH (NINDS 1K99NS089858). I.E.S., S.F.B., and L.D. are supported by a National Health and Medical Research Council of Australia (NHMRC) Program Grant. I.E.S. is supported by an NHMRC Practitioner Fellowship and L.D. is supported by an NHMRC Career Development Fellowship (1032603). K.B.H. is supported by the Gustav Nossal NHMRC Postgraduate Scholarship and the Clifford PhD Scholarship. The work was also supported by the University of Washington Intellectual and Developmental Disabilities Research Center Genetics Core (NIH U54HD083091).
DISCLOSURE
Dr. Carvill is a member of the scientific advisory board of Ambry Genetics and has received grants from the NIH/National Institute of Neurological Disorders and Stroke, the Epilepsy Foundation and Lennox and Lombroso Trust, and Citizens United for Research in Epilepsy. Dr. Crompton has received research support from the National Health and Medical Research Council of Australia. Dr. Regan reports no disclosures. Dr. McMahon has received research support from the National Health and Medical Research Council of Australia. Dr. Saykally, Dr. Zemel, Dr. Schneider, and Dr. Dibbens report no disclosures. Dr. Howell has received research support from the National Health and Medical Research Council Gustav Nossal postgraduate scholarship and her husband holds stock options for General Electric (medical imaging equipment). Dr. Mandelstam has been an employee of the Royal Children's Hospital, the Melbourne Brain Centre, and the University of Melbourne. Dr. Leventer has received honoraria for travel from the Asia Oceania Congress of Child Neurology and has received research support from the National Health and Medical Research Council Project Grant 1068278, the National Health and Medical Research Council Project Grant 1059666, and Campbell Edwards Trust Research Support. Dr. Harvey reports no disclosures. Dr. Mullen has been an employee of the Florey Institute of Neuroscience and Mental Health, the University of Melbourne, Australia Austin Health, and Australia Eastern Health; has received travel funding from the American Epilepsy Society Young Investigator Award; and has received research support from the NHMRC Early Career Fellowship and NHMRC Project Grant. Dr. Berkovic has served on the scientific advisory board of UCB Pharma; has served on the editorial boards of Brain, Epileptic Disorders, and Lancet Neurology; has received research support from the National Health and Medical Research Council of Australia; has received honoraria from UCB; has a patent for PCDH19 testing planned and is one of the inventors listed on a patent held by Bionomics Inc. on diagnostic testing of using the SCN1A gene; has received payment for development of educational presentations from UCB Pharma, Novartis Pharmaceuticals, Sanofi-Aventis, and Janssen-Cilag; and has been a consultant to Bionomics and Athena Diagnostics. Dr. Sullivan has been the chair of SAB for nonprofit entity PCDH19 Alliance; has received honoraria from Zogenix; and has received research support from Marinus Pharmaceuticals. Dr. Scheffer has served on scientific advisory boards for UCB and Janssen-Cilag EMEA; has served on the editorial boards of the Annals of Neurology, Neurology, Epilepsy Currents, Progress in Epileptic Disorders series, Virtual Neuro Centre, and Epileptic Disorders; holds patents for methods of treatment and diagnosis of epilepsy by detecting mutations in the SCN1A gene, a diagnostic method for epilepsy, mutations in ion channels, diagnostic and treatment methods relating to autosomal dominant nocturnal frontal lobe epilepsy (pending), gene and mutations thereof associated with seizure disorders, a gene and mutations thereof associated with seizure and movement disorders, and diagnostic and therapeutic methods for EFMR; may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound; has received speaker honoraria from GlaxoSmithKline, Athena Diagnostics, UCB, Biocodex, and Janssen-Cilag EMEA; has received funding for travel from Athena Diagnostics, UCB, Biocodex, GlaxoSmithKline, Janssen-Cilag EMEA, AOCCN Taiwan, Weizmann Institute, the American Academy of Neurology, IRCCS Oasi Maria SS, Sanofi China, QBI University of Queensland, International League Against Epilepsy, Australian Academy of Science, Commonwealth Department of Industry, Westmead Hospital, Perpetual, University of California, Matthew's Friends, SBS, PTS for NFLE conference, Turkish Child Neurology Association, European Congress on Epileptology, International Child Neurology Association, UCB, Movement Disorder Society, International Epilepsy Congress, University of Auckland, World Congress of Neurology, Epileptic Disorders, Eisai, AOCCN, Sanofi, and Transgenomic; and has received research support from the National Health and Medical Research Council of Australia, NIH, Australian Research Council, Health Research Council of New Zealand, CURE, American Epilepsy Society, US Department of Defense Autism Spectrum Disorder Research Program, the Jack Brockhoff Foundation, the Shepherd Foundation, Perpetual Charitable Trustees, The University of Melbourne, the Epilepsy Association of Tasmania, Melbourne Neurosciences Institute, Weizmann Institute, CURE SUDEP Award, and Perpetual Philanthropic Services. Dr. Mefford has received research support from the NIH/National Institute of Neurological Disorders and Stroke, the Burroughs Wellcome Fund, the American Thoracic Society Foundation Partner Award, and Simon's Foundation for Autism Research; has served on the editorial board of Science Translational Medicine, is a consultant for Sera Prognostics and the Simons Foundation (SFARI Gene Advisory Board); and has served on the scientific advisory boards of the Lennox Gastaut Foundation and Supporting Families with Koolen-de Vries Syndrome. Go to Neurology.org/ng for full disclosure forms.
ONLINE RESOURCES
ExAC: http://exac.broadinstitute.org/
PolyPhen-2: http://genetics.bwh.harvard.edu/pph2/
REFERENCES
- 1.Dibbens LM, de Vries B, Donatello S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet 2013;45:546–551. [DOI] [PubMed] [Google Scholar]
- 2.Ishida S, Picard F, Rudolf G, et al. Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat Genet 2013;45:552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkovic SF, Serratosa JM, Phillips HA, et al. Familial partial epilepsy with variable foci: clinical features and linkage to chromosome 22q12. Epilepsia 2004;45:1054–1060. [DOI] [PubMed] [Google Scholar]
- 4.Xiong L, Labuda M, Li DS, et al. Mapping of a gene determining familial partial epilepsy with variable foci to chromosome 22q11-q12. Am J Hum Genet 1999;65:1698–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callenbach PM, van den Maagdenberg AM, Hottenga JJ, et al. Familial partial epilepsy with variable foci in a Dutch family: clinical characteristics and confirmation of linkage to chromosome 22q. Epilepsia 2003;44:1298–1305. [DOI] [PubMed] [Google Scholar]
- 6.Klein KM, O'Brien TJ, Praveen K, et al. Familial focal epilepsy with variable foci mapped to chromosome 22q12: expansion of the phenotypic spectrum. Epilepsia 2012;53:e151–e155. [DOI] [PubMed] [Google Scholar]
- 7.Scheffer IE, Phillips HA, O'Brien CE, et al. Familial partial epilepsy with variable foci: a new partial epilepsy syndrome with suggestion of linkage to chromosome 2. Ann Neurol 1998;44:890–899. [DOI] [PubMed] [Google Scholar]
- 8.Lal D, Reinthaler EM, Schubert J, et al. DEPDC5 mutations in genetic focal epilepsies of childhood. Ann Neurol 2014;75:788–792. [DOI] [PubMed] [Google Scholar]
- 9.Martin C, Meloche C, Rioux MF, et al. A recurrent mutation in DEPDC5 predisposes to focal epilepsies in the French-Canadian population. Clin Genet 2013;86:570–574. [DOI] [PubMed] [Google Scholar]
- 10.Picard F, Makrythanasis P, Navarro V, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology 2014;86:570–574. [DOI] [PubMed] [Google Scholar]
- 11.Scheffer IE, Heron SE, Regan BM, et al. Mutations in mammalian target of rapamycin regulator DEPDC5 cause focal epilepsy with brain malformations. Ann Neurol 2014;75:782–787. [DOI] [PubMed] [Google Scholar]
- 12.Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol 2015;77:675–683. [DOI] [PubMed] [Google Scholar]
- 13.D'Gama AM, Geng Y, Couto JA, et al. mTOR pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol 2015;77:720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bar-Peled L, Chantranupong L, Cherniack AD, et al. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013;340:1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005–2009. Epilepsia 2010;51:676–685. [DOI] [PubMed] [Google Scholar]
- 16.Osborne JP, Lux AL, Edwards SW, et al. The underlying etiology of infantile spasms (West syndrome): information from the United Kingdom Infantile Spasms Study (UKISS) on contemporary causes and their classification. Epilepsia 2010;51:2168–2174. [DOI] [PubMed] [Google Scholar]
- 17.Widjaja E, Go C, McCoy B, Snead OC. Neurodevelopmental outcome of infantile spasms: a systematic review and meta-analysis. Epilepsy Res 2015;109:155–162. [DOI] [PubMed] [Google Scholar]
- 18.Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 2013;45:825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stromme P, Mangelsdorf ME, Scheffer IE, Gecz J. Infantile spasms, dystonia, and other X-linked phenotypes caused by mutations in Aristaless related homeobox gene, ARX. Brain Dev 2002;24:266–268. [DOI] [PubMed] [Google Scholar]
- 20.Allen AS, Berkovic SF, Cossette P, et al. De novo mutations in epileptic encephalopathies. Nature 2013;501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaud JL, Lachance M, Hamdan FF, et al. The genetic landscape of infantile spasms. Hum Mol Genet 2014;13:4846–4858. [DOI] [PubMed] [Google Scholar]
- 22.Niida Y, Stemmer-Rachamimov AO, Logrip M, et al. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am J Hum Genet 2001;69:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Tsai V, Parker WE, et al. Detection of human papillomavirus in human focal cortical dysplasia type IIB. Ann Neurol 2012;72:881–892. [DOI] [PubMed] [Google Scholar]
- 24.Parker WE, Orlova KA, Parker WH, et al. Rapamycin prevents seizures after depletion of STRADA in a rare neurodevelopmental disorder. Sci Transl Med 2013;5:182ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.