Abstract
Isolates of Fusarium were discovered in peat soil samples collected from peat swamp forest, waterlogged peat soil, and peat soil from oil palm plantations. Morphological characteristics were used to tentatively identify the isolates, and species confirmation was based on the sequence of translation elongation factor-1α (TEF-1α) and phylogenetic analysis. Based on the closest match of Basic Local Alignment Search Tool (BLAST) searches against the GenBank and Fusarium-ID databases, five Fusarium species were identified, namely F. oxysporum (60%), F. solani (23%), F. proliferatum (14%), F. semitectum (1%), and F. verticillioides (1%). From a neighbour-joining tree of combined TEF-1α and β-tubulin sequences, isolates from the same species were clustered in the same clade, though intraspecies variations were observed from the phylogenetic analysis. The Fusarium species isolated in the present study are soil inhabitants and are widely distributed worldwide. These species can act as saprophytes and decomposers as well as plant pathogens. The presence of Fusarium species in peat soils suggested that peat soils could be a reservoir of plant pathogens, as well-known plant pathogenic species such F. oxysporum, F. solani, F. proliferatum, and F. verticillioides were identified. The results of the present study provide knowledge on the survival and distribution of Fusarium species.
Keywords: Fusarium, Peat Soil, Morphology, Phylogenetic
Abstract
Pencilan Fusarium telah dipencilkan daripada sampel tanah gambut yang diperolehi dari hutan paya gambut, tanah gambut takung air, dan tanah gambut ladang kelapa sawit. Pencirian secara morfologi digunakan untuk mengenal pasti pencilan tersebut secara tentatif dan penentuan spesies berdasarkan jujukan gen faktor pemanjangan translasi-1α (TEF-1α) dan analisis filogenetik. Berdasarkan padanan yang terdekat carian Basic Local Alignment Search Tool (BLAST) pangkalan data Genbank dan Fusarium-ID, lima spesies Fusarium telah dikenal pasti iaitu F. oxysporum (60%), F. solani (23%), F. proliferatum (14%), F. semitectum (1%), dan F. verticillioides (1%). Daripada analisis filogenetik penyambung jiran menggunakan gabungan jujukan gen TEF-1α dan β-tubulin, menunjukkan pencilan dari spesies yang sama dikelompokkan dalam klad yang sama walaupun variasi intraspesies diperhatikan melalui analisis filogenetik. Spesies Fusarium yang dipencilkan dalam kajian ini adalah penghuni tanah dan tersebar meluas di seluruh dunia, dan boleh menjadi saprofit dan pengurai. Kehadiran spesies Fusarium dalam tanah gambut mencadangkan tanah gambut boleh menjadi takungan patogen tumbuhan di mana beberapa spesies patogen tumbuhan terkenal seperti F. oxysporum, F. solani, F. proliferatum, and F. verticillioides telah dikenal pasti. Keputusan kajian ini juga memberikan pengetahuan tentang kemandirian dan penyebaran spesies Fusarium.
Keywords: Fusarium, Tanah Gambut, Morfologi, Filogenetik
INTRODUCTION
The genus Fusarium is one of the most important groups of Ascomycetes fungi. They are well-distributed soil inhabitants with the ability to be parasites or saprophytes. The majority of Fusarium survive in plant debris and live close to the soil surface (Nwanma & Nelson 1993). Fusarium species have special characteristics, such as chlamydospores and resistant conidia, which help support their survival in soils and plant debris.
Peat soils can be obtained from peat land in forests, peat land under crops, and waterlogged peat swamps. Peat land in forests and peat land under crops provide dry areas, while waterlogged peat swamps provide wet areas for Fusarium habitation. These three types of peat soils share similar characteristics, such as acidic environments (pH 2–4), high moisture content, high carbon content, and low nitrogen content. Most of the tropical peat lands in Malaysia are geologically recent, as most of the areas were formed over the past 5,000 years (Yule & Gomez 2009).
Peat lands are formed by poor drainage, permanent water logging, and slow decomposition of the organic matter in acidic environments due to rainfall and topography. Most lowland peat contains partially decomposed trunks, branches, and roots of trees (Rieley et al. 1996), which result in low nutrient content in soils due to litter build-up as peat (MacKinnon et al. 1996). Peat soil in peat swamps were formed by the accumulation of partially decaying plant debris in waterlogged conditions, high levels of acidity (pH 2.5–4.7), and lack of oxygen. These conditions prevent microorganisms from rapidly decomposing the plant debris (Pinruan et al. 2007). Peat swamp forests are peat land tropical forest and are rapidly being depleted due to conversion into agricultural land and logging.
The genus Fusarium was one of the fungal genera listed in the compilation of fungi from boreal peatland ecosystems (Thormann & Rice 2007). From a preliminary study conducted by Latiffah et al. (2010), Fusarium species were detected in peat land areas in Perak, Malaysia. Because Fusarium can survive in this extreme environment, the peat ecosystem might be a reservoir for pathogenic Fusarium species. Moreover, many peat land areas are converted to agricultural land, which might provide suitable growth conditions for many species of Fusarium. Information on the presence of various species of Fusarium in peat soil is important, as many Fusarium species are soil-borne pathogens and some plant pathogenic species are cosmopolitan. Therefore, the objectives of this present study are to determine the presence of Fusarium species in peat soils as well as to characterise Fusarium isolates using phylogenetic analysis.
MATERIALS AND METHODS
Soil Sampling and Isolation
Peat soil samples were collected from Perak (Sungai Beriah and Beriah Kecil, Bukit Merah) and Pahang (Sungai Bebar, Pekan) in Peninsular Malaysia. In Perak, soil samples were taken from two waterlogged peat swamp areas and peat soil from an oil palm plantation that was originally peat swamp forest. In Pahang, all the soil samples were collected from peat swamp forest.
Soil samples from the top surface of the soil (10–15 cm) were taken and kept in polyethylene plastic bags. The soil samples were dried at 27±1°C for one week, ground into fine powder using a mortar and pestle, and sieved through a 2 mm strainer.
Three isolation methods, soil dilution plates, debris isolation, and direct isolation, were applied to recover Fusarium isolates from the peat soil samples. The three isolation methods were based on the methods described in The Fusarium Laboratory Manual (Leslie & Summerell 2006) for the isolation of Fusarium from soils.
Morphological Identification
Morphological identification was performed to sort the isolates into their respective groups before molecular identification was conducted. Isolates of Fusarium were tentatively identified based on the shapes of macroconidia, microconidia, chlamydospores, and conidiogenous cells observed on Carnation Leaf Agar, while pigmentation and colony appearance were observed on Potato Dextrose Agar. Species identification was based on the descriptions in The Fusarium Laboratory Manual (Leslie & Summerell 2006).
Molecular Identification
Molecular identification was performed using translation elongation factor-1α (TEF-1α) gene sequences. For phylogenetic analysis both TEF-1α and β-tubulin gene sequences were used. The genomic DNA was extracted using DNeasy® Mini Plant Kit (Qiagen, Hilden, Germany) according to the manufacturers’ instructions.
The primers used to amplify TEF-1α were EF1 (5′-ATG-GGT-AAG-GAG-GAC-AAG-AC-3′) and EF2 (5′-GGA-AGT-ACC-AGT-GAT-CAT-GTT-3′), which were modified from O’Donnell et al. (1998). The primers used to amplify β-tubulin were BT1 (5′- AAC-ATG-CGT-GAG-ATT-GTA-AGT-3′) and BT2 (5′-TAG-TGA-CCC-TTG-GCC-CAG-TTG-3′), as described by O’Donnell and Cigelnik (1997).
PCR reactions for both genes were carried out in 50 µL reaction mixtures containing 4 µL 5× PCR buffer, 4 mM MgCl2, 1.2 µL template DNA, 0.2 mM dNTPs mix (Promega, Madison, WI, USA), 1.25 U Taq DNA polymerase (Promega, Madison, WI, USA), and 0.7 mM of both forward and reverse primers.
PCR was performed in a DNA EngineTM Peltier Thermal Cycler Model PTC-100 (MJ Research Inc., Watertown, MA, USA). PCR cycles for TEF-1α were as follows: an initial denaturation at 94°C for 85 s, 35 cycles of denaturation at 95°C for 35 s, annealing at 59°C for 55 s, and extension at 72°C for 90 s, followed by a final extension at 72°C for 10 min. For PCR cycles for β-tubulin, the cycle started with an initial denaturation at 94°C for 1 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min.
Agarose gel (1%) electrophoresis was used to detect the PCR products. Electrophoresis was carried out using a Wealtec ELITE 300 (Meadowvale Way Sparks, NV, USA) power supply and GES buffer tank with 1× Tris-borate-EDTA (TBE) buffer and was run for 90 min at 90 V and 400 mA. Ten mg/mL ethidium bromide (0.04 µL) was added to the gel to visualise PCR products. The size of the amplified bands was estimated using 1 kb and 100 bp DNA markers (Fermentas, Vilnius, Lithunia). The gel was viewed using a Bio-Rad Molecular Imager® Gel DocTM Xr system (Bio Rad, Hercules, CA, USA) with The Discovery SeriesTM Quantity One® 1-D Analysis software Version 4.6.5 (Bio Rad, Hercules, CA, USA).
The PCR products were purified using a QIAquick® PCR Purification Kit (Qiagen, Germany) according to the manufacturers’ instructions with some modifications to the volume of elution buffer used.
Phylogenetic Analysis
Sequences of TEF-1α and β-tubulin were aligned using Clustal W included in Molecular Evolutionary Genetic Analysis (MEGA5) (Tamura et al. 2011). After pair-wise alignment, the sequences were compared with other sequences in the GenBank and Fusarium-ID databases using Basic Local Alignment Search Tool (BLAST). Two databases were used in this study for comparison and also to give accurate results for species identity based on the closest match of the BLAST search.
Phylogenetic analysis was conducted using a combined dataset of TEF-1α and β-tubulin sequences. The phylogenetic tree was generated based on the Neighbour-Joining (NJ) method using MEGA5. Fusarium denticulatum was chosen as the out-group as this species was not closely related to any of the species identified in this study.
RESULTS
Isolation of Fusarium Isolates
A total of 70 isolates of Fusarium were discovered in the peat soil samples, of which 33 isolates were recovered from Sungai Beriah and Beriah Kecil, Perak, while 37 isolates were recovered from Sungai Bebar, Pahang (Table 1).
Table 1:
Number of isolates recovered from the three sampled locations.
| Locations | Total |
|---|---|
| Beriah Kecil and Sungai Beriah, Perak (oil palm plantation) | 22 |
| Sungai Beriah, Perak (water-logged) | 11 |
| Sungai Bebar, Pahang (peat swamp) | 37 |
| Total | 70 |
Based on morphological characteristics, five species were tentatively identified, namely Fusarium oxysporum (60%), Fusarium solani (23%), Fusarium proliferatum (14%), Fusarium verticillioides (1%), and Fusarium semitectum (1%). The morphological characteristics of the five Fusarium species are presented in Table 2.
Table 2:
Morphological characteristics of Fusarium species isolated from peat soil
| Fusarium species | Microconidia | Microconidia | Conidiogenous cell | Chlamydospore | Pigmentation |
|---|---|---|---|---|---|
| F. oxysporum |
|
|
Short monophialides with false heads | Single or in pairs | Varied, from dark purple, dark red, white to violet, and white to creamy |
| F. solani |
|
|
Long monophialides with false head | Single or in pairs | Varied, from white to yellowish, white to creamy, and white to yellowish |
| F. proliferatum |
|
|
Monophialide and polyphialide with false head | Absent | White to light violet |
| F. semitectum |
|
|
Monophialide and polyphialide | Single or in pairs | Yellow to brownish |
| F. verticillioides |
|
|
Monophialide | Absert | White to dark purple or light purple |
A single band of 750 bp was detected using EF1 and EF2 primer pairs, indicating that TEF-1α was successfully amplified from all 70 isolates recovered from the peat soil samples. A single band of 600 bp was detected using BT1 and BT2 primer pairs, indicating that β-tubulin was also successfully amplified from all 70 Fusarium isolates. All the sequences for both TEF-1α and β-tubulin were deposited in GenBank and the accession numbers are indicated in supplementary files S1 and S2.
Based on the closest match of BLAST searches of TEF-1α sequences, morphologically identified isolates demonstrated the same species identity with isolates deposited in GenBank and Fusarium-ID databases with 93%–99% similarity (Table 3). Morphologically identified F. semitectum isolates showed 98% similarity to F. incarnatum, which is synonymous with F. semitectum (Leslie & Summerell 2006). Ten isolates morphologically identified as F. proliferatum showed 93%–99% similarity to F. proliferatum in both databases. One morphologically identified F. verticillioides (SBP48) isolate showed 99% similarity to F. verticillioides.
Table 3:
Percentage of sequence similarity, based on the TEF-1α gene, of 70 Fusarium isolates from peat soil samples.
| Code | TEF-1α | Morphologically identified species | |
|---|---|---|---|
|
| |||
| GenBank (%) | Fusarium-ID (%) | ||
| BKL24 | F. incarnatum (98) | Fusarium sp.(93) | F. semitectum |
| SBP33 | F. proliferatum (99) | F. proliferatum (99) | F. proliferatum |
| SBP34 | F. proliferatum (98) | F. proliferatum (99) | F. proliferatum |
| SBP35 | F. proliferatum (98) | F. proliferatum (97) | F. proliferatum |
| SBP36 | F. proliferatum (99) | F. proliferatum (99) | F. proliferatum |
| BKL22 | F. proliferatum (99) | F. proliferatum (99) | F. proliferatum |
| BKL31 | F. proliferatum (93) | F. proliferatum (93) | F. proliferatum |
| SBP41 | F. proliferatum (99) | F. proliferatum (99) | F. proliferatum |
| SBP44 | F. proliferatum (98) | F. proliferatum(98) | F. proliferatum |
| SBP47 | F. proliferatum (99) | F. proliferatum (99) | F. proliferatum |
| SBP40 | F. proliferatum (97) | F. proliferatum (99) | F. verticillioides |
| SBP48 | F. verticillioides (99) | F. verticillioides (99) | F. verticillioides |
| SBP39 | F. solani (92) | F. solani (97) | F. solani |
| SBL25 | F. solani (94) | F. solani (98) | F. solani |
| SBL23 | F. solani (96) | F. solani (97) | F. solani |
| SBL21 | F. solani (98) | F. solani (97) | F. solani |
| SBL24 | F. solani (96) | F. solani (98) | F. solani |
| SBL26 | F. solani (92) | F. solani (96) | F. solani |
| BKL12 | F. solani (96) | F. solani (95) | F. solani |
| SBP11 | F. solani (95) | F. solani (98) | F. solani |
| SBL10 | F. solani (93) | F. solani (94) | F. solani |
| SBL11 | F. solani (96) | F. solani (99) | F. solani |
| SBL12 | F. solani (94) | F. solani (96) | F. solani |
| SBL13 | F. solani (94) | F. solani (97) | F. solani |
| SBL14 | F. solani (95) | F. solani (96) | F. solani |
| SBL15 | F. solani (98) | F. solani (99) | F. solani |
| SBL16 | F. solani (95) | F. solani (97) | F. solani |
| SBL17 | F. solani (99) | F. solani (96) | F. solani |
| BKL11 | F. oxysporum (94) | F. oxysporum (97) | F. oxysporum |
| BKL21 | F. oxysporum (96) | F. oxysporum (97) | F. oxysporum |
| BKL26 | F. oxysporum (99) | F. oxysporum (98) | F. oxysporum |
| BKL27 | F. oxysporum (99) | F. oxysporum (98) | F. oxysporum |
| BKL28 | F. oxysporum (98) | F. oxysporum (98) | F. oxysporum |
| SB401 | F. oxysporum (99) | F. oxysporum (99) | F. oxysporum |
| SB404 | F. oxysporum (96) | F. oxysporum (97) | F. oxysporum |
| SB405 | F. oxysporum (96) | F. oxysporum (98) | F. oxysporum |
| SB406 | F. oxysporum (95) | F. oxysporum (98) | F. oxysporum |
| SB911 | F. oxysporum (95) | F. oxysporum (99) | F. oxysporum |
| SB913 | F. oxysporum (97) | F. oxysporum (98) | F. oxysporum |
| SB915 | F. oxysporum (96) | F. oxysporum (98) | F. oxysporum |
| SB916 | F. oxysporum (98) | F. oxysporum (99) | F. oxysporum |
| SB917 | F. oxysporum (97) | F. oxysporum (97) | F. oxysporum |
| SB918 | F. oxysporum (96) | F. oxysporum (97) | F. oxysporum |
| SB919 | F. oxysporum (94) | F. oxysporum (98) | F. oxysporum |
| SB920 | F. oxysporum (98) | F. oxysporum (99) | F. oxysporum |
| SB921 | F. oxysporum (99) | F. oxysporum (99) | F. oxysporum |
| SB922 | F. oxysporum (97) | F. oxysporum (98) | F. oxysporum |
| SB923 | F. oxysporum (98) | F. oxysporum (99) | F. oxysporum |
| SB928 | F. oxysporum (98) | F. oxysporum (97) | F. oxysporum |
| SB929 | F. oxysporum (97) | F. oxysporum (98) | F. oxysporum |
| SB930 | F. oxysporum (96) | F. oxysporum (99) | F. oxysporum |
| SB931 | F. oxysporum (96) | F. oxysporum (97) | F. oxysporum |
| SB933 | F. oxysporum (97) | F. oxysporum (97) | F. oxysporum |
| SB935 | F. oxysporum (94) | F. oxysporum (98) | F. oxysporum |
| SB936 | F. oxysporum (100) | F. oxysporum (98) | F. oxysporum |
| SB937 | F. oxysporum (96) | F. oxysporum (97) | F. oxysporum |
| SB938 | F. oxysporum (96) | F. oxysporum (97) | F. oxysporum |
| SB947 | F. oxysporum (96) | F. oxysporum (99) | F. oxysporum |
| SB949 | F. oxysporum (95) | F. oxysporum (97) | F. oxysporum |
| SB960 | F. oxysporum (97) | F. oxysporum (98) | F. oxysporum |
| SB961 | F. oxysporum (99) | F. oxysporum (99) | F. oxysporum |
| SB971 | F. oxysporum (97) | F. oxysporum (98) | F. oxysporum |
| SB972 | F. oxysporum (94) | F. oxysporum (97) | F. oxysporum |
| SB403 | F. oxysporum (97) | Fusarium sp. (99) | F. oxysporum |
| SB914 | F. oxysporum (99) | Fusarium sp. (99) | F. oxysporum |
| SB939 | F .oxysporum (98) | Fusarium sp. (99) | F. oxysporum |
| SB941 | F. oxysporum (96) | Fusarium sp. (99) | F. oxysporum |
| SB943 | F. oxysporum (95) | Fusarium sp. (99) | F. oxysporum |
| SB944 | F. oxysporum (96) | Fusarium sp. (99) | F. oxysporum |
| SB946 | F. oxysporum (96) | Fusarium sp. (99) | F. oxysporum |
A total of 16 isolates morphologically identified as F. solani showed 93%–99% similarity to F. solani. All 42 isolates morphologically identified as F. oxysporum showed 94%–99% similarity to F. oxysporum in both databases.
Phylogenetic Analysis
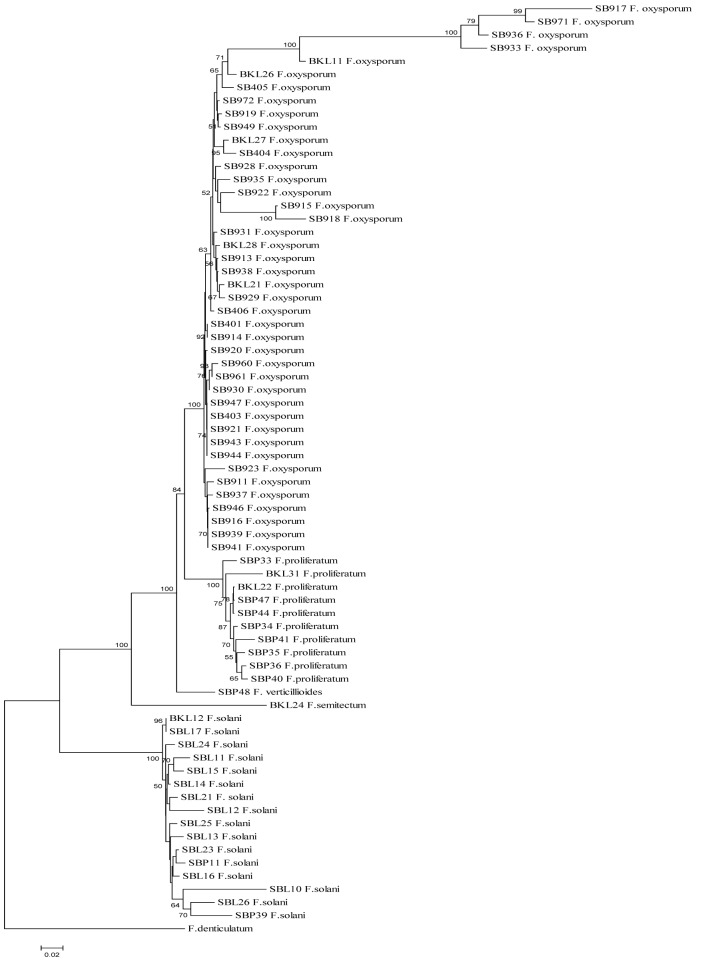
A NJ tree constructed using a combination of TEF-1α and β-tubulin sequences is shown in Figure 1. In the analysis, the sum of the branch length was 1.73 and a total of 1241 nucleotides were used. From the tree, the same isolates from the same species were clustered in the same clade. The outgroup, F. denticulatum, was clustered in a separate clade. The tree can be divided into two main clades, I and II, and main clade I can be further subdivided into sub-clades A, B, and C.
Figure 1:
NJ tree of 70 isolates of Fusarium from peat soil samples based on TEF-1α and β-tubulin sequences using the Jukes-Cantor method.
Notes: The bootstrap values higher than 50% are shown next to the branches. F. denticulatum is the out-group.
Sub-clade A can be divided into sub-sub-clades A1 and A2, in which sub-sub-clade A1 consists of F. oxysporum isolates with 100% bootstrap value, while sub-sub-clade A2 consists of F. proliferatum isolates with 100% bootstrap value. Sub-clades B and C comprise F. verticillioides and F. semitectum, respectively. Main clade II contained all F. solani isolates. Intraspecies variations were observed for isolates of F. oxysporum, F. solani, and F. proliferatum.
DISCUSSION
Five Fusarium species, namely F. oxsyporum, F. solani, F. proliferatum, F. verticillioides, and F. semitectum, were isolated and identified from peat soil samples. Although several isolates showed a low percentage similarity in BLAST results, the information provides a clue regarding the species to which the isolates belong. The same species isolated from different hosts, substrates, and areas may show a few differences in nucleotide sequences compared with the deposited sequences in the databases (Geiser et al. 2004). Moreover, the threshold for species identity, such as the number of sequence differences needed to separate a species, was not set, and there is a high degree of potential variation of TEF-1α alleles within a species (Geiser et al. 2004).
F. oxysporum is a soil inhabitant and one of the most widely distributed soil microorganisms (Gordon & Martyn 1997). F. oxysporum has been isolated from peat soils (Thormann & Rice 2007; Latiffah et al. 2010), from fen peat land (Stenton 1953), and from the rhizophere and roots of smooth cordgrass (Spartina alterniflora) in the Dongtan wetlands in China (Luo et al. 2007). F. oxysporum has been reported in other extreme habitats, such as saline soil habitats (Mandeel 2006) and arid environments (Mandeel & Abbas 1994). Most F. oxysporum isolates were isolated from direct plating from the Sungai Bebar peat swamp forest. In the soil, F. oxysporum produces an abundance of chlamydospores and survives as propagules (Burgess 1981). When the peat soil was plated onto the media, the chlamydospores germinate and produced mycelium. This species has the ability to exist as a saprophyte in the soil and is able to degrade lignin (Sutherland et al. 1983; Rodrigues 1996).
From phylogenetic analysis, F. oxysporum isolates were clustered in several sub-clades, which indicates intraspecies variations. F. oxysporum is regarded as a species complex and has been reported to be a genetically heterogeneous morphospecies by O’Donnell and Cigelnik (1997) and Waalwijk et al. (1996). This species is also considered a monophyletic group and shows a diverse complex of evolutionary lineages (O’Donnell & Cigelnik 1997). The intraspecies variation may be due to its role as a soil inhabitant, as the species can be highly variable in growth characteristics on different substrates (Steinberg et al. 1997) and in ecological characteristics (Alabouvette et al. 1993).
F. solani was recovered from direct isolation and debris plating. This species is also commonly recovered from soil and plant debris and is one of the species that inhabits widely different types of soils, such as soils from subtropical and semi-arid conditions as well as grassland soils (Burgess & Summerell 1992), cultivated soils (Lim & Chew 1970; Latiffah et al. 2007), and sandy soils (Sarquis & Borba 1997). F. solani has also been isolated from peat soil samples in Malaysia (Latiffah et al. 2010) as well as from other extreme environments, such as saline soil habitats (Mandeel 2006) and arid habitats (Mandeel & Abbas 1994). F. solani produces chlamydospores, which will germinate and form mycelia during suitable conditions.
Similar to phylogenetic analysis of F. oxysporum, F. solani isolates were also clustered in the same main clade, with several sub-sub-clades that indicate intraspecies variations. F. solani also represents a species complex of 45 phylogenetic species that form the F. solani species complex (O’Donnell 2000). Within the F. solani species complex, worldwide isolates from soil and plant debris are usually grouped in one or two phylogenetic species, known as FSSC3 and FSSC4 (Zhang et al. 2006; O’Donnell et al. 2008). Intraspecies variations of F. solani have also been reported by Balmas et al. (2010), in which two phylogenetic species, FSSC5 and FSSC9, were identified among the 23 species of the F. solani species complex isolated from Sardinian soils.
F. proliferatum isolates were recovered using debris plating and dilution plating from peat soil samples in Sungai Beriah, Perak. They were also isolated from peat soils in Pondok Tanjung and Sungai Beriah (Latiffah et al. 2010) and from peat soil in Pelitanah in Sarawak (Omar et al. 2012). F. proliferatum has also been detected in non-agricultural soils in Australia (Sangalang et al. 1995; Summerell et al. 2003). Although F. proliferatum does not produce chlamydospores, the fungus survives due to its resistant hyphae and conidia that can also withstand extreme environmental conditions. In this current study, F. proliferatum isolates might be recovered from conidia as fungal colonies derived from soil dilution, a technique commonly used to recover isolates originating from spores or conidia (Griffin 1963). F. proliferatum produced microconidia and macroconidia, which could contribute to the occurrence of this species in peat soils.
F. proliferatum isolates also formed several sub-clades that demonstrated intraspecies variations. This species is a member of the Fusarium fujikuroi species complex and a common plant pathogen, occurring in various climatic regions. Genetic variation of F. proliferatum is commonly reported from isolates associated with crop plants. Genetic and phenotypic variations of F. proliferatum isolates from different hosts have been reported by Stepien et al. (2011). Amplified fragment length polymorphism (AFLP) analysis of F. proliferatum from onion using the TEF-1α gene showed high genetic variation, where the isolates were clustered into two sub-clades with several sub-sub-clades (Galvan et al. 2008).
Although F. semitectum is common inhabitant of different types of soil, such as sandy soils (Sarquis & Borba 1997), grassland (Burgess et al. 1988), arctic (Kommedahl et al. 1988), and desert (Joffe et al. 1973), only one isolate was recovered from the peat soil samples from Beriah Kecil, Perak using the debris plating technique. F. semitectum has been isolated from peat soils in Pondok Tanjung and Sungai Beriah, Perak using direct plating and debris plating (Latiffah et al. 2010) in lower frequency compared to F. oxysporum and F. solani. F. semitectum isolates were clustered in main clade I, but formed a separate clade from other species. F. semitectum is synonymous with F. incarnatum and the species is in the F. incarnatum-equiseti species complex, with high genetic diversity (O’Donnell et al. 2009). Genetic variation in F. semitectum isolates has been reported by Masratul-Hawa et al. (2010) and Ingle and Rai (2011).
Only one isolate of F. verticillioides was isolated using the dilution plating technique. Based on a study by Liddell and Burgess (1985), isolates of F. verticillioides may live in plant residue on the soil surface. Moreover, F. verticillioides produced conidia that will germinate once suitable conditions are available. F. verticillioides has been isolated from some grassland soils in eastern Australia (Summerell et al. 2011) and from mangrove soil in Malaysia (Latiffah et al. 2010). Similar with F. proliferatum, F. verticillioides does not produce chlamydospores, therefore the survival, reproduction, and dispersal of the species are dependent on microconidia and macroconidia (Glenn et al. 2004). The F. verticillioides isolate was clustered in the same main cluster with F. oxysporum and F. proliferatum, but formed a separate clade. F. verticillioides and F. proliferatum are members of F. fujikuroi species complex and thus these two species are closely related. F. oxysporum is grouped in section Elegans but formed a sister group with the F. fujikuroi species complex (Guadet et al. 1989; O’Donnell & Cigelnik 1997; O’Donnell et al. 2000). Genetic variation in F. verticillioides has been reported by Moretti et al. (2004) and Mirete et al. (2004).
The diversity of soil fungi is generally influenced by a large number of factors, such as soil pH, organic contents, and moisture (Rangaswami & Bagyaraj 1998). The acidic nature of the soil is influenced by heavy rainfall resulting in basic cations leaching out of the soil, leaving hydrogen and aluminium cations that make the soil acidic (Swer et al. 2011). Fungal growth favours acidic conditions. Other factors that can contribute to high fungal populations and affect the distribution of many Fusarium species in the soils include the availability of nutrients which promote fungal growth, such as nitrogen, phosphorus, and calcium, the level of organic carbon in the soil (Swer et al. 2011), and environmental factors, such as rainfall, temperature, soil type, and the type of vegetation present (Backhouse et al. 2001; Summerell et al. 2010). Thus, these factors may influence the occurrence and diversity of Fusarium isolates in peat soils.
In peat soils, Fusarium species are most likely saprophytes involved in the degradation of organic materials as well as nutrient cycling in the ecosystem. Fusarium species and other fungi also help in the carbon cycle and interact with plants through exchanging of organic and inorganic compounds (Tiwari et al. 2008). As saprophytes, Fusarium species act as decomposers and are regarded as group three degrading fungi, which degrade simple polymers (Deacon 1997). The simple polymers are mainly cellulose and pectin from plant debris. By secretion of extracellular enzymes such as cellulases, polyphenol oxidases, pectinases, and amylases, these polymers are degraded and nutrients are released into the ecosystems. Complex structural polymers consisting of mainly lignin and lignocellulose in the litter promote the growth of fungi, which in turn decompose the litter (Deacon 1997).
Chlamydospores are survival structures that assist Fusarium in survival in desiccated areas, high and low temperatures, and other harsh conditions (Leslie & Summerell 2006). Among the species isolated from the peat soil samples, F. oxysporum, F. solani, and F. semitectum produce chlamydospores. Fusarium species that do not form chlamydospores have adapted resistant conidia and hyphae for survival in extreme environments, or they remain dormant and are usually found in wetter areas (Burgess 1981). These structures will germinate and form in suitably extreme environmental conditions. The survival of Fusarium species using resistant hyphae and chlamydospores in soil and debris, such as F. oxysporum, has been reported by Vakalounakis and Chalkias (2004), where it was found that F. oxysporum survived in the soil for more than 13 months.
Anamorphic Ascomycetes have been reported to be the largest group of fungi recovered from boreal peat lands (fens and bogs) in temperate regions (Thormann 2006; Thormann & Rice 2007). Among Ascomycetes, species of Penicillium are the main fungal taxon from boreal peat soil, followed by Acremonium spp., Verticillium spp., Aspergillus spp., Trichoderma spp., and Fusarium spp. (Thormann 2006). From bog and peat lands Acremonium spp. were the most common fungal taxon recorded, followed by Aspergillus spp., Oidiodendron spp., Penicillium spp., and Trichoderma spp. (Thormann 2006). Similar scenarios were also observed in tropical peat soils, where Aspergillus spp., Penicillium spp., and Trichoderma spp. were usually isolated. The occurrence of these fungal genera is not surprising as these taxa are regarded as generalists, heavy sporulators, and fast growing fungal groups (Thormann 2006).
Studies on Fusarium in non-agricultural soils, especially peat soil in peat swamp forest and waterlogged areas, provide insights on the survival and distribution of Fusarium species. Peat soil in these two areas could be a reservoir of plant pathogenic species, especially because well-known plant pathogens, such as F. oxysporum, F. solani, and F. proliferatum, were recovered. Moreover, plant pathogens have the potential to evolve in response to strong selection pressures imposed by agricultural ecosystems (Neumann et al. 2004) when the peat swamp areas are converted to agricultural use.
S1:
Accession numbers of TEF-1α sequences of Fusarium species from peat soil.
| Code | Accession number (TEF-1α sequences) |
|---|---|
| BKL24 (F. semitectum) | KC120924 |
| SBP33 (F. proliferatum) | KC120980 |
| SBP34 (F. proliferatum) | KC120981 |
| SBP35 (F. proliferatum) | KC120982 |
| SBP36 (F. proliferatum) | KC120983 |
| BKL22 (F. proliferatum) | KC120923 |
| BKL31 (F. proliferatum) | KC120928 |
| SBP41 (F. proliferatum) | KC120986 |
| SBP44 (F. proliferatum) | KC120987 |
| SBP47 (F. proliferatum) | KC120988 |
| SBP40 (F. proliferatum) | KC120985 |
| SBP48 (F. verticillioides) | KC120989 |
| SBP39 (F. solani) | KC120984 |
| SBL25 (F. solani) | KC120977 |
| SBL23 (F. solani) | KC120975 |
| SBL21 (F. solani) | KC120974 |
| SBL24 (F. solani) | KC120976 |
| SBL26 (F. solani) | KC120978 |
| BKL12 (F. solani) | KC120921 |
| SBP11 (F. solani) | KC120979 |
| SBL10 (F. solani) | KC120966 |
| SBL11 (F. solani) | KC120967 |
| SBL12 (F. solani) | KC120968 |
| SBL13 (F. solani) | KC120969 |
| SBL14 (F. solani) | KC120970 |
| SBL15 (F. solani) | KC120971 |
| SBL16 (F. solani) | KC120972 |
| SBL17 (F. solani) | KC120973 |
| BKL11 (F. oxysporum) | KC120920 |
| BKL21 (F. oxysporum) | KC120922 |
| BKL26 (F. oxysporum) | KC120925 |
| BKL27 (F. oxysporum) | KC120926 |
| BKL28 (F. oxysporum) | KC120927 |
| SB911 (F. oxysporum) | KC120934 |
| SB913 (F. oxysporum) | KC120935 |
| SB915 (F. oxysporum) | KC120937 |
| SB916 (F. oxysporum) | KC120938 |
| SB917 (F. oxysporum) | KC120939 |
| SB919 (F. oxysporum) | KC120941 |
| SB920 (F. oxysporum) | KC120942 |
| SB921 (F. oxysporum) | KC120943 |
| SB922 (F. oxysporum) | KC120944 |
| SB923 (F. oxysporum) | KC120945 |
| SB928 (F. oxysporum) | KC120946 |
| SB929 (F. oxysporum) | KC120947 |
| SB930 (F. oxysporum) | KC120948 |
| SB931 (F. oxysporum) | KC120949 |
| SB933 (F. oxysporum) | KC120950 |
| SB935 (F. oxysporum) | KC120951 |
| SB936 (F. oxysporum) | KC120952 |
| SB937 (F. oxysporum) | KC120953 |
| SB938 (F. oxysporum) | KC120954 |
| SB947 (F. oxysporum) | KC120960 |
| SB949 (F. oxysporum) | KC120961 |
| SB960 (F. oxysporum) | KC120962 |
| SB961 (F. oxysporum) | KC120963 |
| SB971 (F. oxysporum) | KC120964 |
| SB972 (F. oxysporum) | KC120965 |
| SB403 (F. oxysporum) | KC120930 |
| SB914 (F. oxysporum) | KC120936 |
| SB939 (F. oxysporum) | KC120955 |
| SB941 (F. oxysporum) | KC120956 |
| SB943 (F. oxysporum) | KC120957 |
| SB944 (F. oxysporum) | KC120958 |
| SB946 (F. oxysporum) | KC120959 |
S2:
Accession numbers of β-tubulin sequences of Fusarium species from peat soil.
| Code | Accession number (β-tubulin sequences) |
|---|---|
| BKL24 (F. semitectum) | KC161303 |
| SBP33 (F. proliferatum) | KC161319 |
| SBP34 (F. proliferatum) | KC161320 |
| SBP35 (F. proliferatum) | KC161326 |
| SBP36 (F. proliferatum) | KC161327 |
| BKL22 (F. proliferatum) | KC161323 |
| BKL31 (F. proliferatum) | KC161325 |
| BKL31 (F. proliferatum) | KC161325 |
| SBP41 (F. proliferatum) | KC161328 |
| SBP44 (F. proliferatum) | KC161330 |
| SBP47 (F. proliferatum) | KC161331 |
| SBP40 (F. proliferatum) | KC161329 |
| SBP48 (F. verticillioides) | KC161311 |
| SBP39 (F. solani) | KC161321 |
| SBL25 (F. solani) | KC161317 |
| SBL23 (F. solani) | KC161308 |
| SBL21 (F. solani) | KC161307 |
| SBL24 (F. solani) | KC161309 |
| SBL26 (F. solani) | KC161318 |
| BKL12 (F. solani) | KC161302 |
| SBP11 (F. solani) | KC161310 |
| SBL10 (F. solani) | KC161304 |
| SBL11 (F. solani) | KC161305 |
| SBL12 (F. solani) | KC161306 |
| SBL13 (F. solani) | KC161312 |
| SBL14 (F. solani) | KC161313 |
| SBL15 (F. solani) | KC161314 |
| SBL16 (F. solani) | KC161315 |
| SBL17 (F. solani) | KC161316 |
| SB406 (F. oxysporum) | KC161339 |
| SB911 (F. oxysporum) | KC161340 |
| SB913 (F. oxysporum) | KC161341 |
| SB915 (F. oxysporum) | KC161343 |
| SB916 (F. oxysporum) | KC161344 |
| SB918 (F. oxysporum) | KC161345 |
| SB920 (F. oxysporum) | KC161347 |
| SB921 (F. oxysporum) | KC161348 |
| SB922 (F. oxysporum) | KC161349 |
| SB923 (F. oxysporum) | KC161350 |
| SB928 (F. oxysporum) | KC161351 |
| SB929 (F. oxysporum) | KC161352 |
| SB930 (F. oxysporum) | KC161353 |
| SB931 (F. oxysporum) | KC161354 |
| SB935 (F. oxysporum) | KC161355 |
| SB937 (F. oxysporum) | KC161356 |
| SB938 (F. oxysporum) | KC161357 |
| SB947 (F. oxysporum) | KC161360 |
| SB949 (F. oxysporum) | KC161361 |
| SB960 (F. oxysporum) | KC161364 |
| SB961 (F. oxysporum) | KC161366 |
| SB972 (F. oxysporum) | KC161367 |
| SB403 (F. oxysporum) | KC161336 |
| SB914 (F. oxysporum) | KC161342 |
| SB939 (F. oxysporum) | KC161365 |
| SB941 (F. oxysporum) | KC161362 |
| SB943 (F. oxysporum) | KC161363 |
| SB944 (F. oxysporum) | KC161358 |
| SB946 (F. oxysporum) | KC161359 |
Acknowledgments
This work was supported by the Research University Grant, Universiti Sains Malaysia (1001/PBIOLOGI/815039).
REFERENCES
- Alabouvette C, Lemanceau P, Steinberg C. Recent advances in biological control of Fusarium wilts. Pesticide Science. 1993;37(4):365–373. [Google Scholar]
- Backhouse D, Burgess LW, Summerell BA. Biogeography of Fusarium. In: Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burgess L, editors. Fusarium: Paul E. Nelson Memorial Symposium. St. Paul, Minnesota, USA: APS Press; 2001. pp. 122–137. [Google Scholar]
- Balmas V, Migheli Q, Schem B, Garau P, O’Donnell K, Ceccherelli G, Kang S, Geiser DM. Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands) Mycologia. 2010;102(4):803–812. doi: 10.3852/09-201. [DOI] [PubMed] [Google Scholar]
- Burgess LW. General ecology of the Fusaria. In: Nelson PE, Toussoun TA, Cook RJ, editors. Fusarium: Diseases, biology, and taxonomy. University Park, Pennsylvania, USA: The Pennsylvania State University Press; 1981. pp. 225–235. [Google Scholar]
- Burgess LW, Nelson PE, Toussoun TA, Forbes GA. Distribution of Fusarium species in sections Roseum, Arthrosporiella, Gibbosum and Discolor recovered from grassland, pasture and pine nursery soils of eastern Australia. Mycologia. 1988;80(6):815–824. [Google Scholar]
- Burgess LW, Summerell BA. Mycogeography of Fusarium: Survey of Fusarium species in sub-tropical and semi-arid grassland soils from Queensland, Australia. Mycological Research. 1992;96(9):780–784. [Google Scholar]
- Deacon JW. Modern mycology. 3rd ed. Boston, Massachusetts: Blackwell Science Publications; 1997. [Google Scholar]
- Galvan GA, Koning-Bouxorin CFS, Koopman WJM, Burger-Meijer K, González PH, Waalwijk C, Kik C, Scholten OE. Genetic variation among Fusarium isolates from onion and resistance to Fusarium basal rot inrelated Allium species. European Journal of Plant Pathology. 2008;121(4):499–512. [Google Scholar]
- Geiser DM, Jimenz Gasco MM, Kang S, Mkalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. FUSARIUM-IDv.1.0: A DNA sequence database for identifying Fusarium. European Journal of Plant Pathology. 2004;110:473–479. [Google Scholar]
- Glenn AE, Richardson EA, Bacon CW. Genetic and morphological characterization of a Fusarium verticillioides conidiation mutant. Mycologia. 2004;96(5):968–980. [PubMed] [Google Scholar]
- Gordon TR, Martyn RD. The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology. 1997;35:111–128. doi: 10.1146/annurev.phyto.35.1.111. [DOI] [PubMed] [Google Scholar]
- Griffin DM. Soil moisture and the ecology of soil fungi. Biological Reviews. 1963;38(2):141–166. doi: 10.1111/j.1469-185x.1963.tb00781.x. [DOI] [PubMed] [Google Scholar]
- Guadet J, Julien J, Lafay JF, Brygoo Y. Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Molecular Biology Evolution. 1989;6(3):227–242. doi: 10.1093/oxfordjournals.molbev.a040548. [DOI] [PubMed] [Google Scholar]
- Ingle A, Rai M. Genetic diversity among Indian phytopathogenic isolates of Fusarium semitectum Berkeley and Ravenel. Advances in Bioscience and Biotechnology. 2011;2(3):142–148. [Google Scholar]
- Joffe AZ, Palti J, Arbel-Sherman R. Fusarium moniliforme Sheld. in Israel (Gibberella fujikuroi) Mycopathology and Mycology Application. 1973;50(2):85–107. doi: 10.1007/BF02049949. [DOI] [PubMed] [Google Scholar]
- Kommedahl T, Abbas HK, Burnes PM, Mirocha CJ. Prevalence and toxigenicity of Fusarium spp. from soils of Norway near Arctic Circle. Mycologia. 1988;80(6):790–794. [Google Scholar]
- Latiffah Z, Mohd Zariman M, Baharuddin S. Diversity of Fusarium species in cultivated soils in Penang. Malaysian Journal of Microbiology. 2007;3(1):27–30. [Google Scholar]
- Latiffah Z, Nurul Izzati H, Baharuddin S. Fusarium species isolated from peat soil of Pondok Tanjung and Sungai Beriah, Perak. Malaysian Journal Microbiology. 2010;6(1):102–105. [Google Scholar]
- Leslie JF, Summerell BA. The Fusarium laboratory manual. 1st ed. Ames, Iowa, USA: Blackwell Publishing; 2006. [Google Scholar]
- Liddell CM, Burgess LW. Survival of Fusarium moniliforme at controlled temperature and relative humidity. Transaction of British Mycological Society. 1985;84(1):121–130. [Google Scholar]
- Lim G, Chew CH. Fusarium in Singapore soils. Plant and Soil. 1970;33(1):673–677. [Google Scholar]
- Luo JL, Bao K, Weo MN. Cladistic and phenetic analyses of relationships among Fusarium spp. in Dongtan wetland by morphology and isozymes. Biochemical and Systematics Ecology. 2007;35(7):410–420. [Google Scholar]
- MacKinnon K, Hatta G, Halim H, Mangalik A. The ecology of Kalimantan. The ecology of Indonesia series. Singapore: Periplus Editions (HK) Ltd; 1996. [Google Scholar]
- Mandeel QA. Biodiversity of the genus Fusarium in saline soil habitats. Journal of Basic Microbiology. 2006;46(6):480–494. doi: 10.1002/jobm.200510128. [DOI] [PubMed] [Google Scholar]
- Mandeel QA, Abbas JA. Survey of Fusarium species in an arid environment of Bahrain. Mycopathologia. 1994;127(3):167–173. [Google Scholar]
- Masratul-Hawa MM, Salleh B, Latiffah Z. Characterization and intraspecific variation of Fusarium semitectum (Berkeley and Ravenel) associated with red-fleshed dragon fruit (Hylocereus polyrhizus) in Malaysia. African Journal Biotechnology. 2010;9(3):273–284. [Google Scholar]
- Mirete S, Vázquez C, Mulè G, Jurado M, González-Jaén MT. Differentiation of Fusarium verticillioides from banana fruits by IGS and EF-1α sequence analyses. European Journal of Plant Pathology. 2004;110(5):515–523. [Google Scholar]
- Moretti A, Mulè G, Susca A, González-Jaén MT, Logrieco A. Toxin profile, fertility and AFLP analysis of Fusarium verticillioides from banana fruits. European Journal of Plant Pathology. 2004;110(5):601–609. [Google Scholar]
- Neumann MJ, Backhouse D, Carter DA, Summerell BA, Burgess LW. Genetic structure of populations of Fusarium proliferatum in soils associated with Livistona mariae palms in Little Palm Creek, Northern Territory, Australia. Australian Journal of Botany. 2004;52(4):543–550. [Google Scholar]
- Nwanma NO, Nelson PE. The distribution of Fusarium species in soils planted with millet and sorghum in Lesotho, Nigeria and Zimbabwe. Mycopathologia. 1993;121(2):105–114. [Google Scholar]
- O’Donnell K. Molecular phylogeny of the Nectria haematococca–Fusarium solani species complex. Mycologia. 2000;92(5):919–938. [Google Scholar]
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetic and Evolution. 1997;7(1):103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceeding of National Academia of Science USA. 1998;95(5):2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycociences. 2000;41:61–78. [Google Scholar]
- O’Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, Zhang N, Geiser DM. Molecular phylogenetic diversity, multilocus haplotype nomenclature and in vitro antifungal resistance within the Fusarium solani species complex. Journal of Clinical Microbiology. 2008;46:2477–2490. doi: 10.1128/JCM.02371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Sutton DA, Rinaldi MG, Gueidan C, Crous PW, Geiser DM. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. Journal of Clinical Microbiology. 2009;47(12):3851–3861. doi: 10.1128/JCM.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar FN, Ismael NH, Ali SRA. Fungi associated with deep peat soil Sarawak. UMT 11th International Annual Symposium on Sustainability Science and Management; Ri-Yaz Heritage Marina Resort & Spa, Kuala Terengganu, Terengganu. 9–11 July 2012; Kuala Terengganu, Terengganu, Malaysia: Universiti Malaysia Terengganu; 2012. pp. 239–245. [Google Scholar]
- Pinruan U, Hyde KD, Lumyong S, Mckenzie EHC, Jones EBG. Occurrence of fungi on tissues of the peat swamp palm Licuala longicalycata. Fungal Diversity. 2007;25(1):157–173. [Google Scholar]
- Rangaswami G, Bagyaraj DJ. Agricultural microbiology. 2nd ed. New Delhi: Prentice Hall, India Pvt. Ltd; 1998. [Google Scholar]
- Rieley JO, Ahmad-Shah AA, Brady MA. In: Maltby E, Immirzi CP, Safford RJ, editors. The extent and nature of tropical peat swamps; Tropical Lowland Peatlands of Southeast Asia, Proceedings of a Workshop on Integrated Planning and Management of Tropical Lowland Peatlands; Cisarua, Indonesia. 3–8 July 1992; Gland, Switzerland: International Union for Conservation of Nature (IUCN); 1996. pp. 17–53. [Google Scholar]
- Rodrigues KF. Fungal endophytes of palms. In: Redlin SC, Carris LM, editors. Endophytic fungi in grasses and woody plants: Systematics, ecology and evolution. St. Paul, USA: APS Press; 1996. [Google Scholar]
- Sangalang AE, Burgess LW, Backhouse D, Du J, Wurst M. Mycogeography of Fusarium species in soils from tropical, arid and Mediterranean regions of Australia. Mycological Research. 1995;99(5):523–528. [Google Scholar]
- Sarquis MM, Borba CM. Fusarium species in sandy soil from Ipanema Beach. Journal of Basic Microbiology. 1997;37(6):425–429. [Google Scholar]
- Steinberg C, Edel V, Gautheron N, Abadie C, Vallaeys T, Alabouvette C. Phenotypic characterization of natural populations of Fusarium oxysporum in relation to genotypic characterization. FEMS Microbiology Ecology. 1997;24(1):73–85. [Google Scholar]
- Stenton H. The soil fungi of Wicken fen. Transaction of British Mycological Society. 1953;36(4):304–314. [Google Scholar]
- Stepien L, Koczyk G, Waskiewiczl A. Genetic and phenotypic variation of Fusarium proliferatum isolates from different host species. Journal of Applied Genetics. 2011;52:487–496. doi: 10.1007/s13353-011-0059-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerell BA, Salleh B, Leslie JF. A utilitarian approach to Fusarium identification. Plant Disease. 2003;87(2):117–127. doi: 10.1094/PDIS.2003.87.2.117. [DOI] [PubMed] [Google Scholar]
- Summerell BA, Laurence MH, Liew ECY, Leslie JF. Biogeography and phylogeography of Fusarium: A review. Fungal Diversity. 2010;44:3–13. [Google Scholar]
- Summerell BA, Leslie JF, Liew ECY, Laurence MH, Bullock S, Petrovic T, Bentley AR, et al. Fusarium species associated with plants in Australia. Fungal Diversity. 2011;46(1):1–27. [Google Scholar]
- Sutherland JB, Pometto AL, Crawford DL. Lignocellulose degradation by Fusarium species. Canadian Journal Botany. 1983;61(4):1194–1198. [Google Scholar]
- Swer H, Dkhar MS, Kayang H. Fungal population and diversity inorganically amended agricultural soils of Meghalaya, India. Journal of Organic Systems. 2011;6(2):3–12. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thormann MN. Diversity and function of fungi in peatlands: A carbon cycling perspective. Canadian Journal of Soil Science Science. 2006;86(Spl.):281–293. [Google Scholar]
- Thormann MN, Rice AV. Fungi from peatlands. Fungal Diversity. 2007;24:241–299. [Google Scholar]
- Tiwari CK, Verma RK, Ayachi A, Asaiya AJK. Wood decaying fungi of Sal from Madhya Pradesh, India. Science Front. 2008;2(1):13–26. [Google Scholar]
- Vakalounakis DJ, Chalkias J. Survival of Fusarium oxysporum f. sp. Radicis-cucumerinum in soil. Crop Protection. 2004;23(9):871–873. [Google Scholar]
- Waalwijk C, de Koning JRA, Baayen RP, Gams W. Discordant groupings of Fusarium spp. from the sections Elegans, Liseola and Dlaminia based on ribosomal ITS1 and ITS2 sequences. Mycologia. 1996;88(3):361–368. [Google Scholar]
- Yule CM, Gomez LN. Leaf litter decomposition in a tropical peat swamp forest in Peninsular Malaysia. Wetland Ecology and Management. 2009;17(3):231–241. [Google Scholar]
- Zhang N, O’Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye AA, Geiser DM. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. Journal Clinic Microbiology. 2006;44(6):2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1:
Accession numbers of TEF-1α sequences of Fusarium species from peat soil.
| Code | Accession number (TEF-1α sequences) |
|---|---|
| BKL24 (F. semitectum) | KC120924 |
| SBP33 (F. proliferatum) | KC120980 |
| SBP34 (F. proliferatum) | KC120981 |
| SBP35 (F. proliferatum) | KC120982 |
| SBP36 (F. proliferatum) | KC120983 |
| BKL22 (F. proliferatum) | KC120923 |
| BKL31 (F. proliferatum) | KC120928 |
| SBP41 (F. proliferatum) | KC120986 |
| SBP44 (F. proliferatum) | KC120987 |
| SBP47 (F. proliferatum) | KC120988 |
| SBP40 (F. proliferatum) | KC120985 |
| SBP48 (F. verticillioides) | KC120989 |
| SBP39 (F. solani) | KC120984 |
| SBL25 (F. solani) | KC120977 |
| SBL23 (F. solani) | KC120975 |
| SBL21 (F. solani) | KC120974 |
| SBL24 (F. solani) | KC120976 |
| SBL26 (F. solani) | KC120978 |
| BKL12 (F. solani) | KC120921 |
| SBP11 (F. solani) | KC120979 |
| SBL10 (F. solani) | KC120966 |
| SBL11 (F. solani) | KC120967 |
| SBL12 (F. solani) | KC120968 |
| SBL13 (F. solani) | KC120969 |
| SBL14 (F. solani) | KC120970 |
| SBL15 (F. solani) | KC120971 |
| SBL16 (F. solani) | KC120972 |
| SBL17 (F. solani) | KC120973 |
| BKL11 (F. oxysporum) | KC120920 |
| BKL21 (F. oxysporum) | KC120922 |
| BKL26 (F. oxysporum) | KC120925 |
| BKL27 (F. oxysporum) | KC120926 |
| BKL28 (F. oxysporum) | KC120927 |
| SB911 (F. oxysporum) | KC120934 |
| SB913 (F. oxysporum) | KC120935 |
| SB915 (F. oxysporum) | KC120937 |
| SB916 (F. oxysporum) | KC120938 |
| SB917 (F. oxysporum) | KC120939 |
| SB919 (F. oxysporum) | KC120941 |
| SB920 (F. oxysporum) | KC120942 |
| SB921 (F. oxysporum) | KC120943 |
| SB922 (F. oxysporum) | KC120944 |
| SB923 (F. oxysporum) | KC120945 |
| SB928 (F. oxysporum) | KC120946 |
| SB929 (F. oxysporum) | KC120947 |
| SB930 (F. oxysporum) | KC120948 |
| SB931 (F. oxysporum) | KC120949 |
| SB933 (F. oxysporum) | KC120950 |
| SB935 (F. oxysporum) | KC120951 |
| SB936 (F. oxysporum) | KC120952 |
| SB937 (F. oxysporum) | KC120953 |
| SB938 (F. oxysporum) | KC120954 |
| SB947 (F. oxysporum) | KC120960 |
| SB949 (F. oxysporum) | KC120961 |
| SB960 (F. oxysporum) | KC120962 |
| SB961 (F. oxysporum) | KC120963 |
| SB971 (F. oxysporum) | KC120964 |
| SB972 (F. oxysporum) | KC120965 |
| SB403 (F. oxysporum) | KC120930 |
| SB914 (F. oxysporum) | KC120936 |
| SB939 (F. oxysporum) | KC120955 |
| SB941 (F. oxysporum) | KC120956 |
| SB943 (F. oxysporum) | KC120957 |
| SB944 (F. oxysporum) | KC120958 |
| SB946 (F. oxysporum) | KC120959 |
S2:
Accession numbers of β-tubulin sequences of Fusarium species from peat soil.
| Code | Accession number (β-tubulin sequences) |
|---|---|
| BKL24 (F. semitectum) | KC161303 |
| SBP33 (F. proliferatum) | KC161319 |
| SBP34 (F. proliferatum) | KC161320 |
| SBP35 (F. proliferatum) | KC161326 |
| SBP36 (F. proliferatum) | KC161327 |
| BKL22 (F. proliferatum) | KC161323 |
| BKL31 (F. proliferatum) | KC161325 |
| BKL31 (F. proliferatum) | KC161325 |
| SBP41 (F. proliferatum) | KC161328 |
| SBP44 (F. proliferatum) | KC161330 |
| SBP47 (F. proliferatum) | KC161331 |
| SBP40 (F. proliferatum) | KC161329 |
| SBP48 (F. verticillioides) | KC161311 |
| SBP39 (F. solani) | KC161321 |
| SBL25 (F. solani) | KC161317 |
| SBL23 (F. solani) | KC161308 |
| SBL21 (F. solani) | KC161307 |
| SBL24 (F. solani) | KC161309 |
| SBL26 (F. solani) | KC161318 |
| BKL12 (F. solani) | KC161302 |
| SBP11 (F. solani) | KC161310 |
| SBL10 (F. solani) | KC161304 |
| SBL11 (F. solani) | KC161305 |
| SBL12 (F. solani) | KC161306 |
| SBL13 (F. solani) | KC161312 |
| SBL14 (F. solani) | KC161313 |
| SBL15 (F. solani) | KC161314 |
| SBL16 (F. solani) | KC161315 |
| SBL17 (F. solani) | KC161316 |
| SB406 (F. oxysporum) | KC161339 |
| SB911 (F. oxysporum) | KC161340 |
| SB913 (F. oxysporum) | KC161341 |
| SB915 (F. oxysporum) | KC161343 |
| SB916 (F. oxysporum) | KC161344 |
| SB918 (F. oxysporum) | KC161345 |
| SB920 (F. oxysporum) | KC161347 |
| SB921 (F. oxysporum) | KC161348 |
| SB922 (F. oxysporum) | KC161349 |
| SB923 (F. oxysporum) | KC161350 |
| SB928 (F. oxysporum) | KC161351 |
| SB929 (F. oxysporum) | KC161352 |
| SB930 (F. oxysporum) | KC161353 |
| SB931 (F. oxysporum) | KC161354 |
| SB935 (F. oxysporum) | KC161355 |
| SB937 (F. oxysporum) | KC161356 |
| SB938 (F. oxysporum) | KC161357 |
| SB947 (F. oxysporum) | KC161360 |
| SB949 (F. oxysporum) | KC161361 |
| SB960 (F. oxysporum) | KC161364 |
| SB961 (F. oxysporum) | KC161366 |
| SB972 (F. oxysporum) | KC161367 |
| SB403 (F. oxysporum) | KC161336 |
| SB914 (F. oxysporum) | KC161342 |
| SB939 (F. oxysporum) | KC161365 |
| SB941 (F. oxysporum) | KC161362 |
| SB943 (F. oxysporum) | KC161363 |
| SB944 (F. oxysporum) | KC161358 |
| SB946 (F. oxysporum) | KC161359 |