Abstract
Linezolid (LZD) has become increasingly important for the treatment of multidrug-resistant tuberculosis (MDR-TB), but its mechanisms of resistance are not well characterized. We isolated 32 mutants of Mycobacterium tuberculosis with reduced susceptibility to LZD, which was accounted for by rrl and rplC mutations in almost equal proportions, causing lower and higher MICs, respectively. Our findings provide useful information for the rapid detection of LZD resistance for improved treatment of MDR-TB.
TEXT
Tuberculosis (TB) remains a major threat to global public health. Increasing emergence of multidrug-resistant tuberculosis (MDR-TB) and extensively drug-resistant tuberculosis (XDR-TB) calls for urgent development of new drugs to combat drug-resistant TB. Although linezolid (LZD) has been used primarily to treat antibiotic-resistant Gram-positive bacterial infections (1), it has good activity against mycobacteria (2). LZD binds to the 50S ribosomal subunit and inhibits the translation process (3). This mechanism of action is different from that of first- and second-line TB drugs, which indicates no cross-resistance with current TB drugs. Several clinical studies have demonstrated the usefulness of LZD in the treatment of MDR-TB (4, 5), but resistance ranging from 1.9% to 10.8% has been reported in MDR Mycobacterium tuberculosis strains (6, 7). Previous studies have shown that mutations in the 23S rRNA gene rrl encoding the ribosomal L4 protein were present in 5 of 10 (50%) LZD-resistant mutants of M. tuberculosis isolated in vitro, with 4 having mutations at position 2061 and 1 having a mutation at position 2057 in the rrl gene (8). However, the other 5 mutants were not accounted for. A recent study showed that mutations in the rplC gene, encoding the L3 ribosomal protein, were found in 6 of 7 LZD-resistant mutants of M. tuberculosis (9). This was based on a study of Staphylococcus aureus isolates where rplC mutations were found to cause LZD resistance (10). Although the mechanisms of resistance to TB drugs have been well characterized for most drug resistances (11), there is limited information about the genetic analysis of mutations involved in LZD resistance in M. tuberculosis. So far, only 12 resistant mutants isolated in vitro have been studied (8, 9).
To further understand the mechanisms of resistance to LZD, we isolated 32 resistant mutants of M. tuberculosis H37Rv as a parental strain on 7H11 agar plates containing 0, 0.25, 0.5, 1, and 2 μg/ml LZD. We found that the H37Rv control strain did not grow at 0.25 μg/ml, indicating that its MIC (<0.25 μg/ml) is lower than previously reported MIC values of 0.5 μg/ml (12) and 1 μg/ml (6). This may be a reflection of the 7H11 agar solid medium used here compared to the MGIT liquid medium employed in the previous study (6) that reported higher MIC values for the susceptible control strain. The reduced susceptibility of the mutants was confirmed by growing them again on 7H11 agar plates containing LZD ranging from 0 to 2 μg/ml; the degree of growth of each mutant on LZD-containing plates is shown in Table 1. Based on the degree of mutant growth on plates containing LZD compared with that of the sensitive control strain, we chose 0.25 μg/ml LZD as the MIC breakpoint above which strains are considered to have reduced susceptibility. The DNA of these mutants and of the sensitive control strain H37Rv was extracted and used as the template for PCR amplification of known LZD resistance genes rrl and rplC. The primers used for PCR were rrl F1 (5′-CCT GAG GCA ACA CTC GGA CTT-3′) (156 bp before start codon), rrl R1 (5′-ACG GAT TTG CCT ATC GCT CT-3′), rrl F2 (5′-CCT AAG GCG AGG CCG ACA G-3′), rrl R2 (5′-GGC CGC CGT AAC TCT ATG C-3′) (114 bp after stop codon), rplC F (5′-TCCGCTCACCGCATAAGTACA-3′) (167 bp before start codon), and rplC R (5′-CGATGTTGGCCGGGACGT-3′) (112 bp after stop codon). The PCR conditions were initial denaturation at 95°C for 5 min and then 30 cycles of 95°C for 30 s, 58°C for 30 s, 72°C for 2 min, and a final extension at 72°C for 7 min. PCR products were purified and were used for Sanger DNA sequencing. Genomic DNA from the 7 mutants with higher LZD MIC values that did not show mutations in rrl or rplC by Sanger sequencing was subjected to whole-genome sequencing using Illumina MiSeq as described previously (13).
TABLE 1.
Mutations associated with reduced susceptibility to LZD in 32 mutants of M. tuberculosis
| Straina | rrlb | rplCb | RplC amino acid change | Mutation percentage | Growth on various concentrations of LZD (μg/ml)c |
||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.25 | 0.5 | 1 | 2 | |||||
| H37Rv | (-) | (-) | 4+ | - | - | - | - | ||
| L1T5-3 | G2270C | (-) | 4+ | 4+ | 3+ | 2+ | - | ||
| L1T1-5 | G2270T | (-) | 4+ | 4+ | 4+ | 3+ | - | ||
| L1T3-1 | G2270T | (-) | 4+ | 4+ | 3+ | 2+ | - | ||
| *L1T3-4 | G2270C | (-) | 20 | 4+ | 4+ | 2+ | 1+ | - | |
| L1T5-4 | G2270T | (-) | 4+ | 3+ | 3+ | 2+ | - | ||
| *L1T5-5 | G2270T | (-) | 6 | 4+ | 3+ | 2+ | 1+ | - | |
| L1T4-3 | G2270T | (-) | ND | ND | ND | ND | ND | ||
| L1T4-6 | G2746A | (-) | ND | ND | ND | ND | ND | ||
| L1T4-7 | G2746A | (-) | ND | ND | ND | ND | ND | ||
| L1T13-2 | G2746A | (-) | ND | ND | ND | ND | ND | ||
| L1T6-3 | G2746A | (-) | 4+ | 3+ | 3+ | 1+ | - | ||
| L1T4-4 | G2746A | (-) | 4+ | 4+ | 3+ | 2+ | - | ||
| L1T4-5 | G2746A | (-) | 4+ | 4+ | 3+ | 2+ | - | ||
| L1T4-8 | G2746A | (-) | 4+ | 4+ | 2+ | 2+ | - | ||
| L1T4-9 | G2746A | (-) | 4+ | 4+ | 2+ | 1+ | - | ||
| L1T1-6 | A2810T | (-) | 4+ | 3+ | 2+ | 1+ | - | ||
| *L1T3-6 | C2848A | (-) | 13 | 4+ | 2+ | 2+ | 1+ | - | |
| L1T3-2 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 4+ | |
| L1T4-2 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 3+ | |
| L1T5-2 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 4+ | |
| L2T14-1 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 4+ | |
| L2T15-1 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 4+ | |
| L2T15-3 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 3+ | |
| L2T15-4 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 3+ | |
| L2T15-5 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 3+ | |
| L2T15-6 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 3+ | |
| L2T22-1 | (-) | T460C | Cys154Arg | 3+ | 3+ | 3+ | 2+ | 2+ | |
| L2T22-2 | (-) | T460C | Cys154Arg | 4+ | 4+ | 4+ | 4+ | 4+ | |
| *L1T3-3 | (-) | T460C | Cys154Arg | 17 | 4+ | 4+ | 4+ | 4+ | 3+ |
| *L1T5-1 | (-) | T460C | Cys154Arg | 60 | 4+ | 4+ | 4+ | 4+ | 4+ |
| *L1T3-5 | (-) | T460C | Cys154Arg | 27 | 4+ | 4+ | 4+ | 4+ | 3+ |
| *L1T3-7 | (-) | T460C | Cys154Arg | 17 | 4+ | 4+ | 4+ | 4+ | 3+ |
Strains beginning with asterisks were subjected to whole-genome sequencing.
(-), no mutation.
4+, robust growth; 3+, good growth; 2+, moderate growth; 1+, poor growth; -, no growth; ND, not determined.
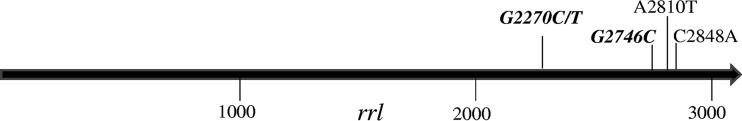
As shown in Table 1, 17 of 32 mutants had a mutation in the rrl gene, and these mutations can be divided into 5 different mutation types, C2848A, A2810T, G2270C, G2270T, and G2746A; mutations at G2746A and G2270 were the most dominant at 47.1% (8/17) and 41.2% (7/17), respectively (Fig. 1). Fifteen mutants without rrl mutations all had the same T460C mutation, which caused substitution of Cys to Arg at amino acid position 154 of the rplC gene. The LZD-susceptible parent strain H37Rv had no mutations in rrl or in rplC.
FIG 1.
Distribution of mutations in the rrl gene of M. tuberculosis mutants with reduced susceptibility to LZD. Bold and italic fonts at G2270 and G2746 indicate dominant mutation sites.
An interesting observation of this study is that mutations in rplC are associated with higher LZD MIC values (MICs of >2 μg/ml), whereas mutations in rrl are correlated with lower MIC values (MICs of 0.5 to 1 μg/ml) (Table 1). It will be of interest to determine if different levels of reduced susceptibility mediated by rplC or rrl have any impact on clinical treatment of drug-resistant TB using LZD in the future.
It has been reported that the G2061T or G2576T mutation in the rrl gene may cause LZD resistance in mutants of M. tuberculosis isolated in vitro (8). In this work, it is noteworthy that we found 5 new mutations (C2848A, A2810T, G2270C, G2270T, and G2746A) in the rrl gene that are associated with reduced susceptibility to LZD (Table 1). The reason why the mutations we identified in rrl differ from those reported in the previous study is unclear but may be due to differences in the mutant selection conditions or to a reflection of different levels of susceptibility. Future structural studies are needed to shed light on how these mutations at different locations can cause LZD resistance. The rplC T460C substitution was found to be a dominant mutation in 6 of 7 LZD-resistant strains isolated from patients in vitro and in vivo (9), and this mutation accounted for all of the remaining 15 mutants with higher MIC values without rrl mutations in this study (Table 1). Thus, the T460C mutation is likely an important mutation for in vitro and in vivo conditions.
An intriguing observation is that 7 mutants with reduced susceptibility initially failed to show any mutation in rrl or rplC by Sanger sequencing (Table 1, strains with an asterisk). However, subsequent Illumina whole-genome sequencing was able to identify a mixed population of mutants having rrl or rplC mutations with sensitive organisms with wild-type rrl or rplC sequences in various proportions (Table 1). Nevertheless, it is of interest to note that the minor proportion of the mutants was able to confer a phenotype of reduced susceptibility (Table 1). This seems unusual, as it is not seen for other drug-resistant mutants conferring resistance to other TB drugs such as pyrazinamide (PZA) (13). This may be due to the nature of LZD in producing a mixed population during mutant selection. Further studies are needed to determine the mechanism and whether this also occurs in vivo during treatment. Alternatively, since efflux has been shown to play a role in LZD resistance in M. tuberculosis (6), it is possible that the reduced susceptibility to LZD in the 7 mutants with only a minor portion of mutants containing rrl or rplC mutations (Table 1) may be due to elevated efflux, which works together with resistance mutations to confer a high level of resistance (14). Future studies are needed to confirm this.
In conclusion, we found that mutants with reduced bacillary susceptibility to LZD harbor rrl and rplC mutations in about equal proportions. In addition, we found new mutations located at the C-terminal part of rrl that have not been previously reported. Furthermore, mutations in rplC are associated with higher MIC values than rrl mutations. These findings provide useful new information on the relative frequency of mutations and should help to develop molecular tests for rapid detection of LZD resistance for more effective treatment of MDR/XDR-TB.
Funding Statement
This work was supported in part by NIH grants AI99512 and AI108535, the Key Technologies Research and Development Program of China (2013ZX10003008-003), National Natural Science Foundation of China (81572046), and Shanghai Natural Science Foundation (13XD1401200, 12ZR1441500).
REFERENCES
- 1.Gu B, Kelesidis T, Tsiodras S, Hindler J, Humphries RM. 2013. The emerging problem of linezolid-resistant Staphylococcus. J Antimicrob Chemother 68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birmingham MC, Rayner CR, Meagher AK, Flavin SM, Batts DH, Schentag JJ. 2003. Linezolid for the treatment of multidrug-resistant, gram-positive infections: experience from a compassionate-use program. Clin Infect Dis 36:159–168. doi: 10.1086/345744. [DOI] [PubMed] [Google Scholar]
- 3.Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother 42:3251–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee M, Lee J, Carroll MW, Choi H, Min S, Song T, Via LE, Goldfeder LC, Kang E, Jin B, Park H, Kwak H, Kim H, Jeon HS, Jeong I, Joh JS, Chen RY, Olivier KN, Shaw PA, Follmann D, Song SD, Lee JK, Lee D, Kim CT, Dartois V, Park SK, Cho SN, Barry CE III. 2012. Linezolid for treatment of chronic extensively drug-resistant tuberculosis. N Engl J Med 367:1508–1518. doi: 10.1056/NEJMoa1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schecter GF, Scott C, True L, Raftery A, Flood J, Mase S. 2010. Linezolid in the treatment of multidrug-resistant tuberculosis. Clin Infect Dis 50:49–55. doi: 10.1086/648675. [DOI] [PubMed] [Google Scholar]
- 6.Richter E, Rusch-Gerdes S, Hillemann D. 2007. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother 51:1534–1536. doi: 10.1128/AAC.01113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Pang Y, Wang Y, Liu C, Zhao Y. 2014. Beijing genotype of Mycobacterium tuberculosis is significantly associated with linezolid resistance in multidrug-resistant and extensively drug-resistant tuberculosis in China. Int J Antimicrob Agents 43:231–235. doi: 10.1016/j.ijantimicag.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Hillemann D, Rusch-Gerdes S, Richter E. 2008. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother 52:800–801. doi: 10.1128/AAC.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, Feuerriegel S. 2012. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother 56:2743–2745. doi: 10.1128/AAC.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locke JB, Hilgers M, Shaw KJ. 2009. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob Agents Chemother 53:5265–5274. doi: 10.1128/AAC.00871-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Yew WW. 2015. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis 19:1276–1289. doi: 10.5588/ijtld.15.0389. [DOI] [PubMed] [Google Scholar]
- 12.Huang TS, Liu YC, Sy CL, Chen YS, Tu HZ, Chen BC. 2008. In vitro activities of linezolid against clinical isolates of Mycobacterium tuberculosis complex isolated in Taiwan over 10 years. Antimicrob Agents Chemother 52:2226–2227. doi: 10.1128/AAC.00414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Chen J, Shi W, Liu W, Zhang WH, Zhang Y. 2013. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microb Infect 2:e34. doi: 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmalstieg AM, Srivastava S, Belkaya S, Deshpande D, Meek C, Leff R, van Oers NS, Gumbo T. 2012. The antibiotic resistance arrow of time: efflux pump induction is a general first step in the evolution of mycobacterial drug resistance. Antimicrob Agents Chemother 56:4806–4815. doi: 10.1128/AAC.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]