Abstract
Using a Galleria mellonella animal model, we compared the virulence of two sequence type 258 (ST258) KPC-producing Klebsiella pneumoniae strains, which were representative of the two clades of this clonal lineage, with that of isogenic colistin-resistant mgrB mutants. With both strains, the mgrB mutants did not exhibit modification in virulence. In the G. mellonella model, the clade 1 strain (capsular type cps-1 [wzi29, producing KPC-2]) was significantly more virulent than the clade 2 strain (capsular type cps-2 [wzi154, producing KPC-3]).
TEXT
Colistin is a backbone component of combination antimicrobial regimens for serious infections caused by carbapenemase-producing strains of Klebsiella pneumoniae (1, 2), which are among the most challenging antibiotic-resistant pathogens spreading globally (3, 4). However, in settings in which carbapenemase-producing K. pneumoniae is endemic, the emergence and rapid dissemination of colistin resistance have been reported, especially among KPC carbapenemase-producing K. pneumoniae strains, including those of the pandemic sequence type 258 (ST258) clonal lineage (5–9).
Colistin resistance is mostly due to modification of the antibiotic target (the lipid A moiety of the bacterial lipopolysaccharide [LPS]), which can be mediated by different mutational events upregulating the endogenous LPS modification systems (10–14), or by the acquisition of exogenous genes encoding LPS modification systems (15). Mutations causing a loss of function in the MgrB protein, a negative-feedback regulator of the PhoP-PhoQ signal transduction system, which controls several biochemical pathways, including those involved in LPS modification, are a frequent mechanism of acquired colistin resistance among KPC carbapenemase-producing K. pneumoniae strains circulating in the clinical setting (14, 16, 17).
We previously showed that colistin resistance associated with inactivation of the mgrB gene is not associated with a significant biological cost and is stably maintained in the absence of selective pressure (18), a finding consistent with the rapid and efficient dissemination of colistin-resistant (COLr) KPC carbapenemase-producing K. pneumoniae strains carrying this resistance mechanism observed in the clinical setting (7, 9).
In this work, we investigated the impact of colistin resistance, mediated by mgrB inactivation, on the virulence of KPC carbapenemase-producing K. pneumoniae strains representative of the two clades of the pandemic ST258 clonal lineage (19); we used the Galleria mellonella infection model, which is considered to be a validated model for testing the virulence of K. pneumoniae (20, 21).
The KPC carbapenemase-producing K. pneumoniae strains used in the experiments are described in Table 1. KK207-1 and KKBO-1 are two previously described strains and are representative of the two clades of ST258 (clades 1 and 2, respectively) (22). The mgrB insertional mutant of KKBO-1 (named mutKKBO-1) was previously described (18), while the mgrB mutant of KK207-1 (named mutKK207-1) was obtained with the same approach as for mutKKBO-1 and carried an ISKpn18 insertion disrupting the mgrB gene. The colistin MICs of the strains were determined by reference broth microdilution (23).
TABLE 1.
Features of the bacterial strains included in this study
| Strain (source or reference) | Sequence type | KPC type | Cladea | Capsular type | LD50 ± standard errorb | Colistin MIC (μg/ml) | mgrB alteration (orientation)c |
|---|---|---|---|---|---|---|---|
| KKBO-1 (10) | 258 | blaKPC-3 | 2 | cps-2 (wzi154) | 6.02 ± 0.09 | 0.125 | Wild type |
| mutKKBO-1 (18) | 258 | blaKPC-3 | 2 | cps-2 (wzi154) | 5.92 ± 0.02 | 8 | IS5-like element at nt 75 (R) |
| KK207-1 (22) | 258 | blaKPC-2 | 1 | cps-1 (wzi29-K41) | 4.68 ± 0.47 | 0.125 | Wild type |
| mutKK207-1 (this study) | 258 | blaKPC-2 | 1 | cps-1 (wzi29-K41) | 4.88 ± 0.13 | 32 | ISKpn18 element at nt 93 (R) |
| NTUH-K2044 (24) | 23 | NAd | wzi1-K1 | 4.57 ± 0.23 | 0.125 | Wild type |
According to reference 19.
Expressed as log CFU LD50.
R, transposase gene is in the opposite orientation of the mgrB gene.
NA, not applicable.
For experiments with G. mellonella, bacteria were grown aerobically in LB broth at 37°C, harvested during exponential phase (optical density at 600 nm [OD600], ≈0.7), and washed once with 10 mM phosphate-buffered saline (PBS) (pH 6.5). Bacteria were then suspended in PBS to an OD600 of 1.5, corresponding to approximately 1 × 109 CFU/ml. Larvae weighing 450 to 600 mg were used for the experiments. For a comparative evaluation of virulence, groups of 10 larvae were injected with 5 × 105 CFU of each strain and with sterile PBS as a control. The larvae were kept at 37°C in the dark under a humidified atmosphere, with food, and examined daily for pigmentation and mobility. Time of death was recorded at 24, 48, and 72 h. Three independent experiments were performed. The 50% lethal dose (LD50) was determined by inoculating larvae with 10-fold serial dilutions of each strain containing 5 × 104 to 5 × 107 CFU. Ten larvae were injected with each dilution. For each strain, data from three independent experiments were combined. LD50s were calculated using the GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA) and were compared using a two-tailed t test. P values of <0.05 were considered statistically significant. K. pneumoniae NTUH-K2044, an ST23 virulent strain with the K1 capsular serotype (24), was included in all experiments as a virulent control.
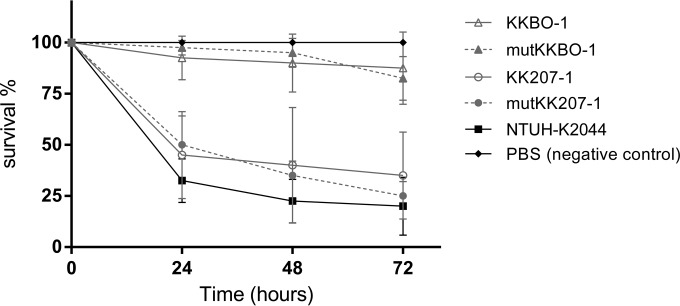
All the studied strains caused a time-dependent death of larvae. In the 5 × 105 CFU challenge, KKBO-1 (clade 2) killed, on average, 10% and 17% of larvae at 24 h and 72 h postinfection, respectively. In contrast, KK207-1 (clade 1) caused higher mortality, with average mortality rates of 55% and 65% at 24 h and 72 h, respectively. No differences were seen between the killing rates caused by the colistin-susceptible ST258 strains and the respective mgrB-inactivated COLr mutants (Fig. 1).
FIG 1.
Results of G. mellonella infection experiments. Larvae were injected with PBS or with 5 × 105 CFU of K. pneumoniae strain NTUH-K2044, KKBO-1, mutKKBO-1, KK207-1, or mutKK207-1 (10 larvae per strain), and survival was monitored over 72 h postinfection. The data are mean values from the results of three independent experiments, and the error bars represent standard deviations from the mean.
In the experiments with scalar inoculum sizes, all tested strains caused larval mortality in a dose-dependent manner. No significant differences were observed in the LD50s of KKBO-1 (colistin susceptible [COLs]) versus mutKKBO-1 (COLr) (P = 0.13), and of KK207-1 (COLs) versus mutKK207-1 (COLr) (P = 0.51). On the other hand, the LD50s of KK207-1 and mutKK207-1 (clade 1) were significantly lower than those of KKBO-1 and mutKKBO-1 (clade 2) (P < 0.05) (Table 1).
The COLr phenotype was checked in isolates obtained from larvae infected with mutKKBO-1 and mutKK207-1 at 24 and 72 h postinfection (60 isolates per experiment) and was confirmed in all tested isolates, revealing stability after the in vivo passage.
Altogether, the results of these experiments indicated that the emergence of colistin resistance mediated by inactivation of the mgrB gene is apparently not associated with significant modifications to the virulence potential of KPC carbapenemase-producing K. pneumoniae representative of the two clades of the ST258 clonal lineage. This finding is notably different from what was observed with COLr mutants of Acinetobacter baumannii, which exhibit a remarkable impairment of virulence potential (25–27), and it is consistent with the observation that COLr KPC carbapenemase-producing K. pneumoniae strains of clonal complex 258 (CC258) are capable of causing severe invasive infections and large outbreaks (8, 9), similar to their colistin-susceptible counterparts. In fact, recent clinical studies have reported that mortality rates among patients with infections caused by COLr KPC carbapenemase-producing K. pneumoniae strains were higher than those among patients infected by colistin-susceptible KPC carbapenemase-producing K. pneumoniae strains (8, 28). The present results suggest that this reported mortality rate excess is likely attributable to the further narrowing of available therapeutic options rather than to an increased virulence potential of COLr KPC carbapenemase-producing K. pneumoniae strains.
Our data also confirmed previous findings that ST258 strains of clade 2 (with a cps-2 capsular gene cluster) exhibit an overall low virulence potential in the G. mellonella model (29) compared with that of highly virulent K. pneumoniae strains, such as NTUH-K2044. On the other hand, for the ST258 strain of clade 1 (with a cps-1 capsular gene cluster), we documented a higher virulence potential than that for clade 2 strains, which was similar to that of NTUH-K2044. This finding apparently differs from that of a previous study that reported a low virulence potential for a KPC-2-producing ST258 strain of clade 1 (deduced by the K41 serotype) in a mouse septicemia model (30). Further studies will be necessary to confirm these discrepancies, which might be dependent on strain-specific differences and/or differences in the animal infection models and their immune systems. In fact, G. mellonella moths lack an adaptive immune response, and the model can be used only to approximate mammals' innate immune response to bacterial infections. However, the difference in virulence among the clade 1 and clade 2 strains observed in this work is consistent with the different prevalences observed for the two clades, with an overall predominance of clade 2 strains in settings of high-level endemicity of KPC carbapenemase-producing K. pneumoniae strains of CC258 (22, 29).
In conclusion, our findings suggest that COLr KPC carbapenemase-producing K. pneumoniae strains of ST258 carrying an inactivated mgrB gene might be as virulent as their COLs ancestors. Efforts focused at containing their dissemination should therefore be maximized. Our findings also suggest that significant differences might exist in the interaction of ST258 strains of KPC carbapenemase-producing K. pneumoniae of clades 1 and 2 with the innate immune response, regardless of their colistin susceptibility status.
ACKNOWLEDGMENTS
We thank Pei-Fang Hsieh and Jin-Town Wang for providing us with the NTUH-K2044 strain.
This work was supported in part by a grant from FP7 project EvoTAR (grant HEALTH-F3-2011-2011-282004) to G.M.R.
REFERENCES
- 1.Petrosillo N, Giannella M, Lewis R, Viale P. 2013. Treatment of carbapenem-resistant Klebsiella pneumoniae: the state of the art. Expert Rev Anti Infect Ther 11:159–177. doi: 10.1586/eri.12.162. [DOI] [PubMed] [Google Scholar]
- 2.Rafailidis PI, Falagas ME. 2014. Options for treating carbapenem-resistant Enterobacteriaceae. Curr Opin Infect Dis 27:479–483. doi: 10.1097/QCO.0000000000000109. [DOI] [PubMed] [Google Scholar]
- 3.Rossolini GM. 2015. Extensively drug-resistant carbapenemase-producing Enterobacteriaceae: an emerging challenge for clinicians and healthcare systems. J Intern Med 277:528–531. doi: 10.1111/joim.12350. [DOI] [PubMed] [Google Scholar]
- 4.Nordmann P, Poirel L. 2014. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 5.Zarkotou O, Pournaras S, Voulgari E, Chrysos G, Prekates A, Voutsinas D, Themeli-Digalaki K, Tsakris A. 2010. Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case-control study. J Clin Microbiol 48:2271–2274. doi: 10.1128/JCM.02301-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y. 2011. Colistin-resistant, Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae belonging to the international epidemic clone ST258. Clin Infect Dis 53:373–376. doi: 10.1093/cid/cir401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, Network EuSCAPE-Italy, Grundmann H, Pantosti A, Rossolini GM. 2014. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill 19:pii=20939. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20939. [DOI] [PubMed] [Google Scholar]
- 8.Giacobbe DR, Del Bono V, Trecarichi EM, De Rosa FG, Giannella M, Bassetti M, Bartoloni A, Losito AR, Corcione S, Bartoletti M, Mantengoli E, Saffioti C, Pagani N, Tedeschi S, Spanu T, Rossolini GM, Marchese A, Ambretti S, Cauda R, Viale P, Viscoli C, Tumbarello M, ISGRI-SITA (Italian Study Group on Resistant Infections of the Società Italiana Terapia Antinfettiva). 2015. Risk factors for bloodstream infections due to colistin-resistant KPC-producing Klebsiella pneumoniae: results from a multicenter case-control-control study. Clin Microbiol Infect 21:1106.e1–1106.e8. doi: 10.1016/j.cmi.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, De Angelis LH, Fornaini R, Grazzini M, Niccolini F, Pecile P, Rossolini GM. 2015. Large nosocomial outbreak of colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae by clonal expansion of an mgrB deletion mutant. J Clin Microbiol 53:3341–3344. doi: 10.1128/JCM.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain JM. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, D'Andrea MM, Rossolini GM. 2014. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother 58:4399–4403. doi: 10.1128/AAC.02555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Y-H, Lin T-L, Pan Y-J, Wang Y-P, Lin Y-T, Wang J-T. 2015. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother 59:2909–2913. doi: 10.1128/AAC.04763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y-Y, Wang Y, Walsh TR, Yi L- X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2015. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 15:424–427. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 16.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 18.Cannatelli A, Santos-Lopez A, Giani T, Gonzalez-Zorn B, Rossolini GM. 2015. Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob Agents Chemother 59:2898–2900. doi: 10.1128/AAC.04998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, Bonomo RA, Musser JM, Kreiswirth BN. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A 111:4988–4993. doi: 10.1073/pnas.1321364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Insua JL, Llobet E, Moranta D, Pérez-Gutiérrez C, Tomás A, Garmendia J, Bengoechea JA. 2013. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect Immun 81:3552–3565. doi: 10.1128/IAI.00391-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavanagh K, Reeves EP. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol Rev 28:101–112. doi: 10.1016/j.femsre.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 22.D'Andrea MM, Amisano F, Giani T, Conte V, Ciacci N, Ambretti S, Santoriello L, Rossolini GM. 2014. Diversity of capsular polysaccharide gene clusters in KPC-producing Klebsiella pneumoniae clinical isolates of sequence type 258 involved in the Italian epidemic. PLoS One 9:e96827. doi: 10.1371/journal.pone.0096827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.CLSI. 2015. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically; approved standard, 10th ed CLSI document M07-A10. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.Fang C-T, Chuang Y-P, Shun C-T, Chang S-C, Wang J-T. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López-Rojas R, Domínguez-Herrera J, McConnell MJ, Docobo-Peréz F, Smani Y, Fernández-Reyes M, Rivas L, Pachón J. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 203:545–548. doi: 10.1093/infdis/jiq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. 2014. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 58:518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pournaras S, Poulou A, Dafopoulou K, Chabane YN, Kristo I, Makris D, Hardouin J, Cosette P, Tsakris A, Dé E. 2014. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother 58:828–832. doi: 10.1128/AAC.01439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capone A, Giannella M, Fortini D, Giordano A, Meledandri M, Ballardini M, Venditti M, Bordi E, Capozzi D, Balice MP, Tarasi A, Parisi G, Lappa A, Carattoli A, Petrosillo N, SEERBIO-GRAB Network. 2013. High rate of colistin resistance among patients with carbapenem-resistant Klebsiella pneumoniae infection accounts for an excess of mortality. Clin Microbiol Infect 19:E23–E30. doi: 10.1111/1469-0691.12070. [DOI] [PubMed] [Google Scholar]
- 29.Diago-Navarro E, Chen L, Passet V, Burack S, Ulacia-Hernando A, Kodiyanplakkal RP, Levi MH, Brisse S, Kreiswirth BN, Fries BC. 2014. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J Infect Dis 210:803–813. doi: 10.1093/infdis/jiu157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzouvelekis LS, Miriagou V, Kotsakis SD, Spyridopoulou K, Athanasiou E, Karagouni E, Tzelepi E, Daikos GL. 2013. KPC-producing, multidrug-resistant Klebsiella pneumoniae sequence type 258 as a typical opportunistic pathogen. Antimicrob Agents Chemother 57:5144–5146. doi: 10.1128/AAC.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]