Abstract
Protease inhibitors (PIs) are used as a first-line regimen in HIV-1-infected children. Here we investigated the phenotypic consequences of amino acid changes in Gag and protease on lopinavir (LPV) and ritonavir (RTV) susceptibility among pediatric patients failing PI therapy. The Gag-protease from isolates from 20 HIV-1 subtype C-infected pediatric patients failing an LPV and/or RTV-based regimen was phenotyped using a nonreplicative in vitro assay. Changes in sensitivity to LPV and RTV relative to that of the matched baseline (pretherapy) sample were calculated. Gag and protease amino acid substitutions associated with PI failure were created in a reference clone by site-directed mutagenesis and assessed. Predicted phenotypes were determined using the Stanford drug resistance algorithm. Phenotypic resistance or reduced susceptibility to RTV and/or LPV was observed in isolates from 10 (50%) patients, all of whom had been treated with RTV. In most cases, this was associated with protease resistance mutations, but substitutions at Gag cleavage and noncleavage sites were also detected. Gag amino acid substitutions were also found in isolates from three patients with reduced drug susceptibilities who had wild-type protease. Site-directed mutagenesis confirmed that some amino acid changes in Gag contributed to PI resistance but only in the presence of major protease resistance-associated substitutions. The isolates from all patients who received LPV exclusively were phenotypically susceptible. Baseline isolates from the 20 patients showed a large (47-fold) range in the 50% effective concentration of LPV, which accounted for most of the discordance seen between the experimentally determined and the predicted phenotypes. Overall, the inclusion of the gag gene and the use of matched baseline samples provided a more comprehensive assessment of the effect of PI-induced amino acid changes on PI resistance. The lack of phenotypic resistance to LPV supports the continued use of this drug in pediatric patients.
INTRODUCTION
Protease inhibitors (PIs) are potent antiretroviral drugs which inhibit the function of the HIV-1 protease enzyme, thereby preventing viral maturation (1). In South Africa, the use of ritonavir (RTV)-boosted lopinavir (LPV/r) is recommended as a first-line regimen for pediatric patients <3 years of age and as a second-line regimen for adults and older children (2). Prior to 2008, RTV was used as a single PI in infants <6 months of age and also for those receiving rifampin for the cotreatment of tuberculosis (3, 4). It is estimated that over half a million HIV-1-infected infants are being treated with PIs, and 140,000 of these infants reside in South Africa (5). Current guidelines recommend treatment of infants immediately upon a diagnosis of HIV infection, and treatment according to these guidelines has been associated with good clinical outcomes (6, 7). Nevertheless, an analysis of children treated in South Africa showed that the probability of virological suppression at age 12 months was only 56% (8). This may be due to the higher viral loads and challenges associated with accurate dosing in infants, placing them at a potentially greater risk for developing PI drug resistance than adult patients (9).
Resistance to PIs is characterized by the gradual accumulation of major mutations in the protease gene, including M46I, I54V, and V82A, as well as accessory mutations that can enhance resistance but appear to have no effect individually (10). However, many adults and children fail PI-based therapies in the absence of any protease resistance-associated amino acid substitutions (11–13). While such cases are often attributed to poor adherence, it has been suggested that regions outside protease may contribute to PI resistance (1, 9, 14–16). HIV-1 protease recognizes and cleaves the Gag and Gag-Pol polyproteins at specific cleavage sites (CSs) to produce infectious virions. Since PIs inhibit the cleavage of viral proteins, this prevents the formation of mature infectious particles (17). PI resistance mutations on their own reduce viral fitness; however, amino acid substitutions at the Gag CS and non-CS that restore viral fitness in the presence of protease substitutions have been identified (1, 18–22). Furthermore, Gag CS substitutions at positions 431, 436, and 437 have been associated with a reduction in PI susceptibility and virological failure during PI therapy in the absence of protease substitutions (23, 24). One study showed the interdependency between a nelfinavir-resistant protease (D30N/N88D) and P1/P6Gag CS substitutions (L449F and S451N) (25). This association likely strengthens the intermolecular interactions between the Gag substrate and the active site of protease to allow the coevolution of mutant protease and Gag. Analysis of HIV-1 Gag sites associated with PI resistance has identified the CS substitutions S451G and A431V in subtype B and circulating recombinant form (CRF) 01_AE isolates and the non-CS substitution K415R in subtype C isolates (26). Furthermore, analysis of the sequences of subtype C isolates from drug-naive patients has shown variability in the Gag CS which may impact viral fitness and/or PI efficacy (27). The CS at p2 in the nucleocapsid was found to be the most highly variable, followed closely by P6pol/PR, while the remaining CSs were relatively conserved. These data suggest that genetic diversity in Gag may contribute to PI resistance.
We previously reported genotypic changes in protease and Gag among isolates from 20 HIV-1 subtype C-infected pediatric patients failing a PI-based regimen. In addition to protease mutations, we identified amino acid substitutions in Gag in some patient isolates that had the wild-type protease (28). Routine drug resistance assays and algorithms analyze changes in protease only when PI resistance is assessed and do not take Gag into consideration. We therefore used a phenotypic assay to examine whether Gag amino acid changes in the presence and absence of protease substitutions impact susceptibility to RTV and LPV and, in so doing, contribute to PI failure.
MATERIALS AND METHODS
Samples from HIV-1-infected children.
Twenty matched baseline (pretherapy) and posttreatment plasma samples from children who were less than 2 years of age and who had failed a PI-based regimen were studied (29–31). Children were initiated on first-line treatment and followed as part of a randomized clinical trial between 2005 and 2009. Those children who were <6 months of age or who were receiving rifampin for concomitant tuberculosis were treated with a RTV-based regimen according to standard practice in South Africa between 2005 and 2009. The use of RTV in these cases was as a singular PI and not as a pharmacological booster. Once the children had passed 6 months of age or the tuberculosis had cleared, they were switched onto a ritonavir-boosted lopinavir (LPV/r) regimen. The nucleoside reverse transcriptase inhibitors (NRTIs) lamivudine and stavudine formed the backbone of the RTV and LPV/r-based regimens (30). This study was approved by the institutional review boards of Columbia University and the University of the Witwatersrand.
Construction of patient-derived test vectors.
Phenotypic susceptibility to LPV and RTV was determined using a retroviral vector system capable of a single infectious event, as previously described (19, 22). For this, an HIV-1 expression vector, p8.9NSX+ (22, 32), containing a NotI restriction site at the beginning of Gag (isolate HXB2 position 772) and a XhoI restriction site at the end of protease (HXB2 position 2563) was used. The vector was digested with NotI and XhoI restriction enzymes to enable cloning of patient-derived gag-protease genes. The gag-protease gene from the p8.MJ4 (33) subtype C reference clone was used to create p8.9MJ4GP. The latter functioned as a wild-type control in the PI phenotypic assay.
Viral RNA was extracted from 200 μl patient plasma using a QIAamp viral RNA minikit (Qiagen, Belgium) and reverse transcribed using a ThermoScript reverse transcription-PCR kit (Invitrogen, CA, USA) and gene-specific primer 3′InProt (5′-CCTGGCTTTAATTTTACTGGTACAG-3′). A 1.8-kb Gag-protease fragment spanning HXB2 positions 792 to 2549 was amplified by nested PCR using an Expand high-fidelity PCR kit (Roche Applied Science, Basel, Switzerland). Primers BKT03 (5′-CGCAGGACTCGGCTTGC-3′) and ProOutR (5′-TTGGGCCATCCATTCCTGG-3′) were used for the first-round PCR, while primers GagNot+ (5′-GCGGCGGCCGCAAGGAGAGAGATGGGTGCG-3′) and ProXhoR2 (5′-CTGGTACAGTCTCGAGRGGACTRATKGG-3′) were used during the second-round PCR to introduce the NotI and XhoI restriction sites (underlined) into the 1.8-kb Gag-protease. The second-round products were cloned into the pCR-2.1 TOPO cloning vector (TOPO TA cloning kit; Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Clones containing Gag-protease inserts of the correct size were subcloned into the p8.9NSX+ expression vector by NotI/XhoI restriction (Roche, Basel, Switzerland) and subsequent ligation with a Rapid DNA Dephos and Ligation dephosphorylation and ligation kit (Roche, Basel, Switzerland). Ligation products were transformed into Escherichia coli XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA, USA) by a standard heat shock method. All pCR-2.1 TOPO and p8.9NSX+ clones were sequenced, as described below, to confirm the presence of the relevant mutations and polymorphisms. Single clones from the baseline and PI treatment failure time points whose sequences were the most representative of the sequences of the population at that time point were selected.
Site-directed mutagenesis.
A 3.6-kb Gag-protease-reverse transcriptase fragment (bp 775 to 4,403 of HXB2) in the p8.9NSX+ expression plasmid was replaced with the corresponding fragment from the pMJ4 subtype C reference strain (GenBank accession number AF321523.1) to create p8.MJ4 (33). Site-directed mutants were created in the p8.MJ4 expression plasmid by use of a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) as previously described (31). The mutagenesis primers were designed using the QuikChange Primer Design online tool and synthesized by Integrated DNA Technologies (Coralville, IA, USA). Clones with multiple mutations were created by successive rounds of site-directed mutagenesis, and the presence of mutations was confirmed by population-based sequencing. p8.MJ4 was used as a control for the site-directed mutants in the PI susceptibility assays.
Phenotypic assessment of protease inhibitor susceptibility.
A phenotypic assay which allows only a single infectious event per virion was used, as previously described (19, 22), with minor modifications. Briefly, 1.25 × 106 HEK293T cells were transfected with 300 ng of plasmid pMDG (carrying the gene for the vesicular stomatitis virus G protein), 500 ng of plasmid pCSFLW (carrying the gene for the firefly luciferase protein), and 300 ng the HIV-1 expression vector (containing patient-derived inserts or site-directed mutations) using 3.3 μg polyethylenimine (PEI; Polysciences, Inc., Warrington, PA, USA). The transfected cells were harvested after 18 h and seeded in the presence of serial dilutions of LPV or RTV. After 24 h, the supernatants were transferred to the corresponding wells of 96-well culture plates that contained fresh HEK293T cells. The degree of infection was determined 48 h after supernatant transfer by measuring the expression of firefly luciferase with a BrightGlo luciferase assay system (Promega, Madison, WI, USA). For each drug-virus combination, the 50% effective concentration (EC50) was calculated. For patient-derived clones, phenotypic susceptibility was expressed as the fold change (FC) in the EC50 relative to that for the baseline virus. The assay-specific lower FC cutoff value for each drug was determined using the 99th percentile of the average EC50 for p8.MJ4GP and p8.MJ4 assessed in multiple repeat screens of each drug. For LPV, we used the upper cutoff (55-FC) of the Monogram Biosciences PhenoSense assay. There are no clinical cutoffs available for RTV. On the basis of this, phenotypes were classified as sensitive (S; for LPV, <3.6-FC; for RTV, <3.8-FC), reduced susceptibility (RS; for LPV, 3.6- to 55-FC; for RTV, >3.8-FC), or resistant (R; for LPV, >55-FC). These classifications served to rank the phenotypic responses to the PIs and are not linked to clinical outcomes.
Sequence analysis.
Amplicons were analyzed for changes in the gag and protease genes by population-based sequencing using a BigDye Terminator (v3.1) cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA). Polymorphisms in the protease and gag genes were defined as changes in amino acid sequence between the sequences obtained at the time of PI treatment failure and those obtained at the baseline. Protease sequences were submitted to the Stanford HIV drug resistance database (hivdb.stanford.edu/) to identify major and accessory protease resistance mutations and predict phenotypic susceptibility to LPV/r. The predicted phenotype for RTV could not be determined through the Stanford HIV drug resistance database.
Statistical analysis.
Two-way analysis of variance (ANOVA) and Bonferroni's posttest were used to identify significant differences in FC values between site-directed mutants using GraphPad Prism (v5.00) software for Windows (GraphPad Software, San Diego, CA, USA).
RESULTS
Phenotypic susceptibility of patient-derived protease and Gag.
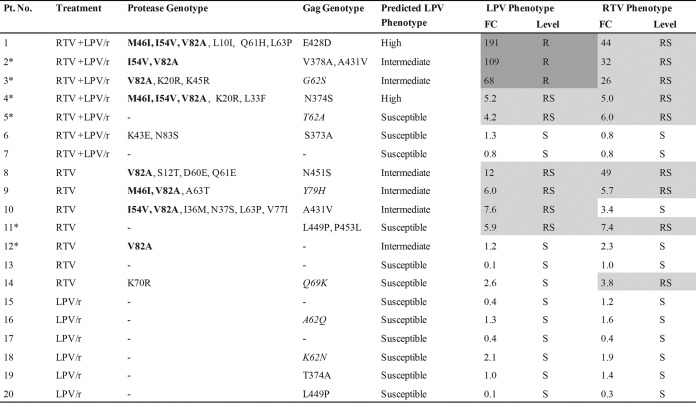
We previously reported the genotypic changes in the gag and protease genes among isolates from 20 HIV-1 subtype C-infected pediatric patients failing a PI-based regimen (28). Of these, 8 were found to have major protease amino acid changes, while 9 had Gag CS changes and 17 had non-CS changes. In the current study, we sought to examine the impact of these amino acid changes on phenotypic resistance. The LPV and RTV phenotypes of samples from all 20 children together with their treatment history, genotypic profiles, and predicted phenotypes are shown in Fig. 1. All genotypic changes in the protease gene and gag CSs, as well as the phenotypic fold changes in EC50s, between the samples obtained at the time of PI treatment failure and the corresponding baseline samples are reported.
FIG 1.
Genotypic and phenotypic resistance of pediatric patient samples to LPV and RTV at treatment failure. The treatment histories of the patients are shown, where RTV + LPV/r indicates the receipt of both RTV as a single PI and LPV/r at some point, RTV indicates the receipt of RTV as a single PI only, and LPV/r refers to the receipt of LPV/r only. The Stanford HIV drug resistance database was used to predict the phenotype. Asterisks, the six patients for whom discordance between the predicted and the actual phenotype was found. Levels of drug resistance were classified as follows: sensitive (S; no shading), reduced susceptibility (RS; light gray shading), and resistant (R; dark gray shading). The assay cutoffs used are as follows: for sensitive, a <3.6-FC in the EC50 for LPV and a <3.8-FC in the EC50 for RTV; for reduced susceptibility, a 3.6- to 55-FC in the EC50 for LPV and a >3.8-FC in the EC50 for RTV; and for resistant, a >55-FC in the EC50 for LPV. Protease amino acid changes in bold represent major PI mutations. Gag amino acid changes in italics represent those at non-CSs that were previously shown to be associated with PI exposure (28).
Among the isolates from eight patients with major resistance-associated protease amino acid changes, seven had high-level resistance or reduced susceptibility to LPV and six had reduced susceptibility to RTV (Fig. 1). The isolate from one patient (patient 12) with the major protease amino acid change V82A was found to be susceptible to both drugs in our assay, despite having a predicted phenotype of intermediate resistance to LPV. Of the isolates from 12 patients that lacked major protease amino acid substitutions, only 10 were susceptible to LPV and 9 were susceptible to RTV. The isolates from three patients with wild-type protease (patients 5, 11, and 14) showed a reduced susceptibility to RTV, and two of the isolates also showed a reduced susceptibility to LPV. The isolate from patient 11 contained two Gag CS amino acid changes (L449P, P453L) in conjunction with Gag non-CS changes (A67S, P339Q, K410R, and K411G) (data not shown in Fig. 1), as previously reported (28). The isolates from patients 5 and 14 did not show changes at the Gag CS but had Gag non-CS changes: A55T, T62A, K69R, and S148L (patient 5) and Q69K (patient 14) (only those previously shown to be associated with PI exposure are shown in Fig. 1). In addition, the isolate from patient 14 also had an accessory protease amino acid change, K70R.
Comparison of phenotypic resistance levels with the predicted phenotype.
We next compared the LPV resistance phenotype predicted by the Stanford drug resistance algorithm (http://hivdb.stanford.edu/) to the in vitro phenotype. The Stanford algorithm uses data from protease only and does not include predictions for RTV. We noted four patient samples where phenotypic resistance (or reduced susceptibility) to LPV was underpredicted by the Stanford drug resistance algorithm. The isolates from two of these patients (patients 2 and 3) had changes in Gag, in addition to major protease amino acid changes, which may have contributed to the higher levels of resistance detected in our Gag-protease assay. The isolates from two other patients (patients 5 and patient 11) that had reduced susceptibility but that were predicted to be susceptible by the Stanford drug resistance algorithm had changes in Gag in the absence of major protease mutations. The isolate from patient 5 had one Gag non-CS change (T62A), and the isolate from patient 11 had both Gag CS and non-CS substitutions, as discussed above. The isolates from two additional patients (patients 4 and 12) showed levels of resistance lower than those predicted by the Stanford drug resistance algorithm, despite having one or more major protease mutations.
Baseline phenotypic susceptibility relative to that of the p8.MJ4GP control.
Since our assay measures the level of phenotypic resistance relative to that for the matched sample obtained at the baseline (before antiretroviral therapy[pre-ART]), the contribution of natural polymorphisms to resistance levels is negated. We therefore determined if genotypic variations at the baseline influenced PI susceptibility by comparing the EC50s for the isolates in the baseline samples to those for the p8.MJ4GP control. None of the isolates in the baseline samples had major protease resistance-associated amino acid changes, but all harbored protease polymorphisms and Gag CS changes compared to the sequence of p8.MJ4GP.
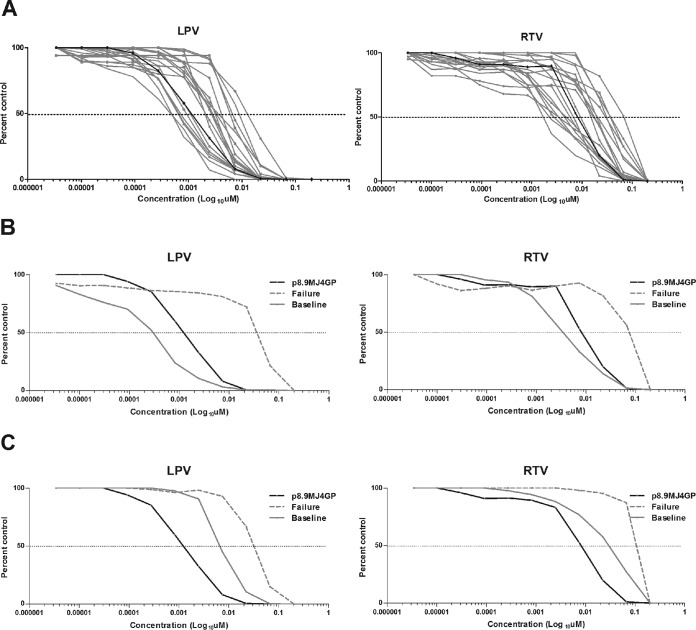
A range of EC50s was observed among the baseline Gag-protease products from the 20 pediatric patients (Fig. 2A). For LPV the mean EC50 was 0.00345 μM (range, 0.0003 to 0.0142 μM), while for RTV it was 0.0174 μM (range, 0.0019 to 0.0687 μM), giving 47-fold and 36-fold ranges of the EC50s, respectively. The p8.MJ4GP control plasmid had an EC50 of 0.0014 μM for LPV and an EC50 of 0.0087 μM for RTV, which placed the values for p8.MJ4GP approximately in the middle of the values for the 20 patient samples. Relative to the control plasmid, the isolates from five patients showed a reduced susceptibility to RTV and the isolates from four showed a reduced susceptibility to LPV (see Table S1 in the supplemental material). Conversely, a number of patients had baseline isolates that were more sensitive than p8.9MJ4GP. This interpatient variation had a major impact on the fold change calculations. As such, the use of p8.9MJ4GP rather than the matched baseline sample underestimated the phenotypic fold change for patient 1 (Fig. 2B), while it overestimated the fold change for patient 10 (Fig. 2C).
FIG 2.
(A) Phenotypic susceptibility to LPV and RTV of Gag-protease from the baseline samples. Dose-response curves of baseline (pre-ART) samples from 20 pediatric patients (gray) relative to the dose-response curve for control plasmid p8.MJ4GP (black) for both LPV and RTV. Dotted line, 50% inhibition. (B) Dose-response curves of the baseline sample and a sample obtained at the time of treatment failure from patient 1. The baseline sample from patient 1 showed increased susceptibility to LPV and RTV relative to the assay control, p8.MJ4GP. The fold change in the EC50 relative to that for the sample obtained at the time of treatment failure is therefore higher than that for the baseline sample rather than the control plasmid. (C) Dose-response curves of samples obtained at the baseline and the time of treatment failure from patient 10. The baseline sample from patient 10 showed reduced susceptibility to LPV and RTV relative to that of the assay control, p8.MJ4GP. The fold change in the EC50 relative to that for the sample obtained at the time of treatment failure is therefore lower than that for the baseline sample rather than the control plasmid. For actual values, see Fig. 1 and Table S1 in the supplemental material.
In order to determine if the use of different reference plasmids accounted for the discordance with the phenotype predicted by the Stanford drug resistance algorithm that was seen, we reassessed the fold changes relative to the EC50 for p8.9MJ4GP. In the reassessment, the phenotypes of five of the six samples with discordant phenotypes were in agreement with the predicted phenotype (see Table S2 in the supplemental material). Only the phenotype of the isolate from patient 4, predicted to have resistance, remained discordant with an intermediate level of resistance relative to the matched baseline sample or the p8.9MJ4GP reference control. This highlights the important contribution of patient-specific Gag-protease polymorphisms to PI phenotypic resistance levels and the value of using of baseline samples when available.
Phenotypic susceptibility of site-directed mutants with protease and/or Gag amino acid changes.
In order to evaluate the contribution of specific Gag and protease amino acid changes to PI resistance, site-directed mutagenesis was used to recreate the amino acid substitution patterns seen in the isolates from the 10 pediatric patients in whom PI treatment had failed. Individual and combinations of protease changes (M46I, I54V, and V82A) and/or Gag CS substitutions (N374S, A431V, L449P, N451S, and P453L) were introduced into the p8.MJ4 backbone through multiple rounds of mutagenesis. In addition, the Gag non-CS substitutions Q62S and F79H were included, as we and others have previously shown these to be under positive selection pressure and/or selected during PI exposure (34, 35).
The fold change in susceptibility to LPV and RTV of the site-directed mutants is shown in Table 1. In general, the site-directed mutants showed lower fold changes than their respective patient samples (Fig. 1). Gag-protease clones from patients 1, 2, 3, 8, and 9 showed resistance to LPV (FC range, 6 to 191) and RTV (FC range, 5.7 to 49), but the corresponding site-directed mutants showed a maximum fold change of 5.9 for LPV and 16 for RTV. The isolate from patient 4 and its matched site-directed mutant clone with N374S, M46I, I54V, and V82A substitutions showed comparable levels of LPV resistance, but for RTV the mutant showed a 4-fold higher level of resistance. The two Gag-CS substitutions L449P and P453L present in the isolate from patient 11, which showed reduced susceptibility to both RTV and LPV in the absence of protease amino acid changes, had no effect on susceptibility to either drug when they were tested in site-directed mutants. Protease and Gag CS/non-CS amino acid changes on their own were unable to confer resistance, similar to what was seen in the isolates from patients 6, 12, 19, and 20.
TABLE 1.
Fold change in LPV and RTV sensitivity for patient-derived clones and the corresponding Gag-protease mutants
| Patient no. | Site-directed mutation(s) in p8.MJ4 backbonea | Fold change in sensitivity of site-directed mutant clones |
|
|---|---|---|---|
| LPV | RTV | ||
| 1 | M46I + I54V + V82A + E428D | 5.9 | 16 |
| 2 | I54V + V82A + A431V | 3.2 | 11 |
| 3 | V82A + Q62S | 0.7 | 1.1 |
| 4 | M46I + I54V + V82A + N374S | 5.9 | 22 |
| 6 | S373A | 1.0 | 1.9 |
| 8 | V82A + N451S | 1.4 | 1.0 |
| 9 | M46I + V82A + F79H | 1.7 | 3.0 |
| 10 | I54V + V82A + A431V | 3.2 | 4.0 |
| 11 | L449P + P453L | 0.8 | 1.5 |
| 12 | V82A | 1.3 | 1.2 |
| 19 | T374A | 1.4 | 1.1 |
| 20 | L449P | 0.7 | 0.6 |
Protease amino acid changes are highlighted in bold, Gag CSs are in normal font, and non-CS amino acid changes are italicized.
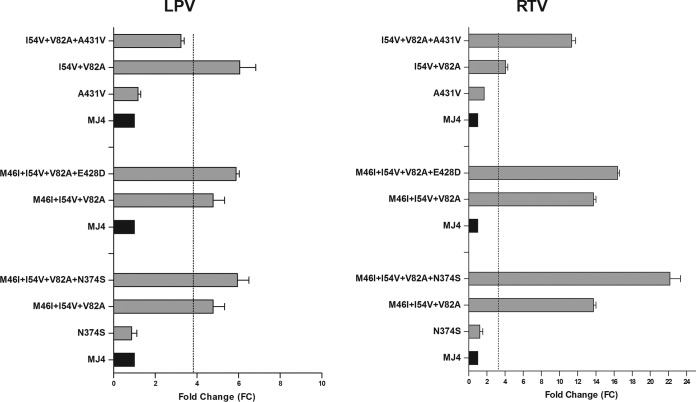
We also examined the effects of adding Gag substitutions to existing major protease amino acid changes. The results for the Gag CS mutants in which the greatest additive effects were found are shown in Fig. 3. Isolates with individual Gag CS substitutions (N374S, E428D, and A431V) showed susceptibility to LPV and RTV. Addition of N374S to the M46I, I54V, and V82A substitutions had a minimal effect on susceptibility to LPV but caused increased resistance (FC, 22.1) to RTV (P < 0.001). Addition of E428D to the M46I, I54V, and V82A substitutions did not alter susceptibility to LPV but caused a slight increase in resistance (FC, 16.4) to RTV (P < 0.05). Gag CS amino acid change A431V reversed the effects of the I54V and V82A substitutions and in doing so increased susceptibility to LPV, while it significantly increased RTV resistance (FC, 11.3) (P < 0.001).
FIG 3.
Impact of Gag amino acid changes on phenotypic resistance to LPV and RTV. The results for Gag CS mutations A431V, E428D, and N374S in various combinations with major protease amino acid changes (M46I, I54V, and V82A) are shown. Black, p8.MJ4 (MJ4), the positive control for the assay; vertical dotted lines, lower assay cutoff values; error bars, standard deviations of the means.
Frequency of Gag and protease amino acid changes in global HIV-1 subtype C sequences.
The frequency of protease amino acid changes identified in the 20 patient samples compared to the sequences in the Stanford HIV drug resistance database (http://hivdb.stanford.edu/cgi-bin/MutPrevBySubtypeRx.cgi) was determined. An analysis of 10,466 subtype C protease sequences from untreated patients and 1,208 subtype C protease sequences from PI-treated patients showed that major amino acid changes (M46I, I54V, and V82A) were rare in isolates from patients that had not been treated with PIs but occurred at frequencies of 9.1%, 11%, and 12%, respectively, in isolates from PI-treated patients (Table 2). This was somewhat lower than the levels seen in our study, particularly for V82A, where 40% of the isolates from pediatric patients harbored this amino acid change, probably due to the use of RTV and/or LPV/r in this cohort.
TABLE 2.
Frequency of PI-associated amino acid changes in pediatric samples and global databases
| Amino acid change | Frequency (%) of PI-associated amino acid changes in isolates from: |
|||
|---|---|---|---|---|
| Pediatric cohort |
HIV-1 subtype C database |
|||
| Baseline (n = 20) | Failure (n = 20) | PI naivea | PI treated (n = 1,208) | |
| Protease | ||||
| M46I | 0 | 15 | 0.2 | 9.1 |
| I54V | 0 | 20 | 0.0 | 11 |
| V82A | 0 | 40 | 0.1 | 12 |
| Gag | ||||
| N374S | 25 | 5 | 21 | NAb |
| A431V | 0 | 10 | 0.1 | NA |
| L449P | 5 | 15 | 0.0 | NA |
| P453L | 10 | 15 | 3.2 | NA |
| Q62Sc | 0 | 5 | 0.8 | NA |
| F79Hc | 0 | 5 | 2.2 | NA |
Data are for 10,466 protease sequences (obtained from Stanford) and 2,152 Gag sequences (obtained from Los Alamos) for PI-naive samples.
NA, not available.
Gag non-CS amino acid changes.
Since the Gag region is not generally sequenced in treatment failures, we were limited to a comparison with subtype C sequences from drug-naive individuals retrieved from the Los Alamos database. The Gag CS amino acid change N374S occurred in 21% of subtype C sequences from PI-naive patients in the database and in 25% of sequences from isolates obtained at the baseline from patients in our cohort, although only one isolate from a patient failing treatment developed this amino acid change (28). Gag CS amino acid changes L449P and P453L were observed at the baseline and increased among the isolates from the pediatric patients obtained at the treatment failure time points. L449P occurred in the isolates from three patients (15%) at the time of treatment failure, and in the isolate from one of these patients (5%), it was also present at the baseline. P453L arose in the isolates from three patients (15%) at the time of treatment of failure and was present in the isolates from two of these patients at the baseline (10%). Gag non-CS changes Q62S and F79H occurred at low frequencies in isolates from PI-naive individuals from the database. Among the isolates from our cohort, these changes were present only in isolates from patients failing PI treatment.
DISCUSSION
This study examined the levels of phenotypic resistance to LPV and RTV in isolates from 20 HIV-1-infected children who had virological failure while they were receiving a first-line PI-containing regimen. In addition to the protease gene, we also studied the gag gene, which has previously been implicated in PI resistance (18, 19, 36, 37). We found that the isolates from the majority of the patients who had received RTV-based therapy had high-level phenotypic resistance or reduced susceptibility to one or both drugs, including isolates from three patients that had changes in Gag in the absence of major protease amino acid changes. Site-directed mutagenesis revealed that the Gag amino acid changes were able to augment protease-mediated resistance in some cases. All samples from children treated with LPV/r-only, however, were phenotypically fully susceptible to both drugs and displayed no PI resistance-associated amino acid changes.
Among the isolates from these 20 pediatric patients, in vitro phenotypic susceptibility to LPV and RTV varied with treatment history. Samples from 10 of the 14 patients treated with RTV or RTV plus LPV/r showed phenotypic resistance or reduced susceptibility, while those from patients treated with LPV/r remained susceptible to both drugs. This generally correlated with the presence or absence of resistance-associated amino acid changes (28, 30). RTV-based regimens have been associated with the selection of protease amino acid changes, and these changes are one of the main reasons why such regimens are no longer recommended (38). These results highlight some of the complex mutation patterns associated with the prior use of RTV-based regimens. Further investigation of the effects of these mutations on PI cross-resistance is warranted to identify effective PIs for use in future regimens. LPV/r is now the mainstay of pediatric treatment regimens and, as shown by us and others, does not commonly select for protease resistance-associated amino acid changes (12, 38). As shown here, LPV/r was found to select for Gag amino acid changes, including those at CS sites; however, these samples were all phenotypically susceptible. Thus, failure on LPV/r is unlikely to select for high levels of genotypic or phenotypic HIV-1 drug resistance, and this finding supports the continued use of LPV in the first-line treatment of pediatric patients.
Some differences between the levels of phenotypic resistance determined experimentally and those predicted by the Stanford HIV drug resistance database were observed. This may be in part due to the inclusion of Gag in our assay, whereas the database predictions consider only substitutions in the protease gene. Currently, Gag is not part of standard genotypic or phenotypic assays for resistance to PIs. Such a step would require the identification of all PI resistance-associated Gag amino acid changes, including those that differ between subtypes (26). Additionally, the predicted phenotype is based on mutation penalty scores determined using published phenotypic data that invariably use standard reference virus controls, many of which are from subtype B viruses. In contrast, our assay made use of a single clone whose sequence was most representative of the sequence in the patient's population. The cloned Gag-protease obtained from the isolate from each patient at the baseline was used as a reference to calculate the EC50. As shown here, baseline samples varied in the levels of phenotypic resistance relative to those of the reference virus, similar to the findings of another recent study (36). While a comparison to a reference strain standardizes the evaluation of levels of resistance between samples, the use of matched baseline samples provides an opportunity to measure the evolutionary advantage conferred by selected mutations within a single patient, which is more clinically relevant. Since no major resistance-associated protease amino acid changes were present at the baseline, this indicates that polymorphisms in protease and Gag determine this variability. The discordance between the actual and predicted phenotypes seen was largely accounted for by these patient isolate-specific polymorphisms and highlights the value of including matched baseline pre-ART samples when possible. Although the sequences of the selected clones were representative of the sequences of the isolate population at the baseline and the treatment failure time points, the impact of minority variants could not be accounted for in this study.
When tested individually by site-directed mutagenesis in the p8.MJ4 backbone, Gag CS and non-CS amino acid changes did not cause a reduction in PI susceptibility. However, in some cases isolates with the combination of protease and Gag amino acid changes showed increased resistance, similar to what was seen with the pediatric patient samples. Thus, the addition of the Gag CS amino acid changes N374S, E428D, and A431V to major resistance-associated protease substitutions caused an increase in phenotypic resistance to RTV. However, in most cases we were unable to recreate the levels of resistance seen with the patient samples. This is likely due to background polymorphisms which, in combination with other changes in Gag and protease, may influence PI susceptibility. Since the recreation of all mutation combinations and/or the inclusion of all Gag non-CS changes is not practical, a possible way to examine the impact of Gag on PI resistance would be to generate chimeras using the entire gag gene and a standard protease backbone. Individual PI resistance-associated amino acid changes could then be introduced into the backbone to assess the impact of Gag polymorphisms on PI resistance.
The cause of PI treatment failure in half the patients in this study remains unresolved. Recent studies have implicated amino acid changes in the envelope gene as contributors to PI treatment failure, although this requires additional study (15). The sharp dose-response curves and short half-lives of PIs, which provide only a limited period for the selection of resistance, account for why PI resistance-associated amino acid changes are less common (39). Even though our data suggest a role for Gag in PI resistance, this is unlikely to be the major reason for treatment failure in most of these pediatric patients. According to standard practice at the time of the study, infants and children receiving concomitant treatment for tuberculosis received a double dose of RTV. Since many of the children in this cohort were receiving treatment for tuberculosis, incorrect dosing of RTV may have led suboptimal drug levels to contribute to treatment failure and resistance. However, it is more likely that suboptimal adherence to ART was responsible for the virological treatment failure. Therefore, increasing adherence by improving the tolerability and palatability of drugs and developing long-acting formulations for pediatric patients will have the greatest impact on maintaining suppression rates on a PI regimen.
In conclusion, our study has demonstrated the utility of phenotypic testing for assessing PI resistance in pediatric patients. The use of the matched baseline (pre-ART) sample and the inclusion of analysis of the gag gene may provide further information on resistance in patients failing PI-based therapy compared to that obtained by genotypic analysis and predicted phenotypic scores based merely on protease. LPV/r-based treatment is unlikely to select for genotypic or phenotypic HIV-1 drug resistance, and our data support its continued use in the first-line treatment of pediatric patients.
Supplementary Material
ACKNOWLEDGMENTS
We thank Elaine J. Abrams and Renate Strehlau for access to the specimens. We acknowledge Didier Trono (EPFL, Lausanne, Switzerland) for the pMDG plasmid and Nigel Temperton (Medway School of Pharmacy, Universities of Kent and Greenwich, United Kingdom) for the pCSFLW plasmid carrying the luciferase gene.
LPV and RTV were supplied by the National Institutes of Health (NIH) AIDS Reagent and Reference Program. We thank Public Health London (formerly the Health Protection Agency) for providing J. Giandhari with training on the protease phenotypic assay. We thank the South African Medical Research Council (MRC) Self-Initiated Research Program, the Poliomyelitis Research Foundation (PRF), the KwaZulu-Natal Research Institute for Tuberculosis and HIV (K-RITH), and the Faculty of Health Sciences Research Council of the University of the Witwatersrand (FRC) for funding this study. J. Giandhari was funded by bursaries from the PRF and the National Research Foundation (NRF) of South Africa. J. Giandhari received funding from the Health Protection Agency Global Health Fund for the training that she received on the protease phenotypic assay. The parent study that collected these specimens was funded by Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) grant HD 47177.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02682-15.
REFERENCES
- 1.Fun A, Wensing AM, Verheyen J, Nijhuis M. 2012. Human immunodeficiency virus Gag and protease: partners in resistance. Retrovirology 9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Department of Health. 2013. The South African antiretroviral treatment guidelines. Department of Health, Pretoria, South Africa. [Google Scholar]
- 3.Ren Y, Nuttall JJ, Egbers C, Eley BS, Meyers TM, Smith PJ, Maartens G, McIlleron HM. 2008. Effect of rifampicin on lopinavir pharmacokinetics in HIV-infected children with tuberculosis. J Acquir Immune Defic Syndr 47:566–569. doi: 10.1097/QAI.0b013e3181642257. [DOI] [PubMed] [Google Scholar]
- 4.van Zyl GU, Rabie H, Nuttall JJ, Cotton MF. 2011. It is time to consider third-line options in antiretroviral-experienced paediatric patients? J Int AIDS Soc 14:55. doi: 10.1186/1758-2652-14-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. 2013. Global update on HIV treatment 2013: results, impact and opportunities. WHO, Geneva, Switzerland. [Google Scholar]
- 6.Violari A, Cotton MF, Gibb DM, Babiker AG, Steyn J, Madhi SA, Jean-Philippe P, McIntyre JA. 2008. Early antiretroviral therapy and mortality among HIV-infected infants. N Engl J Med 359:2233–2244. doi: 10.1056/NEJMoa0800971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palumbo P, Lindsey JC, Hughes MD, Cotton MF, Bobat R, Meyers T, Bwakura-Dangarembizi M, Chi BH, Musoke P, Kamthunzi P, Schimana W, Purdue L, Eshleman SH, Abrams EJ, Millar L, Petzold E, Mofenson LM, Jean-Philippe P, Violari A. 2010. Antiretroviral treatment for children with peripartum nevirapine exposure. N Engl J Med 363:1510–1520. doi: 10.1056/NEJMoa1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porter M, Davies MA, Mapani MK, Rabie H, Phiri S, Nuttall J, Fairlie L, Technau KG, Stinson K, Wood R, Wellington M, Haas AD, Giddy J, Tanser F, Eley B. 2015. Outcomes of infants starting antiretroviral therapy in Southern Africa, 2004-2012. J Acquir Immune Defic Syndr 69:593–601. doi: 10.1097/QAI.0000000000000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RK, Gibb D, Pillay D. 2009. Management of paediatric HIV-1 resistance. Curr Opin Infect Dis 22:256–263. doi: 10.1097/QCO.0b013e3283298f1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyidogan P, Anderson KS. 2014. Current perspectives on HIV-1 antiretroviral drug resistance. Viruses 6:4095–4139. doi: 10.3390/v6104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levison JH, Orrell C, Gallien S, Kuritzkes DR, Fu N, Losina E, Freedberg KA, Wood R. 2012. Virologic failure of protease inhibitor-based second-line antiretroviral therapy without resistance in a large HIV treatment program in South Africa. PLoS One 7:e32144. doi: 10.1371/journal.pone.0032144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallis CL, Mellors JW, Venter WD, Sanne I, Stevens W. 2011. Protease inhibitor resistance is uncommon in HIV-1 subtype C infected patients on failing second-line lopinavir/r-containing antiretroviral therapy in South Africa. AIDS Res Treat 2011:769627. doi: 10.1155/2011/769627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartlett JA. 2013. Lack of protease inhibitor resistance following treatment failure—too good to be true? J Clin Invest 123:3704–3705. doi: 10.1172/JCI71784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orrell C, Levison J, Ciaranello A, Bekker LG, Kuritzkes DR, Freedberg KA, Wood R. 2013. Resistance in pediatric patients experiencing virologic failure with first- and second-line antiretroviral therapy. Pediatr Infect Dis J 32:644–647. doi: 10.1097/INF.0b013e3182829092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabi SA, Laird GM, Durand CM, Laskey S, Shan L, Bailey JR, Chioma S, Moore RD, Siliciano RF. 2013. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest 123:3848–3860. doi: 10.1172/JCI67399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siliciano JD, Siliciano RF. 2013. Recent trends in HIV-1 drug resistance. Curr Opin Virol 3:487–494. doi: 10.1016/j.coviro.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali A, Bandaranayake RM, Cai Y, King NM, Kolli M, Mittal S, Murzycki JF, Nalam MN, Nalivaika EA, Ozen A, Prabu-Jeyabalan MM, Thayer K, Schiffer CA. 2010. Molecular basis for drug resistance in HIV-1 protease. Viruses 2:2509–2535. doi: 10.3390/v2112509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dam E, Quercia R, Glass B, Descamps D, Launay O, Duval X, Kräusslich H-G, Hance AJ, Clavel F, ANRS 109 Study Group. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog 5:e1000345. doi: 10.1371/journal.ppat.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RK, Kohli A, McCormick AL, Towers GJ, Pillay D, Parry CM. 2010. Full-length HIV-1 Gag determines protease inhibitor susceptibility within in-vitro assays. AIDS 24:1651–1655. doi: 10.1097/QAD.0b013e3283398216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verheyen J, Litau E, Sing T, Daumer M, Balduin M, Oette M, Fatkenheuer G, Rockstroh JK, Schuldenzucker U, Hoffmann D, Pfister H, Kaiser R. 2006. Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients. Antivir Ther 11:879–887. [PubMed] [Google Scholar]
- 21.Kolli M, Stawiski E, Chappey C, Schiffer CA. 2009. Human immunodeficiency virus type 1 protease-correlated cleavage site mutations enhance inhibitor resistance. J Virol 83:11027–11042. doi: 10.1128/JVI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry CM, Kohli A, Boinett CJ, Towers GJ, McCormick AL, Pillay D. 2009. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. J Virol 83:9094–9101. doi: 10.1128/JVI.02356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nijhuis M, van Maarseveen NM, Lastere S, Schipper P, Coakley E, Glass B, Rovenska M, de Jong D, Chappey C, Goedegebuure IW, Heilek-Snyder G, Dulude D, Cammack N, Brakier-Gingras L, Konvalinka J, Parkin N, Kräusslich HG, Brun-Vezinet F, Boucher CA. 2007. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med 4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fun A, van Maarseveen NM, Pokorna J, Maas RE, Schipper PJ, Konvalinka J, Nijhuis M. 2011. HIV-1 protease inhibitor mutations affect the development of HIV-1 resistance to the maturation inhibitor bevirimat. Retrovirology 8:70. doi: 10.1186/1742-4690-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolli M, Özen A, Kurt-Yilmaz N, Schiffer CA. 2014. HIV-1 protease-substrate coevolution in nelfinavir resistance. J Virol 88:7145–7154. doi: 10.1128/JVI.00266-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Verheyen J, Theys K, Piampongsant S, Van Laethem K, Vandamme AM. 2014. HIV-1 Gag C-terminal amino acid substitutions emerging under selective pressure of protease inhibitors in patient populations infected with different HIV-1 subtypes. Retrovirology 11:79. doi: 10.1186/s12977-014-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Oliveira T, Engelbrecht S, Janse van Rensburg E, Gordon M, Bishop K, zur Megede J, Barnett SW, Cassol S. 2003. Variability at human immunodeficiency virus type 1 subtype C protease cleavage sites: an indication of viral fitness? J Virol 77:9422–9430. doi: 10.1128/JVI.77.17.9422-9430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giandhari J, Basson AE, Coovadia A, Kuhn L, Abrams EJ, Strehlau R, Morris L, Hunt G. 2015. Genetic changes in HIV-1 Gag-protease associated with protease inhibitor-based therapy failure in paediatric patients. AIDS Res Hum Retroviruses 31:776–782. doi: 10.1089/aid.2014.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coovadia A, Abrams EJ, Stehlau R, Meyers T, Martens L, Sherman G, Hunt G, Hu CC, Tsai WY, Morris L, Kuhn L. 2010. Reuse of nevirapine in exposed HIV-infected children after protease inhibitor-based viral suppression: a randomized controlled trial. JAMA 304:1082–1090. doi: 10.1001/jama.2010.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor B, Hunt G, Abrams EJ, Coovadia A, Meyers T, Sherman G, Strehlau R, Morris L, Kuhn L. 2011. Rapid development of antiretroviral drug resistance mutations in HIV-infected children less than two years of age initiating protease inhibitor-based therapy in South Africa. AIDS Res Hum Retroviruses 27:945–956. doi: 10.1089/aid.2010.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reitz C, Coovadia A, Ko S, Meyers T, Strehlau R, Sherman G, Kuhn L, Abrams EJ. 2010. Initial response to protease-inhibitor-based antiretroviral therapy among children less than 2 years of age in South Africa: effect of cotreatment for tuberculosis. J Infect Dis 201:1121–1131. doi: 10.1086/651454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutherland KA, Mbisa JL, Cane PA, Pillay D, Parry CM. 2014. Contribution of Gag and protease to variation in susceptibility to protease inhibitors between different strains of subtype B human immunodeficiency virus type 1. J Gen Virol 95(Pt 1):190–200. doi: 10.1099/vir.0.055624-0. [DOI] [PubMed] [Google Scholar]
- 33.Ndung'u T, Renjifo B, Essex M. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J Virol 75:4964–4972. doi: 10.1128/JVI.75.11.4964-4972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parry CM, Kolli M, Myers RE, Cane PA, Schiffer C, Pillay D. 2011. Three residues in HIV-1 matrix contribute to protease inhibitor susceptibility and replication capacity. Antimicrob Agents Chemother 55:1106–1113. doi: 10.1128/AAC.01228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koh Y, Das D, Leschenko S, Nakata H, Ogata-Aoki H, Amano M, Nakayama M, Ghosh AK, Mitsuya H. 2009. GRL-02031, a novel nonpeptidic protease inhibitor (PI) containing a stereochemically defined fused cyclopentanyltetrahydrofuran potent against multi-PI-resistant human immunodeficiency virus type 1 in vitro. Antimicrob Agents Chemother 53:997–1006. doi: 10.1128/AAC.00689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutherland KA, Mbisa JL, Ghosn J, Chaix ML, Cohen-Codar I, Hue S, Delfraissy JF, Delaugerre C, Gupta RK. 2014. Phenotypic characterization of virological failure following lopinavir/ritonavir monotherapy using full-length Gag-protease genes. J Antimicrob Chemother 69:3340–3348. doi: 10.1093/jac/dku296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maguire MF, Guinea R, Griffin P, Macmanus S, Elston RC, Wolfram J, Richards N, Hanlon MH, Porter DJT, Wrin T, Parkin N, Tisdale M, Furfine E, Petropoulos C, Snowden BW, Kleim J-P. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J Virol 76:7398–7406. doi: 10.1128/JVI.76.15.7398-7406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Zyl GU, van der Merwe L, Claassen M, Cotton MF, Rabie H, Prozesky HW, Preiser W. 2009. Protease inhibitor resistance in South African children with virologic failure. Pediatr Infect Dis J 28:1125–1127. doi: 10.1097/INF.0b013e3181af829d. [DOI] [PubMed] [Google Scholar]
- 39.Rosenbloom DI, Hill AL, Rabi SA, Siliciano RF, Nowak MA. 2012. Antiretroviral dynamics determines HIV evolution and predicts therapy outcome. Nat Med 18:1378–1385. doi: 10.1038/nm.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.