Abstract
Hepatitis E virus (HEV) is the causative agent of hepatitis E in humans and a member of the genus Orthohepevirus in the family Hepeviridae. HEV infections are the common cause of acute hepatitis but can also take chronic courses. Ribavirin is the treatment of choice for most patients, and type I interferon (IFN) has been evaluated in a few infected transplant patients in vivo. In this study, the antiviral effects of different exogenously administered interferons were investigated by using state-of-the-art subgenomic replicon and full-length HEV genome cell culture models. Hepatitis C virus (HCV) subgenomic replicons based on the genotype 2a JFH1 isolate served as the reference. The experiments revealed that HEV RNA replication was inhibited by the application of all types of IFN, including IFN-α (type I), IFN-γ (type II), and IFN-λ3 (type III), but to a far lesser extent than HCV replication. Simultaneous determination of interferon-stimulated gene (ISG) expression levels for all IFN types demonstrated efficient downregulation by HEV. Furthermore, different IFN-α subtypes were also able to block viral replication in combination with ribavirin. The IFN-α subtypes 2a and 2b exerted the strongest antiviral activity against HEV. In conclusion, these data demonstrate for the first time moderate anti-HEV activities of types II and III IFNs and different IFN-α subtypes. As HEV employed a potent anti-interferon mechanism by restricting ISG expression, exogenous application of IFNs as immunotherapy should be carefully assessed.
INTRODUCTION
Hepatitis E virus (HEV) is the causative agent of numerous cases of viral hepatitis in humans worldwide with a case fatality rate of 1 to 4% (1). For pregnant women infected during the third trimester, mortality rates of up to 30% were reported (2). In developing countries, HEV (genotypes 1 and 2) is a major cause of acute hepatitis, transmitted via the fecal-oral route and mainly associated with ingestion of contaminated drinking water. In industrialized countries, HEV (genotypes 3 and 4) has been found to be more prevalent in the human population than originally thought with a significant number of people experiencing an HEV infection during their lifetime (2). While genotypes 1 and 2 solely infect humans, genotypes 3 and 4 are zoonotic pathogens with major virus reservoirs in pigs, wild boars, and deer (3, 4). HEV genotype 3 infections in humans can be associated with prolonged viremia, leading to chronic infection in immunocompromised patients such as organ transplant recipients, patients with HIV infection, or patients with hematological malignancies (5). Treatment options for chronically infected patients include either ribavirin (RBV) for at least 3 months or pegylated alpha 2a/alpha 2b interferon (IFN-α2a/α2b) which, however, can be associated with severe side effects and can cause graft rejection (6).
HEV was classified recently by the International Committee on Taxonomy of Viruses (ICTV) in the new genus Orthohepevirus, comprising all mammalian and avian HEV isolates within the family of Hepeviridae (7). The HEV genome is single stranded with an RNA genome size of 7.2 kb (8). The capped positive-sense genome encodes three open reading frames (ORFs): the nonstructural polyprotein required for RNA replication (ORF1), the capsid protein of the HEV virion (ORF2), and a small multifunctional protein with a molecular mass of 13 kDa (ORF3) (9).
IFNs are signaling proteins belonging to the class of cytokines and are named for their ability to “interfere” with viral replication (10). Mammalian IFNs are divided into three types, each using their unique receptor complex: type I is mainly represented by IFN-α and -β, type II by IFN-γ, and type III comprising IFN-λ (11). IFN-α can be further divided into 13 subtypes which exhibit a high degree of amino acid similarity (>75%); all bind to the same receptor but still display a unique activity profile (12) and differ in their biological activities (13). IFN-γ is the sole representative of type II IFN, while the IFN type III family is the most recently discovered group of IFNs, comprising IFN-λ1 to -4 (14). Viruses utilize a variety of mechanisms to evade the host interferon response, and for HEV it was shown that the ORF1 polyprotein can antagonize type I IFN induction, whereas ORF3 was reported to inhibit IFN-α signaling (15). However, the effects of other types of IFN (types II and III) on HEV have not been investigated so far.
In this study, we first compared the antiviral activities of all IFN types (I, II, and III) against HEV using HCV as a reference pathogen. HEV was susceptible to all three types of IFN, but the antiviral activity was moderate in comparison to that of HCV. At the same time, HEV employed a potent restriction of interferon gene induction by all types of interferon. Next, we tested all different IFN-α subtypes for their ability to inhibit HEV replication in a HEV subgenomic model and a full-length infection system. These results should help in understanding the HEV virus-host interactions in liver cells and the potential clinical applications of exogenous IFNs as immunotherapies against HEV infections.
MATERIALS AND METHODS
Compounds and reagents.
All of the recombinant IFN-α subtypes (α1, α2a, α4, α5, α6, α7, α8, α10, α14, α16, α17, α21) were obtained from the human IFN sampler (PBL Biomedical Laboratories, Piscataway, NJ, USA). IFN-α2b (Intron A) was purchased from Schering Corporation (Kenilworth, NJ, USA), and IFN-γ was acquired from PeproTech (Rocky Hill, NJ, USA). Recombinant IFN-λ3 was produced and purified as previously described (16, 17). Ribavirin (RBV) was received from Sigma-Aldrich, St. Louis, MO, USA. All compounds were stored and diluted according to the manufacturer's recommendations.
Cell culture.
Human hepatoma cells (Huh7.5 and HepG2 cells) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (Invitrogen), 1% nonessential amino acids (Invitrogen), 100 μg/ml of streptomycin (Invitrogen), and 100 IU/ml of penicillin. Cells were kept at 37°C in a 5% (vol/vol) CO2 incubator.
HEV constructs and in vitro transcription.
A plasmid construct containing the full-length HEV genome (Kernow-C1 p6 clone, genotype 3; GenBank accession number JQ679013) and a construct harboring a subgenomic HEV sequence coupled with a Gaussia luciferase reporter gene (p6-Gluc) were used to generate HEV in vitro transcripts as previously described with an additional capping step (m7G Cap analog; Promega, Madison, WI, USA) (18, 19).
HEV replication assay.
For transfection, we used the electroporation technique in accordance with previous reports (20). In brief, Huh7.5 or HepG2 cells at 1.2 × 107 cells/ml (12-well assay) or 5 × 106 cells/ml (96-well assay) in 400 μl of Cytomix containing 2 mM ATP and 5 mM glutathione were mixed with 3 μg of p6-Luc subgenomic or p6 full-length HEV RNA. Electroporation was carried out with a Gene Pulser system (Bio-Rad, Munich, Germany). Cells were immediately transferred to 11.6 ml (12-well assay) or 12.1 ml (96-well assay) of DMEM complete and the cell suspension was seeded in the respective plates (5 × 105 cells/well for 12-well plates or 2 × 104 cells/well for 96-well plates). After 4 h, DMEM containing compounds or reagents at indicated concentrations were added. Viral replication was determined by measuring luciferase activity in the supernatant of 96-well plates 72 h after transfection or by real-time quantitative PCR (qRT-PCR) for the p6 full-length genome (12-well plates).
Luciferase assay.
A 20-μl aliquot of supernatant (Gaussia luciferase) or lysed cells (firefly luciferase, lysis performed in passive lysis buffer 5×; Promega) was added per well of a 96-well white, flat-bottom microplate followed by the detection of luminescence using a microplate reader (Centro XS3 LB960; Berthold Technologies, Bad Wildbad, Germany) using coelenterazine (for HEV encoding Gaussia luciferase) or luciferine (HCV encoding firefly luciferase) as a substrate.
qRT-PCR.
Cellular total RNAs of HEV p6 full-length or yeast tRNA (Sigma-Aldrich) transfected HepG2 cells were extracted using a NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) with the involvement of RNase-free DNase, following the manufacturer's protocol. Subsequently, the total RNAs were reverse transcribed into cDNA using a PrimeScript 1st strand cDNA synthesis kit (TaKaRa Bio, Otsu, Japan). Quantitative PCR was carried out using 400 nmol of primers together with SYBR Premix Ex Taq (TaKaRa Bio) and the LightCycler 480 system (Roche, Basel, Switzerland). The primer sequences for amplification of HEV RNA (21) and each interferon-stimulated gene (ISG) product have been described previously: hMX1 (22), hIFIT1 (23), hCXCL10 (24), and hIFIT3 (25). hGAPDH was considered the reference gene to normalize gene expression (26). The relative gene expression was determined using the cycle threshold (ΔΔCT) method.
Statistical analysis.
GraphPad Prism 6 software was used for data analysis employing either analysis of variance (ANOVA) followed by Dunnett's corrected unpaired t test or the Holm-Sidak method. P values of <0.05, <0.01, and <0.001 were considered significant, and P values of ≥0.05 were considered not significant.
RESULTS
Antiviral activities of types I, II, and III IFN against HEV replication.
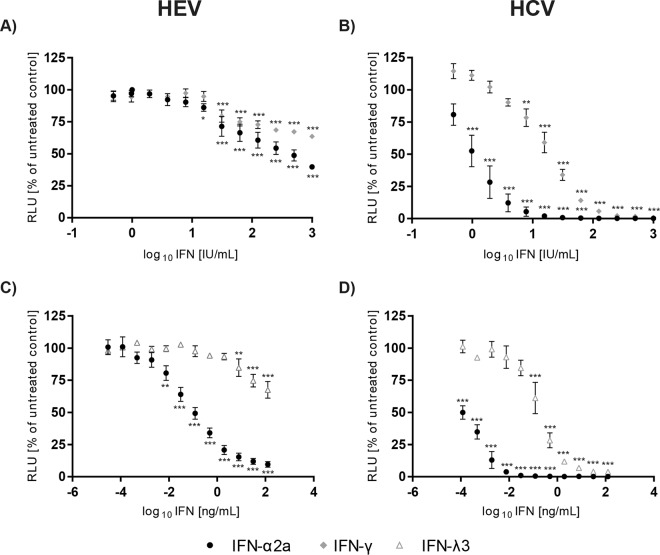
To test the effects of the different IFN types against HEV replication, the human hepatoma cell line Huh7.5, which is highly permissive for HCV replication, was transfected with a subgenomic HEV replicon in which a part of the ORF2 is replaced by a Gaussia luciferase as a reporter gene (18). As HCV is susceptible to exogenous IFNs and type I IFN has been used for decades in HCV therapy (27), we employed a subgenomic HCV replicon based on the JFH1 isolate (28) as the reference. The different IFNs were added 4 h after electroporation of each viral construct to the cell culture medium, and replication activity was determined by reporter assay 72 h later as HEV replication levels peaked at that time point (see Fig. S1 in the supplemental material). As depicted in Fig. 1A, HEV replication can be moderately inhibited by treatment with exogenous IFN-α2a (50% inhibitory concentration [IC50], 359.7 IU/ml) and IFN-γ (IC50, indefinable) in a dose-dependent manner based on international units (Fig. 1A). In the case of HCV, the antiviral effects for type I and type II IFNs were much more pronounced with an inhibition to background levels for IFN-α2a at 32 IU/ml (IC50, 1.096 IU/ml) and about 256 IU/ml for IFN-γ (IC50, 19.77 IU/ml) (Fig. 1B). At present, no international unit definition of type III IFNs exists, and therefore the concentration of IFN-λ3 is given as weight per volume (ng/ml). HEV was significantly inhibited when concentrations of 8 ng/ml IFN-λ3 (IC50, indefinable) were exceeded (Fig. 1C). As previously described (17), IFN-λ3 reduced HCV replication in a dose-dependent manner with concentrations already lower than 1 ng/ml (IC50, 0.1938 ng/ml) (Fig. 1C), indicating that an HEV replicon is less sensitive to type III IFN than an HCV replicon (Fig. 1C and D). Nonnormalized relative light units (RLUs) of the IFN-based inhibition of HEV replication by IFN types I and III are depicted in Fig. S2A and B in the supplemental material. In summary, these results demonstrate weak to moderate antiviral activities of IFN types I, II, and III against HEV.
FIG 1.
Antiviral activities of different IFN types against HEV replication. The dose-dependent inhibition rates of HEV subgenomic replicon (SGR) (A, C) and HCV SGR (B, D) replication in Huh7.5 cells by representatives of all three IFN types are depicted. IFN-α2a as the type I representative, IFN-γ as the sole type II interferon, and IFN-λ3 as the IFN type III were added 4 h posttransfection. The y axes display the relative light units (RLUs) normalized to those of the untreated control, assessed 72 h posttreatment; the x axes reflect the IFN concentrations utilized. IFN-γ was used as international units per milliliter (A, B), and IFN-λ3 was implemented as nanograms per milliliter (C, D). IFN-α2a concentrations were applied based on international units per milliliter and nanograms per milliliter. Shown are the means (± standard errors of the mean [SEM]) from at least three independent experiments; asterisks represent significant reductions in RLUs tested against untreated controls (Dunnett's t test; *P < 0.05; **P < 0.01; ***P < 0.001).
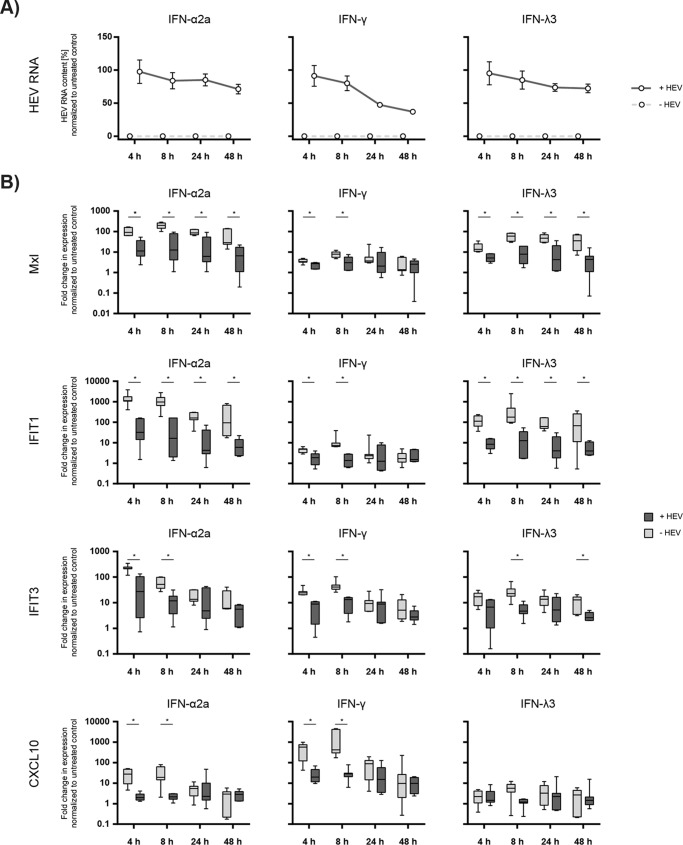
Interferon-stimulated gene expression induced by different types of IFN is downregulated by HEV.
Next, we confirmed the antiviral activities of the different types of IFN using an HEV full-length infection model based on the p6 clone in HepG2 cells (19). This human liver cell was transfected with full-length HEV and viral RNA and interferon-stimulated gene (ISG) expression was assessed at different time points after addition of IFN-α2a (1,000 IU/ml), IFN-γ (1,000 IU/ml), or IFN-λ3 (200 ng/ml). As depicted in Fig. 2A, HEV RNA copy numbers (ranging from 1.53 × 107 to 1.84 × 107 RNA copies per well 4 h after transfection) normalized to the untreated control were only moderately reduced by all three types of IFN, which is similar to the only modest suppression of HEV-encoded luciferase expression in the replicon assay (Fig. 2A). To investigate if this weak IFN-dependent antiviral activity was due to the virus-dependent blunting of IFN signaling, we determined the effect of HEV infection on the induction of a panel of ISGs known to be specifically stimulated by different types of IFN: Mx1 and IFIT1 (types I and III), IFIT3 (types I and II), and CXCL10 (type II). HEV blocked IFN type I responses by a significant downregulation of Mx1 and IFIT1 at all time points and for IFIT3 between 4 and 8 h (Fig. 2B). For the IFN type II specific marker gene CXCL10, a significant blockage of IFN-dependent gene expression in the presence of HEV was observed at the early time points after administration of IFN (Fig. 2B). Although IFN type I and type III signal via distinct receptor complexes, they activate the same intracellular signaling pathway and similar to the restriction of IFN-α-induced Mx1, IFIT1, and IFIT3, we also detected a significant downregulation of the expression of these genes after IFN-λ3 stimulation in HEV-replicating cells, which was not the case for CXCL10 (Fig. 2B). In conclusion, HEV exerted a potent anti-interferon mechanism by downregulating ISG expression.
FIG 2.
Interferon-stimulated gene expression levels induced by different types of IFN can be blocked by HEV. HepG2 cells were transfected with full-length HEV or yeast tRNA, and 24 h later were treated with interferons. Viral RNA and ISG expression levels were assessed at 4 h, 8 h, 24 h, and 48 h after the addition of IFN-α2a (1,000 IU/ml, left panels), IFN-γ (1,000 IU/ml, middle panels), or IFN-λ (200 ng/ml, right panels) and compared to those in tRNA control transfected cells. (A) Total RNA was extracted, and the amounts of viral RNA in the cells were measured via qRT-PCR over time and normalized to an untreated control (y axes). The solid lines display the course of the HEV-transfected cells; in control cells, HEV RNA always resided below the detection limits (dashed lines). (B) Expression levels of the ISGs MxI, IFIT1, IFIT3, and CXCL10 in HepG2 cells with and without replicating full-length HEV were determined at the indicated time points after addition of IFNs via qRT-PCR. The y axes represent the fold changes in the expression levels normalized to a respective control without IFN treatment. Statistical significance was determined using the Holm-Sidak method (n = 7; α = 0.05, *, statistically significant).
Effect of different IFN-α subtypes against HEV replication and their combinatory activity with ribavirin.
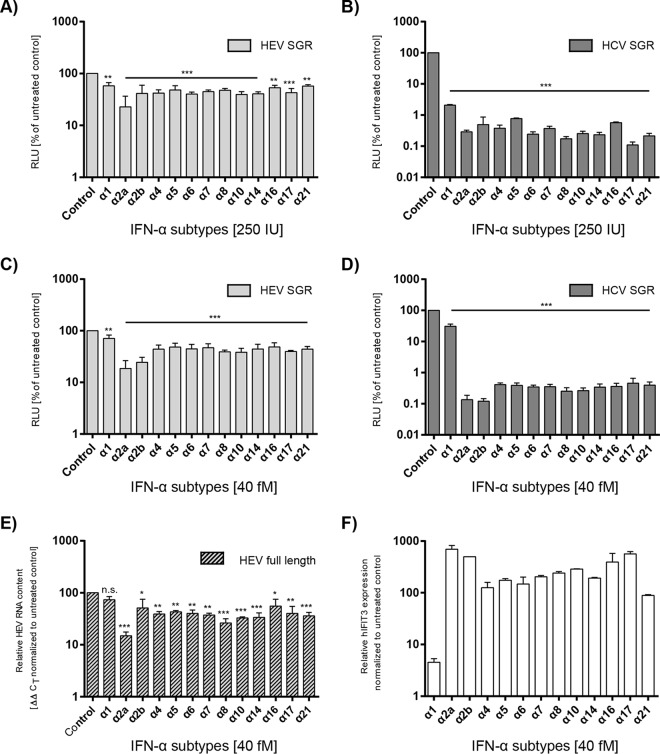
Although the 13 known IFN-α subtypes display unique activity profiles (12), only the IFN-α2a and IFN-α2b subtypes have been licensed for the treatment of viral infections (13). To evaluate the potency of the different IFN-α subtypes against HEV, subgenomic HEV replicon cells were incubated with either 250 IU (Fig. 3A) or 40 fM (Fig. 3C) exogenous IFNs. RNA replication levels were determined by Gaussia luciferase reporter assays as previously described. All IFN-α subtypes significantly inhibited HEV replication by at least 20% (Fig. 3A and C). IFN-α1 showed the lowest antiviral activity, whereas IFN-α2a and -α2b exerted the highest inhibitions. The antiviral activity against HCV as the reference virus was much more pronounced with a 1,000-fold inhibition in comparison to that for HEV, and IFN-α2a and -2b also showed the strongest antiviral effects (Fig. 3B and D). The nonnormalized results of the IFN subtype inhibition against HEV and HCV are displayed as Fig. S2C and D in the supplemental material. These results were also confirmed with a full-length HEV-replicating genome that does not harbor a reporter gene by determination of HEV RNA copy numbers in Huh7.5 cells (Fig. 3E) or HepG2 cells (data not shown). To demonstrate that the cells were in general responsive to the different subtypes, IFIT3 mRNA was quantified in parallel. This ISG was induced in correlation with the antiviral activities of the different IFNs, as for IFN-α2a and -2b the highest induction was observed, whereas subtype α1 upregulated IFIT3 only 5-fold (Fig. 3F).
FIG 3.
Antiviral activities of different IFN-α subtypes in HEV and HCV single transfection assays. The inhibition of SGR replication by 13 IFN-α subtypes is depicted for HEV SGR (A, C) and HCV SGR (B, D) in Huh7.5 cells as a change in RLUs compared to those of untreated controls (y axes) and their antiviral effects on full-length HEV (E), shown as a reduction in HEV RNA measured via qRT-PCR. Interferon subtypes were applied 4 h posttransfection with concentrations of either 250 IU/ml (A, B) or 40 fM (C to F). Reporter activity or HEV RNA was assessed 72 h posttreatment. Depicted are the means (±SEM) from at least three independent experiments. Significance was tested against untreated controls using ANOVA followed by Dunnett's corrected t test (n.s., not significant; *P < 0.05; **P < 0.01; ***P < 0.001). (F) The functional integrity of the IFN-α subtypes and the responsiveness of the Huh7.5 cells to the treatment was tested via qRT-PCR measurement of IFIT3 gene induction 6 h after the addition of IFNs. Error bars represent SEM for two technical replicates.
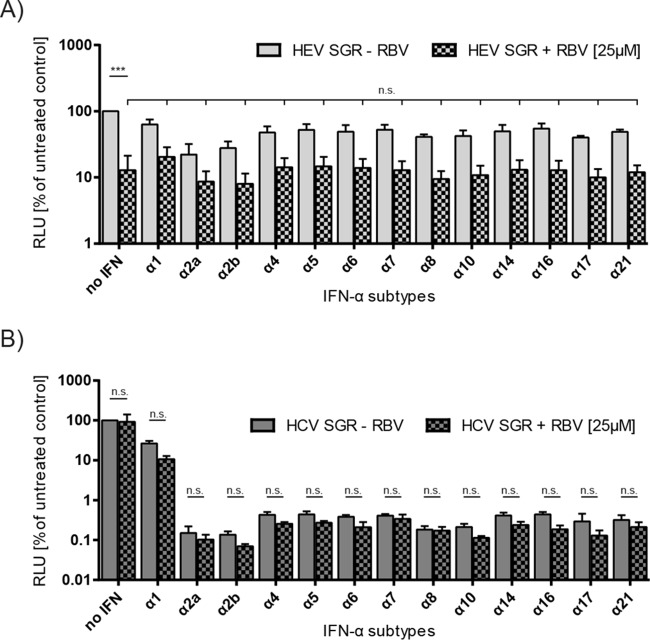
Ribavirin monotherapy has been demonstrated to be a successful treatment option for HEV (8) and has been combined with IFN-α for decades in HCV therapies. Therefore, we next tested the combinatory antiviral activity of the different IFN-α subtypes together with ribavirin. As seen in Fig. 4A, ribavirin alone had a strong antiviral effect against HEV replication, which was not significantly increased by any of the IFN-α subtypes when a dose of 40 fM IFN was administered (Fig. 4A). In the case of HCV, the ribavirin treatment did not reduce the JFH1 RNA replication but as also seen in Fig. 3D, the IFN-α subtypes exerted a strong antiviral effect with the exception of IFN-α1 (Fig. 4B). The combination with ribavirin did not significantly increase the antiviral activities compared to that with IFN treatment alone (Fig. 4B). In summary, these results demonstrate that IFN-α subtypes 2a and 2b exerted the strongest antiviral activity against HEV, and there is no additive effect in combination with ribavirin.
FIG 4.
Combinatory effects of different IFN-α subtypes with ribavirin against HEV replication. The inhibitory effects on HEV SGR (A) and HCV SGR (B) replication of IFN-α subtypes (40 fM) combined with ribavirin (25 μM) compared to those of the IFN-only treated samples is depicted. The means (±SEM) from three independent experiments in Huh7.5 cells are plotted. Compounds were added 4 h posttransfection; RLUs were assessed 72 h posttreatment. The significance of the reduction in RLUs was tested for the dually treated samples against the RBV-only treated sample for HEV SGR or for each IFN-α subtype plus RBV against the subtype-only treated sample for HCV SGR (n.s., not significant; ***P < 0.001).
DISCUSSION
In this study, we characterized the antiviral properties of type I, type II, and type III IFNs, including 13 IFN-α subtypes, against HEV replication in tissue culture to evaluate the therapeutic options by the exogenous addition of IFNs. In contrast to the efficient blockage of HCV, we observed only weak to moderate inhibition of HEV replication by types I, II, and III IFN (Fig. 1 and 2). In the case of type I IFN, these findings are in line with previous results from nonliver A549 cells where only a low response to IFN-α was also reported and suppression of interferon signaling through the regulation of STAT1 phosphorylation by ORF3 was postulated (29). Furthermore, Debing et al. showed antiviral activity of IFN-α in liver cells using the p6 clone in a subgenomic and full-length model (30), and very recently, Zhou et al. also observed a moderate and delayed anti-HEV effect of IFN-α in vitro and in patients in contrast to the effect against HCV (31). By further investigating the mRNA levels of different ISGs (Mx1, IFIT1, IFIT3, and CXCL10) in the presence or absence of HEV after IFN treatment, we showed that HEV was able to counteract the ISG responses induced by all types of IFNs (Fig. 2). As antagonists of IFN type I induction, two domains in the HEV ORF1, X and papain-like cysteine protease domain (PCP), were identified by Nan et al. (32). Interestingly, in vivo studies with HEV- and HCV-infected chimpanzees were also conducted to compare the innate immune responses of these two viruses. In comparison to the genes in the HCV-infected chimpanzees, HEV induced only one-third of differentially expressed genes in liver biopsy specimens, but almost all of these genes overlapped with those in HCV-induced cells (33). In addition, HEV infection led to a smaller number of IFN-induced genes with a lower magnitude of expression levels than HCV infection, indicating that HEV also restricts innate immune responses efficiently in vivo (33).
When testing the 13 different IFN-α subtypes against HEV, we observed antiviral activities for all 13 IFNs, with IFN-α2a and -2b showing the highest inhibitory activities against HEV, which also correlated with an upregulation of IFIT3 (Fig. 3). All IFN-α subtypes bind to the same type I IFN receptor (IFNAR), and so far no unique function has been attributed to any given subtype (34, 35), but differences in their binding affinities to their receptors were reported (34). Only IFN-α subtypes 10 and 17 had lower binding affinities than 2a, whereas all other subtypes showed capacities for tighter binding to their receptors (34). The authors further showed that these different binding affinities were associated with an antiproliferative effect but not with the antiviral potency against vesicular stomatitis virus (VSV) or encephalomyocarditis virus (EMCV) (34). Very few previous studies have analyzed the antiviral activities of the different IFN-α subtypes against virus infection systems (13). Similar to our finding, Dubois et al. showed the anti-HCV effects of all IFN-α subtypes and reported an enhanced effect of IFN-α17 compared to that of IFN-α2a in a genotype 2a infection setup (36). By using a subgenomic genotype 2a replicon system, we observed high susceptibility of HCV for all subtypes with no distinction for IFN-α17, which could be due to the different experimental setup and viral constructs. For herpes simplex virus (HSV), it was shown that the murine IFN-α homologs 1, 4, 5, 6, and 9 were inhibiting viral replication (37) and in a Friend virus in vivo infection model, therapeutic application with murine IFN-α1, -4, -9, and -11 was able to reduce viral loads significantly in contrast to IFN-α2, -5, and -6 (38, 39). HEV replication could be efficiently blocked by ribavirin treatment and a significant benefit of a combination of any IFN-α subtype with ribavirin could not be observed using one constant ribavirin dose of 25 μM (Fig. 4). This guanosine analog was earlier reported to inhibit HEV replication through depletion of cellular GTP pools and a moderate but statistically significant synergy was reported with 4 IU/ml IFN-α subtype 2a and 0.4 μM ribavirin (30). Ribavirin and pegylated IFN-α are the only available compounds for treatments of acute and chronic HEV infections so far, and ribavirin is considered the first-choice therapy (6, 40). Pegylated IFN-α resulted in sustained virological responses in five individuals with liver transplants who were infected with HEV (41, 42). However, therapy with IFN-α2a, which exerted the highest antiviral activity of the known IFN-α subtypes, can be associated with side effects in organ transplant patients so that only well-selected patients with chronic HEV infections may be candidates for IFN-α treatment options. Future clinical trials are necessary for comparison of the efficiencies and side effects of these antiviral agents and their combination. Interestingly, one study reported successful IFN-α and ribavirin therapy for a chronically HEV-infected patient coinfected with HIV (43).
In summary, we showed that in comparison to HCV, HEV was moderately susceptible to IFN type I, type II, and type III and at the same time downregulated ISG expression levels. The different IFN-α subtypes exerted antiviral activities against HEV to distinct extents with IFN-α2a and -2b being the most potent agents. An additive effect in combination with ribavirin could not be observed.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Takaji Wakita (Department of Virology II, National Institute of Infectious Diseases, Tokyo, Japan) for the JFH1 isolate, to Charles Rice (Center for the Study of Hepatitis C, The Rockefeller University, New York, NY, USA) for Huh7.5 cells, and to Suzanne Emerson (Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA) for the hepatitis E virus p6 clone. We also thank Stephanie Pfaender (Twincore, Hannover, Germany) for critical reading of the manuscript and all members of the Institute of Experimental Virology, Twincore, for helpful support, suggestions, and discussions.
Funding Statement
Eike Steinmann was funded by a Helmholtz Centre for Infection Research intramural young investigator award and Deutsche Forschungsgemeinschaft (DFG) grant STE 1954/1-1, as well as by Bundeministerium für Bildung und Forschung (BMBF) GINACIO grant 16GW0105. Thomas Pietschmann has received consulting fees from Biotest AG and from Janssen Global Services.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02427-15.
REFERENCES
- 1.Purcell RH, Emerson SU. 2008. Hepatitis E: an emerging awareness of an old disease. J Hepatol 48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Kamar N, Dalton HR, Abravanel F, Izopet J. 2014. Hepatitis E virus infection. Clin Microbiol Rev 27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayed IM, Vercouter AS, Abdelwahab SF, Vercauteren K, Meuleman P. 2015. Is hepatitis E virus an emerging problem in industrialized countries? Hepatology 62:1883−1892. doi: 10.1002/hep.27990. [DOI] [PubMed] [Google Scholar]
- 4.Wedemeyer H, Pischke S, Manns MP. 2012. Pathogenesis and treatment of hepatitis e virus infection. Gastroenterology 142:1388−1397. doi: 10.1053/j.gastro.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara S, Yokokawa Y, Morino K, Hayasaka K, Kawabata M, Shimizu T. 2014. Chronic hepatitis E: a review of the literature. J Viral Hepat 21:78–89. doi: 10.1111/jvh.12156. [DOI] [PubMed] [Google Scholar]
- 6.Behrendt P, Steinmann E, Manns MP, Wedemeyer H. 2014. The impact of hepatitis E in the liver transplant setting. J Hepatol 61:1418–1429. doi: 10.1016/j.jhep.2014.08.047. [DOI] [PubMed] [Google Scholar]
- 7.Purdy MA, Smith DB, Simmonds P, Emerson SU, Harrison T, Meng X-J, Okamoto H, Van der Poel WHM, Jameel S, Hepeviridae Study Group. New classification scheme for Hepeviridae. http://www.ictvonline.org/proposals/2014.008a-hV.A.v6.Hepeviridae.pdf.
- 8.Debing Y, Neyts J. 2014. Antiviral strategies for hepatitis E virus. Antiviral Res 102:106–118. doi: 10.1016/j.antiviral.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao D, Meng XJ. 2012. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect 1:e17. doi: 10.1038/emi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs A, Lindenmann J. 1957. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 11.Egli A, Santer DM, O'Shea D, Tyrrell DL, Houghton M. 2014. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg Microbes Infect 3:e51. doi: 10.1038/emi.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestka S, Krause CD, Walter MR. 2004. Interferons, interferon-like cytokines, and their receptors. Immunol Rev 202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibbert K, Schlaak JF, Yang D, Dittmer U. 2013. IFN-alpha subtypes: distinct biological activities in anti-viral therapy. Br J Pharmacol 168:1048–1058. doi: 10.1111/bph.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wack A, Terczynska-Dyla E, Hartmann R. 2015. Guarding the frontiers: the biology of type III interferons. Nat Immunol 16:802–809. doi: 10.1038/ni.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C, Nan Y, Zhang Y. 2015. New insights into hepatitis E virus virus-host interaction: interplay with host interferon induction. Future Virol 10:439–448. doi: 10.2217/fvl.15.17. [DOI] [Google Scholar]
- 16.Dellgren C, Gad HH, Hamming OJ, Melchjorsen J, Hartmann R. 2009. Human interferon-lambda3 is a potent member of the type III interferon family. Genes Immun 10:125–131. doi: 10.1038/gene.2008.87. [DOI] [PubMed] [Google Scholar]
- 17.Hamming OJ, Terczynska-Dyla E, Vieyres G, Dijkman R, Jorgensen SE, Akhtar H, Siupka P, Pietschmann T, Thiel V, Hartmann R. 2013. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J 32:3055–3065. doi: 10.1038/emboj.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU. 2012. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86:5697–5707. doi: 10.1128/JVI.00146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus-host recombinant. Proc Natl Acad Sci U S A 108:2438–2443. doi: 10.1073/pnas.1018878108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinmann E, Doerrbecker J, Friesland M, Riebesehl N, Ginkel C, Hillung J, Gentzsch J, Lauber C, Brown R, Frentzen A, Pietschmann T. 2013. Characterization of hepatitis C virus intra- and intergenotypic chimeras reveals a role of the glycoproteins in virus envelopment. J Virol 87:13297–13306. doi: 10.1128/JVI.01708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, Wang Y, Metselaar HJ, Janssen HL, Peppelenbosch MP, Pan Q. 2014. Rapamycin and everolimus facilitate hepatitis E virus replication: revealing a basal defense mechanism of PI3K-PKB-mTOR pathway. J Hepatol 61:746–754. doi: 10.1016/j.jhep.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Calmon MF, Rodrigues RV, Kaneto CM, Moura RP, Silva SD, Mota LD, Pinheiro DG, Torres C, de Carvalho AF, Cury PM, Nunes FD, Nishimoto IN, Soares FA, da Silva AM, Kowalski LP, Brentani H, Zanelli CF, Silva WA Jr, Rahal P, Tajara EH, Carraro DM, Camargo AA, Valentini SR. 2009. Epigenetic silencing of CRABP2 and MX1 in head and neck tumors. Neoplasia 11:1329–1339. doi: 10.1593/neo.91110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuno T, Mejido J, Zhao T, Schmeisser H, Morrow A, Zoon KC. 2009. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha. J Immunother 32:803–816. doi: 10.1097/CJI.0b013e3181ad4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Joo H, Clayton S, Dullaers M, Herve MC, Blankenship D, De La Morena MT, Balderas R, Picard C, Casanova JL, Pascual V, Oh S, Banchereau J. 2012. Macrophages induce differentiation of plasma cells through CXCL10/IP-10. J Exp Med 209:1813−1823. doi: 10.1084/jem.20112142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metz P, Dazert E, Ruggieri A, Mazur J, Kaderali L, Kaul A, Zeuge U, Windisch MP, Trippler M, Lohmann V, Binder M, Frese M, Bartenschlager R. 2012. Identification of type I and type II interferon-induced effectors controlling hepatitis C virus replication. Hepatology 56:2082–2093. doi: 10.1002/hep.25908. [DOI] [PubMed] [Google Scholar]
- 26.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. 2010. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 11:1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manns MP, von Hahn T. 2013. Novel therapies for hepatitis C-one pill fits all? Nat Rev Drug Discov 12:595–610. doi: 10.1038/nrd4050. [DOI] [PubMed] [Google Scholar]
- 28.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong C, Zafrullah M, Mixson-Hayden T, Dai X, Liang J, Meng J, Kamili S. 2012. Suppression of interferon-alpha signaling by hepatitis E virus. Hepatology 55:1324–1332. doi: 10.1002/hep.25530. [DOI] [PubMed] [Google Scholar]
- 30.Debing Y, Emerson SU, Wang Y, Pan Q, Balzarini J, Dallmeier K, Neyts J. 2014. Ribavirin inhibits in vitro hepatitis E virus replication through depletion of cellular GTP pools and is moderately synergistic with alpha interferon. Antimicrob Agents Chemother 58:267–273. doi: 10.1128/AAC.01795-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou X, Xu L, Wang W, Watashi K, Wang Y, Sprengers D, de Ruiter PE, van der Laan LJ, Metselaar HJ, Kamar N, Peppelenbosch MP, Pan Q. Disparity of basal and therapeutically activated interferon signalling in constraining hepatitis E virus infection. J Viral Hepat, in press. [DOI] [PubMed] [Google Scholar]
- 32.Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. 2014. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol 88:11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu C, Boon D, McDonald SL, Myers TG, Tomioka K, Nguyen H, Engle RE, Govindarajan S, Emerson SU, Purcell RH. 2010. Pathogenesis of hepatitis E virus and hepatitis C virus in chimpanzees: similarities and differences. J Virol 84:11264–11278. doi: 10.1128/JVI.01205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, Pestka S, Schreiber G. 2011. Binding and activity of all human alpha interferon subtypes. Cytokine 56:282–289. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 35.Uzé G, Schreiber G, Piehler J, Pellegrini S. 2007. The receptor of the type I interferon family. Curr Top Microbiol Immunol 316:71–95. [DOI] [PubMed] [Google Scholar]
- 36.Dubois A, Francois C, Descamps V, Fournier C, Wychowski C, Dubuisson J, Castelain S, Duverlie G. 2009. Enhanced anti-HCV activity of interferon alpha 17 subtype. Virol J 6:70. doi: 10.1186/1743-422X-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Härle P, Cull V, Agbaga MP, Silverman R, Williams BR, James C, Carr DJ. 2002. Differential effect of murine alpha/beta interferon transgenes on antagonization of herpes simplex virus type 1 replication. J Virol 76:6558–6567. doi: 10.1128/JVI.76.13.6558-6567.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerlach N, Gibbert K, Alter C, Nair S, Zelinskyy G, James CM, Dittmer U. 2009. Anti-retroviral effects of type I IFN subtypes in vivo. Eur J Immunol 39:136–146. doi: 10.1002/eji.200838311. [DOI] [PubMed] [Google Scholar]
- 39.Gibbert K, Joedicke JJ, Meryk A, Trilling M, Francois S, Duppach J, Kraft A, Lang KS, Dittmer U. 2012. Interferon-alpha subtype 11 activates NK cells and enables control of retroviral infection. PLoS Pathog 8:e1002868. doi: 10.1371/journal.ppat.1002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamar N, Izopet J, Tripon S, Bismuth M, Hillaire S, Dumortier J, Radenne S, Coilly A, Garrigue V, D'Alteroche L, Buchler M, Couzi L, Lebray P, Dharancy S, Minello A, Hourmant M, Roque-Afonso AM, Abravanel F, Pol S, Rostaing L, Mallet V. 2014. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N Engl J Med 370:1111–1120. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- 41.Haagsma EB, Riezebos-Brilman A, van den Berg AP, Porte RJ, Niesters HG. 2010. Treatment of chronic hepatitis E in liver transplant recipients with pegylated interferon alpha-2b. Liver Transpl 16:474–477. doi: 10.1002/lt.22014. [DOI] [PubMed] [Google Scholar]
- 42.Kamar N, Rostaing L, Abravanel F, Garrouste C, Lhomme S, Esposito L, Basse G, Cointault O, Ribes D, Nogier MB, Alric L, Peron JM, Izopet J. 2010. Ribavirin therapy inhibits viral replication on patients with chronic hepatitis e virus infection. Gastroenterology 139:1612–1618. doi: 10.1053/j.gastro.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Dalton HR, Keane FE, Bendall R, Mathew J, Ijaz S. 2011. Treatment of chronic hepatitis E in a patient with HIV infection. Ann Intern Med 155:479–480. doi: 10.7326/0003-4819-155-7-201110040-00017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.