Abstract
The hepatitis C virus (HCV) RNA-dependent RNA-polymerase NS5B is essentially required for viral replication and serves as a prominent drug target. Sofosbuvir is a prodrug of a nucleotide analog that interacts selectively with NS5B and has been approved for HCV treatment in combination with ribavirin. Although the emergence of resistance to sofosbuvir is rarely seen in the clinic, the S282T mutation was shown to decrease susceptibility to this drug. S282T was also shown to confer hypersusceptibility to ribavirin, which is of potential clinical benefit. Here we devised a biochemical approach to elucidate the underlying mechanisms. Recent crystallographic data revealed a hydrogen bond between S282 and the 2′-hydroxyl of the bound nucleotide, while the adjacent G283 forms a hydrogen bond with the 2′-hydroxyl of the residue of the template that base pairs with the nucleotide substrate. We show that DNA-like modifications of the template that disrupt hydrogen bonding with G283 cause enzyme pausing with natural nucleotides. However, the specifically introduced DNA residue of the template reestablishes binding and incorporation of sofosbuvir in the context of S282T. Moreover, the DNA-like modifications of the template prevent the incorporation of ribavirin in the context of the wild-type enzyme, whereas the S282T mutant enables the binding and incorporation of ribavirin under the same conditions. Together, these findings provide strong evidence to show that susceptibility to sofosbuvir and ribavirin depends crucially on a network of interdependent hydrogen bonds that involve the adjacent residues S282 and G283 and their interactions with the incoming nucleotide and complementary template residue, respectively.
INTRODUCTION
Hepatitis C virus (HCV) has a single-stranded RNA genome of positive polarity and belongs to the Flaviviridae family (1). Chronic HCV infection is associated with severe liver disease, including cirrhosis, and an increased risk of hepatocellular carcinoma (2). If adequately treated, HCV can be cured. Previously, treatment options for HCV infected persons were limited to combination therapy with pegylated alpha interferon (IFN-α) and ribavirin (3). Ribavirin is a nucleoside analog, with a guanine-like base moiety, that can be incorporated by the HCV RNA-dependent RNA polymerase (RdRp) opposite cytosine or uracil, although the detailed mechanism of action remains elusive (4). In 2011, the first direct-acting antivirals (DAAs) targeting the viral protease, or nonstructural protein 3 (NS3), were approved as a component of IFN-based therapies (5); however, the clinical use of the narrow-spectrum protease inhibitors boceprevir and telaprevir is limited by a narrow coverage of HCV genotypes and a low barrier to the selection of resistance (6). Nucleoside or nucleotide inhibitors (NIs) that target the HCV RdRp, or nonstructural protein 5B (NS5B), address these weaknesses (7). Sofosbuvir, a nucleotide prodrug of 2′-deoxy-2′-α-fluoro-β-C-methyluridine, was approved by the U.S. Food and Drug Administration (FDA) in December 2013 in IFN-free combination therapies for chronic HCV infection (8, 9).
NIs compete with the natural ribonucleoside 5′-triphosphates (NTPs) for binding to the highly conserved HCV polymerase active site and, once incorporated, interfere with subsequent nucleotide additions (10). These compounds generally show pangenotypic activity and exhibit a high barrier to the development of resistance (11). HCV NS5B can initiate RNA synthesis de novo or in the presence of a dinucleotide primer that represents the initial product following phosphodiester bond formation (12, 13). This stage of RNA synthesis is fragile and accompanied by frequent dissociation events (12, 13). Conversely, RNA synthesis is highly processive following the incorporation of two to three nucleotides, which defines the elongation stage (14). Since the HCV RNA genome contains approximately 9,600 residues, the elongation stage provides theoretically thousands of opportunities for NIs to incorporate and inhibit viral RNA synthesis (15).
The crystal structure of the HCV NS5B polymerase has been determined in the absence or presence of nucleic acid substrates, with or without a bound nucleotide (16–18). The fold is reminiscent of a right hand with fingers, palm, and thumb subdomains that form a fist-like conformation without sufficient space for the RNA primer template to bind (19). The HCV NS5B polymerase contains a β-hairpin loop that appears to interfere with binding to double-stranded RNA (20). Recent structures of NS5B with a bound primer template show that the thumb domain opens relative to the fingers, which helps to accommodate the nucleic acid substrate (18). Structures of NS5B that have a bound nucleotide and mimic the elongation phase shed light on binding of nucleotides and nucleotide analogues (16). The 2′-hydroxyl group of the ribonucleotide substrate is involved in a hydrogen bonding network that includes position S282 that is associated with resistance to sofosbuvir (21). The S282T mutation confers decreased susceptibility to most 2′-C-methylated compounds, including sofosbuvir (22). This mutation is rarely seen in the clinic, which can be ascribed to a fitness deficit and a high genetic barrier (15, 23). It has been suggested that this mutation may discriminate against the inhibitor, causing a steric clash with the 2′-C-methyl motif; however, the detailed mechanism of resistance has yet to be elucidated (23). The S282T mutation was also shown to confer hypersusceptibility to ribavirin, which adds another layer of complexity to this problem (32, 33). Here, we devised a biochemical approach and identified complex interactions between the template strand and the adjacent residues S282 and G283 that affect susceptibility to sofosbuvir and ribavirin.
MATERIALS AND METHODS
Nucleic acids, nucleotides, and inhibitors.
The following 20-mer heteropolymeric RNA template sequences were used in this study: 5′-AACAGUXUCCUUUUCUCUCC-3′ (T20-X14), where the base X represents uridine (U, T20-U14), thymidine (dT, T20-dT14), 2′-deoxyguanosine (dG, T20-dG14), 2′-deoxyuridine (dU, T20-dU14), or 2′-fluorouridine (2′-FU, T20-2′F-U14) (Trilink); and 5′-AACAGUUUCCUUUUCUCUCC-3′ (T20-G16), where the base G represents guanine (G, T20-G16), 2′-deoxyguanosine (dG, T20-dG16), 2′-deoxy-2′-fluoro-guanosine (2′-FG, T20-2′F-G16), or 2′-O-methyl-guanosine (2′-O-Me-G, T20-2′OMe-G16). The modified nucleic acids were provided by Masad Damha (McGill University): 5′-CUCGAUUUCCUUUUCUCUCC-3′ (T20-A16), where the base A was modified to adenosine (A, T20-A16) or deoxyadenosine (dA, T20-dA16) (Trilink); and 5′-AAAUCGAGAAGGAGAAAGCC-3′ (T20-C16), where the base C was modified to cytosine (C, T20-C16) or 2′-deoxycytidine (dC, T20-dC16) (Trilink). All of the templates were purified by polyacrylamide gel electrophoresis (PAGE). The 5′-GG-3′ dinucleotide (Trilink) was used as a primer. Labeling of the 5′ end of the dinucleotide primer with [γ-32P]ATP was carried out with T4 polynucleotide kinase (Thermo Scientific). NTPs were purchased from Thermo Scientific. The nucleotide analog 2′-C-methyl-CTP (2′-C-methyl-CTP) was provided by Raymond Schinazi (Emory University); the triphosphate form of the active uridine metabolite of sofosbuvir (2′-deoxy-2′-α-fluoro-β-C-methyluridine-TP or 2′-C-Me-2′-F-UTP) was provided by Gilead, and ribavirin-TP was purchased from Jena Bioscience (Jena, Germany).
Expression and purification of HCV NS5B.
The HCV NS5B sequence derived from genotype 1b (Con-1) without the C-terminal, hydrophobic tail of 21 amino acids was inserted into the expression vector pET-21 (Novagen). The plasmid encoding the truncated enzyme with a C-terminally added His tag was transformed into Escherichia coli BL21(DE3) cells. The protein was purified using a Ni-chelating Sepharose column. Mutant enzymes were generated by site-directed mutagenesis by using a Stratagene QuikChange kit according to the manufacturer's instructions (24). All mutations were confirmed by sequencing at the Genome Quebec Innovation Center.
Full-length RNA synthesis.
The standard reaction mixture contained 500 nM RNA template (T20-X14), 1 μM HCV NS5B, 0.2 μM radiolabeled 5′-GG-3′ primer, and 5 μM NTPs. Reactions were carried out in a buffer containing 40 mM HEPES (pH 8.0), 15 mM NaCl, 1 mM dithiothreitol, and 0.5 mM EDTA. RNA synthesis was initiated at room temperature using 6 mM MgCl2 and stopped at specific time points by the addition of 0.5 M EDTA in 95% formamide containing xylene cyanol and bromophenol blue. The samples were then heat denatured at 95°C for 5 min and resolved on a 20% polyacrylamide–7 M urea gel. Products were visualized using a Bio-Rad phosphorimager.
Dose-dependent incorporation of nucleotides and nucleotide analogues.
A solution of 500 nM RNA template (T20-G16, T20-dG16, T20-2′OMe-G16), 1 μM HCV NS5B, and 5 μM ATP and GTP was incubated for 45 min to halt RNA synthesis at position +15. Increasing concentrations of CTP were then added to determine the incorporation efficiency against G, dG, and 2′-O-Me-G at position +16. Reactions were allowed to proceed for 1 min. For 2′C-Methyl-CTP, the standard reaction mixture contained RNA template (T20-G16, T20-dG16, T20-2′F-G16), 5 μM GTP, ATP, and UTP, and a 0.5 μM concentration of competing nucleotide CTP, with increasing concentrations of 2′-C-methyl-CTP. For the active metabolite of sofosbuvir-TP, the standard reaction mixture contained RNA template (T20-A16, T20-dA16), 5 μM GTP, ATP, and CTP, and a 0.5 μM concentration of the competing nucleotide UTP, with increasing concentrations of sofosbuvir-TP. The reactions were allowed to proceed at room temperature for 45 min. The concentration of 2′-C-methyl-CTP or 2′-C-methyl-2′-fluoro-UTP required to inhibit 50% of the full-length product formation (IC50) was calculated using Prism software (GraphPad, Inc.). For the ribavirin assays, the standard reaction mixture contained RNA template (T20-C16, T20-dC16) and 5 μM CTP and UTP. Reactions were allowed to proceed at room temperature for 45 min up to position +15. Increasing concentrations of GTP or ribavirin were then added to determine incorporation efficiency against C or dC at position +16. GTP was allowed to incorporate for 1 min and ribavirin was allowed to incorporate for 5 min before the reactions were stopped. Samples were analyzed as described above. All assays were repeated a minimum of three times.
RESULTS
Experimental strategy.
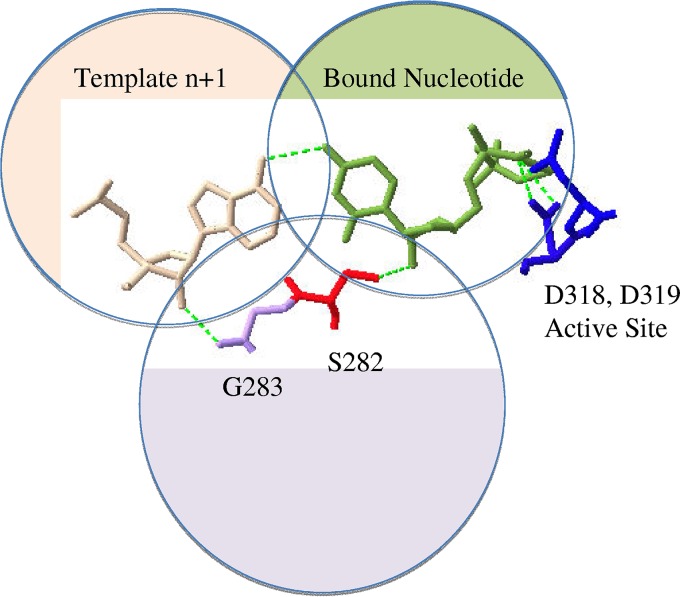
The recent structures of binary and ternary complexes of HCV NS5B revealed that the side chain of S282 is involved in a hydrogen bond network that involves the 2′-hydroxyl of the bound nucleotide (Fig. 1). The structures further show that the complementary residue of the template forms a hydrogen bond with its 2′-hydroxyl group and the backbone of the conserved G283 (Fig. 1). In light of these structural observations, it is tempting to hypothesize that the precise arrangement between the ultimate base pair and the adjacent residues S282 and G283 is crucial for nucleotide binding and/or incorporation. The functional role of the hydrogen bond between G283 and the 2′ hydroxyl of the template is of potential relevance in this regard and remains to be elucidated. To address this problem, we studied NS5B-mediated RNA synthesis with chimeric templates with strategically engineered modifications at the site of incorporation.
FIG 1.
Hydrogen bonding network at the active site of NS5B. The network involves three components of a ternary complex of NS5B, primer/template, and bound nucleotide. The nucleotide (UDP) is shown in green, and the complementary residue of the template is shown in beige. The 2′-hydroxyl of the template forms a hydrogen bond with backbone oxygen of G283 (purple), and the covalently linked S282 (red) forms a hydrogen bond with the 2′-hydroxyl of the bound nucleotide. Active site residues D318 and D319 (blue) and the diphosphate moiety of the bound nucleotide interact with the catalytic metal ions. The figure was generated with SwissPdbViewer software (PDB 4WTA).
RNA synthesis by wild-type NS5B with RNA and RNA-DNA chimeric templates.
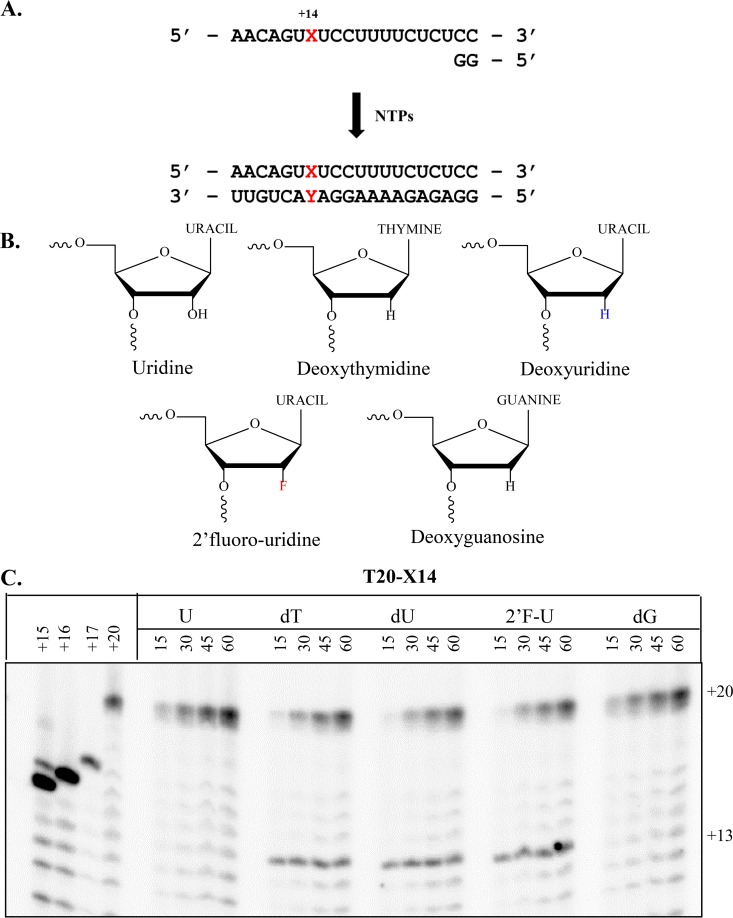
Previous studies have shown that HCV NS5B is able to utilize DNA templates, albeit with reduced efficiency (25). The reduction in RNA synthesis can be ascribed to missing hydrogen bonds between the template and specific amino acids of the RNA binding channel, including G283. To better understand the specific contribution of G283, we synthesized chimeric RNA-DNA templates with chemical modification introduced at the site of incorporation opposite to the incoming nucleotide substrate. The modifications are strategically engineered at position +14 of a model 20-mer heteropolymeric template (T20X14) (Fig. 2A). Modifications at position +14 include U, dT, dG, dU, or 2′-F-U (Fig. 2B). Hence, the lack of the 2′-hydroxyl group is studied in the context of different bases dT, dG, and dU. In 2′F-U the 2′-hydroxyl group is substituted with the bioisosteric 2′-fluoro, which locks the sugar pucker in an RNA-like North conformation.
FIG 2.
NS5B-mediated RNA synthesis and effects of DNA-like modifications on the template. (A) Experimental design for RNA synthesis with a short model template. The modification (X) in the template and its complementary residue Y of the newly synthesized RNA are indicated in red. The reaction is initiated with a dinucleotide primer and yields a 20mer full-length product after exposure to all four NTPs. (B) Structures of modifications at template position +14: dT, dG, dU (South sugar pucker conformation), and U, 2′F-U (80 to 90% North sugar pucker conformation). (C) RNA synthesis with modified templates as described in panel B with RNA length markers at positions +15, +16, +17, and +20 are shown on the left. Enzyme pausing is seen at position +13.
RNA synthesis with the regular RNA template shows full-length product formation in the absence of specific pausing sites. The replacement of U by dT or dU resulted in pausing at position +13 (Fig. 2C). This could either be due to the lack of 2′-hydroxyl group at template position +14, when RNA is replaced by DNA, or to potential conformational differences in the sugar pucker. The DNA residue of the template may adopt the South conformation that is typical for DNA residues (26). However, pausing is also evident with 2′-F-U at position +14, which suggests that the lack of the 2′-hydroxyl group rather than putative differences in the sugar conformation causes pausing of the polymerase. The nature of the base seems to play an additional role in this regard, given that pausing is not seen with a dG modification under the same reaction conditions.
CTP dose-dependent elongation by WT NS5B.
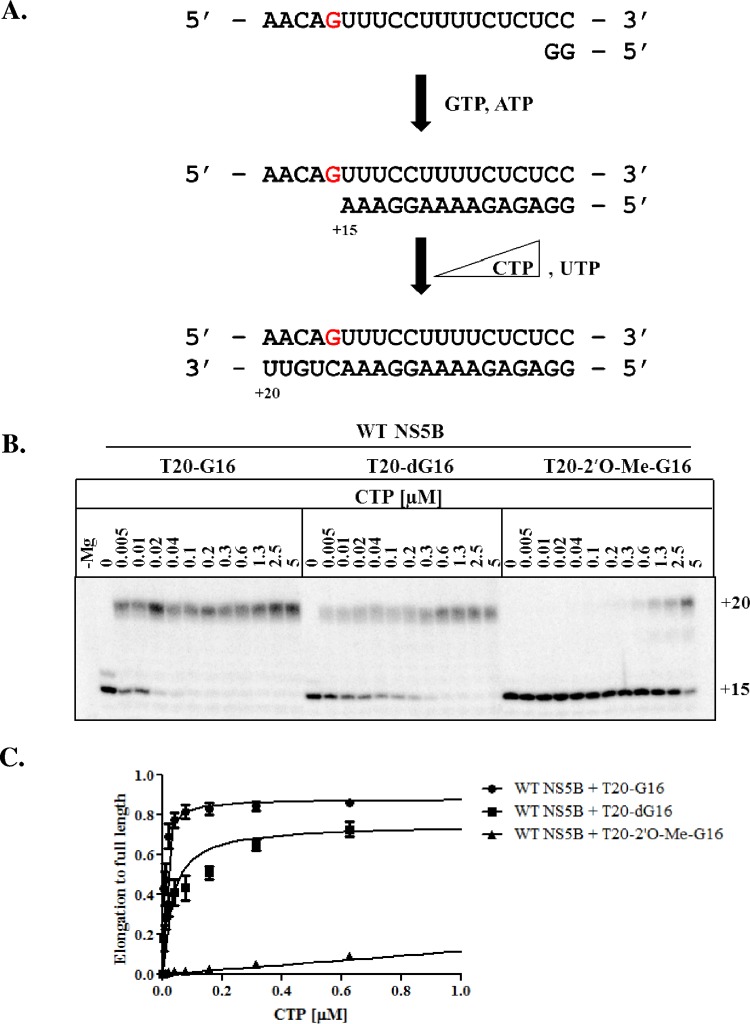
To further assess the importance of the 2′-hydroxyl group of the template opposite the incoming nucleotide, we included the 2′-O-Me-G and compared the efficiency of CTP incorporation with the regular RNA template (G) and the DNA chimeric variant (dG) that showed no significant pausing. Pre-elongation by NS5B was conducted up to position +15 by the omission of CTP and UTP from the reaction mixture. The natural CTP substrate, along with UTP, was then added at increasing concentrations to monitor the efficiency of nucleotide incorporation at the modified template position (Fig. 3A). Templates containing G and dG gave rise to full-length product formation with similar efficiencies (Fig. 3B). However, templates containing the 2′-O-Me-G modification required markedly higher substrate concentrations for synthesis of the full-length product (Fig. 3B). The bulky nature of the 2′-O-Me group can interfere with several processes, including nucleotide binding, nucleotide incorporation, and/or translocation of NS5B relative to the bound nucleic acid. Formation of the posttranslocated conformation is required for nucleotide binding; hence, a bias to the pretranslocated state of the complex would also reduce access to the nucleotide binding site (24).
FIG 3.
Incorporation efficiency on modified templates. (A) Schematic of NS5B elongation on T20-G16 templates modified at position +16 (red). The addition of GTP and ATP to the reaction mixture leads to a 15mer RNA product, and the subsequent addition of UTP and increasing concentrations of CTP yields a 20mer full-length product. (B) WT NS5B-mediated RNA synthesis performed with increasing concentrations of CTP in the presence of RNA templates containing G, dG, and 2′-O-Me-G at position +16. (C) Graphical representation of the data in panel B.
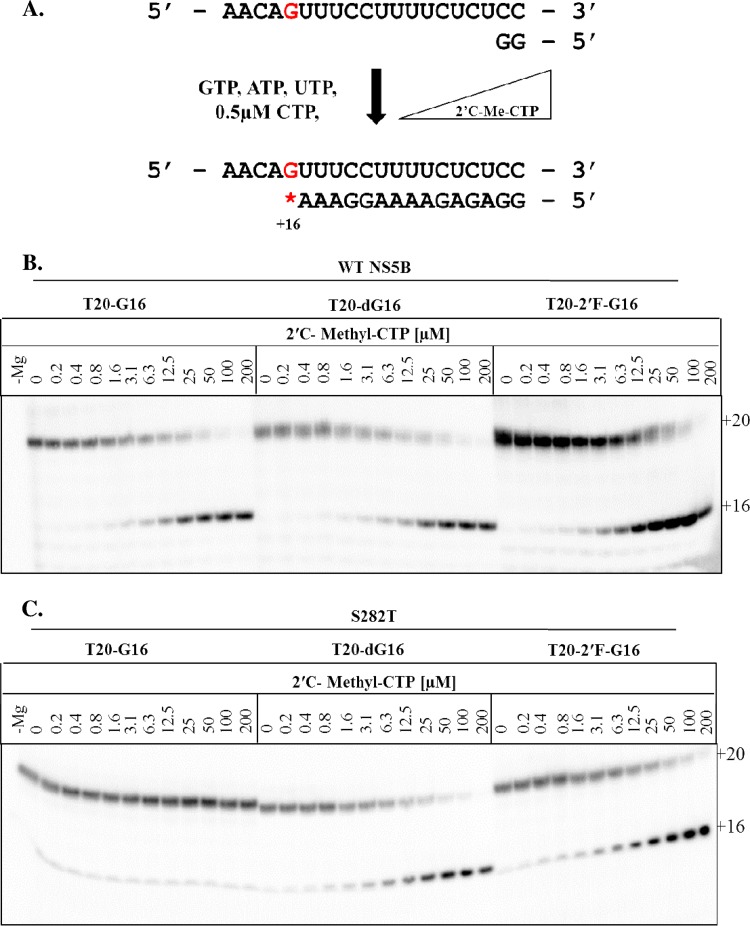
Incorporation of 2′-C-Me-CTP and 2′-C-Me-2′-F-UTP by WT NS5B and resistance-conferring mutants.
We next evaluated the potential impact of changes of the interaction between G283 and the 2′-hydroxyl group of the RNA template with respect to the incorporation of NIs. For this purpose, we compared the efficiency of incorporation of 2′-C-methyl-CTP by WT NS5B to templates containing G, dG, or 2′-F-G at position +16 (Fig. 4A). Full-length RNA synthesis by WT NS5B with the natural template was sensitive to inhibition by 2′-C-Me-CTP, which causes chain-termination at position +16 (Fig. 4B). The modifications of the template had now a significant effect on the inhibition pattern. The S282T mutant enzyme was unable to efficiently incorporate the inhibitor under these conditions, which is consistent with the expected, resistant phenotype (Fig. 4C). WT NS5B showed an IC50 of approximately 12 μM, whereas the S282T mutant prevented incorporation of the inhibitor under these conditions (Table 1). However, when tested with a template containing dG at position +16, both WT NS5B and S282T efficiently incorporated the inhibitor with IC50s of ∼10 μM. Similar results were obtained with templates containing 2′-F-G at position +16. WT NS5B and S282T are inhibited by 2′-C-Me-CTP with IC50s of 10.6 and 21.5 μM, respectively (Fig. 4B and C and Table 1). Together, these data provide strong evidence to suggest that the 2′-hydroxyl group of the template nucleotide also plays an important role in establishing the resistant phenotype.
FIG 4.
RNA synthesis performed in the presence of increasing concentrations of 2′C-Methyl-CTP. (A) Schematic of chain termination by 2′-C-Me-CTP on T20-G16 modified at position +16 to G, dG, and 2′-F-G (red). (B and C) RNA synthesis was performed in the presence of increasing concentrations of 2′-C-Me-CTP with WT NS5B (B) and the S282T mutant enzyme (C).
TABLE 1.
IC50s for 2′-C-Me-CTP incorporation by WT NS5B and S282T on modified templates
| Template | Mean IC50 ± SD (μM) and/or fold increasea |
||
|---|---|---|---|
| NS5B WT (IC50) | S282T |
||
| IC50 | Fold increase | ||
| T20-G16 | 11.3 ± 1.3 | >200 | NA |
| T20-dG16 | 10.3 ± 1.7 | 8.0 ± 1.4 | 0.77 |
| T20-2′F-G16 | 10.6 ± 1.1 | 21.5 ± 2.9 | 2 |
The IC50 is the concentration of 2′-C-Me-CTP that inhibits 50% of the enzyme. Values were calculated by fitting 12 data points to a sigmoidal dose-response equation using GraphPad Prism (version 5.0). Standard deviation values were determined on the basis of three independent experiments. The fold increase was calculated as the ratio of the IC50 of the mutant enzyme to that of the wild-type enzyme. NA, not applicable (we were not able to calculate a value).
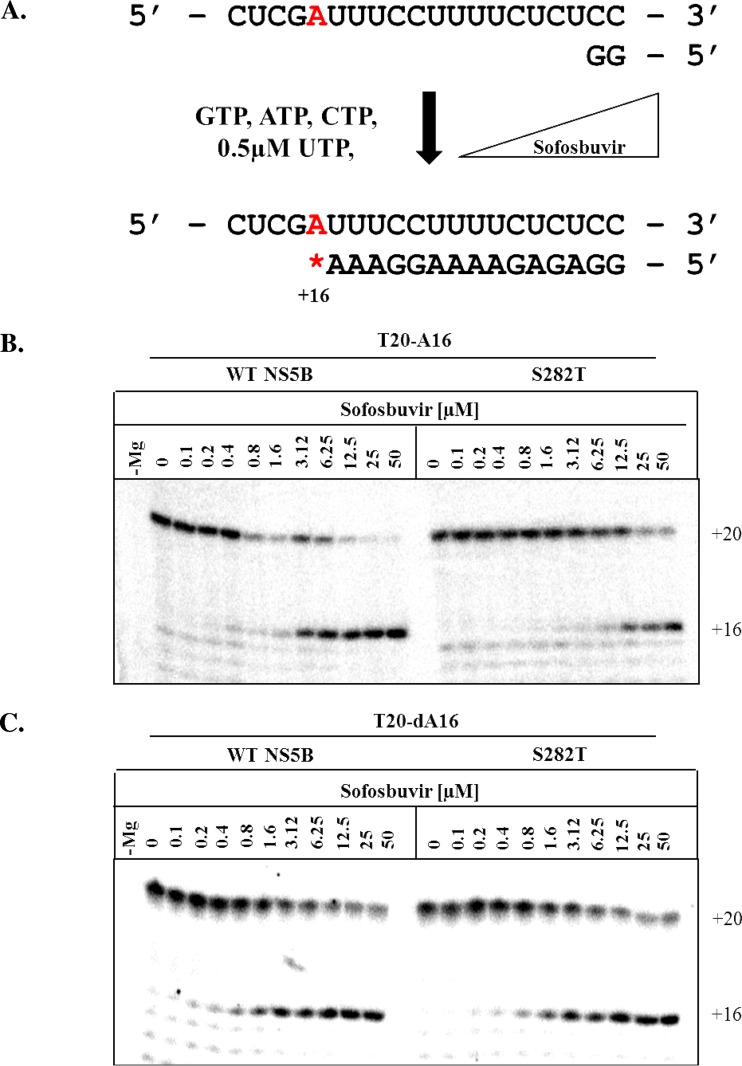
Very similar patterns are seen with the uridine analog 2′-C-Me-2′-F-UTP. WT NS5B was sensitive to 2′-C-Me-2′-F-UTP, whereas S282T had reduced rates of incorporation of this inhibitor (Fig. 5B and Table 2). With WT NS5B and a template containing A at position +16, we measured an IC50 of 2 μM, while S282T had an IC50 of 19.5 μM. When tested with a template containing dA at position +16, S282T showed sensitivity to sofosbuvir, resulting in a lower IC50 of 2 μM as measured with WT NS5B (Table 2). The lack of a hydrogen bond between G283 and the template at the site of incorporation seems to neutralize the effect of the 2′-C-Me group, which is a common structural feature of 2′-C-Me-2′-F-UTP and 2′-C-Me-CTP.
FIG 5.
RNA synthesis performed in the presence of increasing concentrations of sofosbuvir-TP. (A) Schematic of chain termination by 2′-C-Me-2′-F-UTP on T20-A16 modified at position +16 to A or dA (red). (B and C) RNA synthesis performed in the presence of increasing concentrations of 2′-C-Me-2′-F-UTP with WT NS5B and the S282T mutant on the natural template T20-AU16 (B) and the modified template T20-dA16 (C).
TABLE 2.
IC50s for sofosbuvir incorporation by WT NS5B and the S282T mutant on T20-A16 and T20-dA16
| Enzyme | Mean IC50 ± SD (μM) and fold increasea |
|||
|---|---|---|---|---|
| T20-A16 |
T20-dA16 |
|||
| IC50 | Fold increase | IC50 | Fold increase | |
| WT NS5B | 2.1 ± 0.1 | 1.4 ± 0.2 | 0.6 | |
| S282T | 19.5 ± 2 | 9.2 | 2.1 ± 0.5 | 9.2 |
IC50 is the inhibitory concentration of 2′-C-Me-2′-F-UTP that inhibits 50% of the enzyme. Values were calculated by fitting 12 data points to a sigmoidal dose-response equation using GraphPad Prism (version 5.0). Standard deviation values were determined on the basis of three independent experiments. The fold increase was calculated as the ratio of the IC50 of the mutant enzyme to that of the wild-type enzyme.
Incorporation of ribavirin by WT NS5B and S282T.
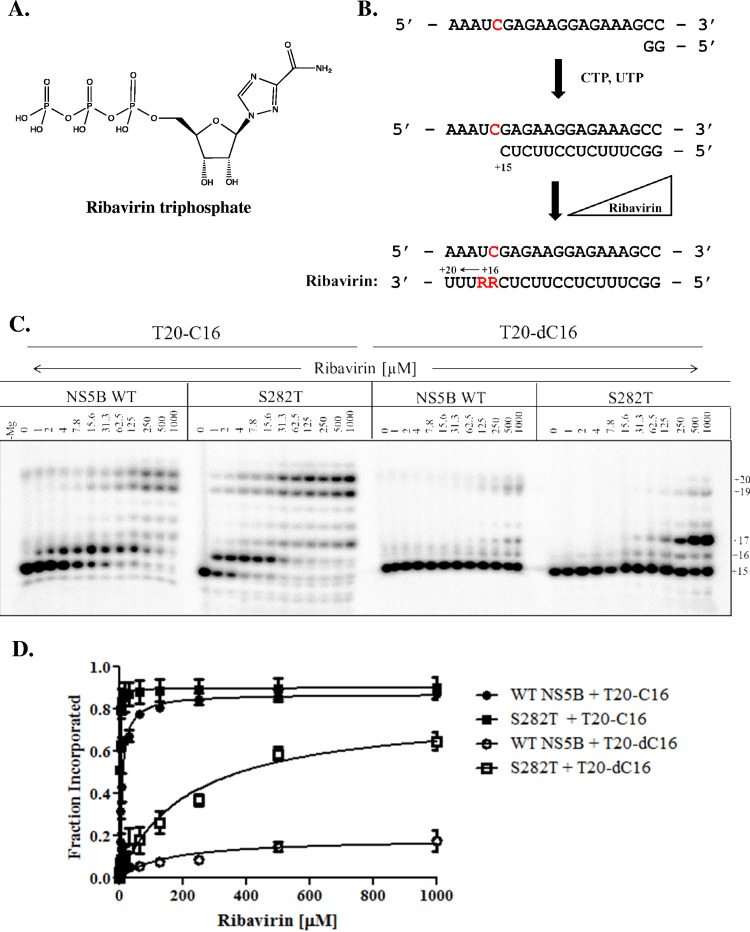
Finally, we measured the efficiency of incorporation of ribavirin-TP by WT NS5B and S282T (Fig. 6). Pre-elongation by WT NS5B or S282T was conducted up to position +15, followed by the addition of ribavirin-TP in a dose-dependent manner to monitor incorporation opposite C or dC at position +16 (Fig. 6A). When tested with the natural RNA template, we measured higher levels of RNA synthesis with the S282T mutant compared to WT NS5B (Fig. 6B and C). Note that both template positions +16 (C) and +17 (U) allow the incorporation of ribavirin-TP, which together with the preexisting NTP pool yields a full-length 20-mer product. The incorporation of ribavirin-TP is negligible when tested with a template containing dC, indicating that the loss of the hydrogen bond between G283 and the sugar moiety of the template prevents the reaction in this context. However, modest levels of ribavirin-TP incorporation are seen with the S282T mutant enzyme, which supports the trend seen with the natural RNA template.
FIG 6.
RNA synthesis performed in the presence of increasing concentrations of ribavirin-TP. (A) Chemical structure of ribavirin-5′-triphosphate. (B) A scheme depicts the incorporation of ribavirin (R), indicated in red, on T20-C16 templates modified at position +16. Note that ribavirin can get incorporated opposite C and U. (C) RNA synthesis performed by WT NS5B and S282T with increasing concentrations of ribavirin on T20-C16 and T20-dC16. (D) Graphical representation of the data shown in panel C.
DISCUSSION
Sofosbuvir and ribavirin are currently the only FDA-approved NIs for HCV treatment, although ribavirin is not classified as a DAA (27). Clinical trials revealed sustained virological response rates of >90% in the context combination therapies with the two drugs (28). The emergence of resistance to regimens containing sofosbuvir is rare, although a few cases are documented (29). The NS5B S282T variant can emerge in vitro and in vivo under the selective pressure of 2′-C-methylated nucleotides, including sofosbuvir. Specific mutations conferring resistance to ribavirin have not been identified (30). However, Svarovskaia and coworkers reported subtle increases in susceptibility to ribavirin in the context of the S282T mutation (31, 32). Such a pattern may translate in clinical benefits. Moreover, the observation that a mutation in close proximity to the active site of HCV NS5B can affect susceptibility to ribavirin suggests that the viral RdRp is the drug target. Potential underlying mechanisms include competitive inhibition with cellular nucleotide pools, reductions in nucleotide incorporations following ribavirin-terminated primers, or decreases in fidelity; however, ribavirin's mechanism of action remains elusive, and indirect effects are also being considered. In contrast, biochemical data have shown that sofosbuvir acts as a nonobligate chain terminator that prevents further incorporation events (31, 32). Modeling studies suggested that that S282T may cause a steric conflict with the 2′-C-methyl group, which provides a possible mechanism for resistance. Based on structures of NS5B with a bound primer/template, Mosley et al. pointed to more complex mechanisms that can involve adjacent amino acids, such as G283, which forms a hydrogen bond with the 2′-hydroxyl of the template (18). Here, we utilized templates with DNA-like modifications to disrupt this hydrogen bond and studied the consequences on the rates of incorporation of sofosbuvir and ribavirin, respectively. The data provide a model that helps to explain the effect of S282T on changes in susceptibility to nucleotide analogue inhibitors (Fig. 7).
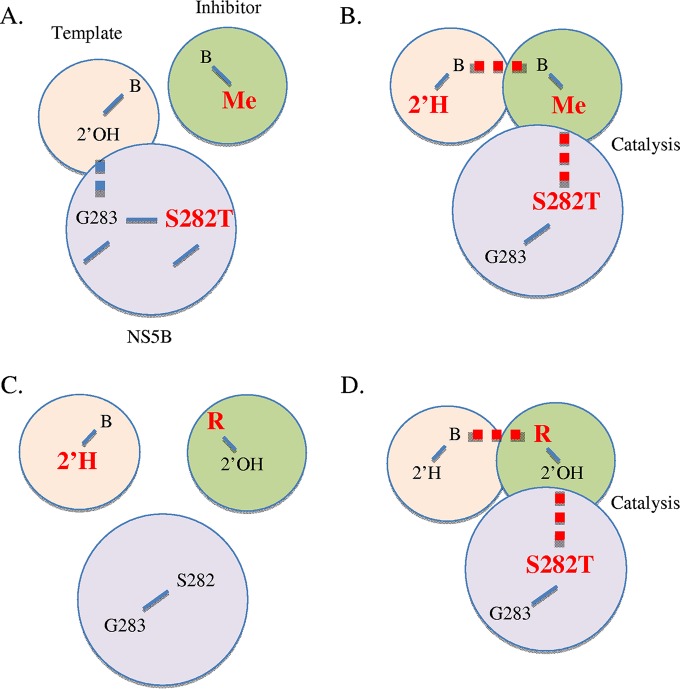
FIG 7.
Model of interdependent hydrogen bonding affects susceptibility to 2′C-methylated compounds and ribavirin. (A) S282T prevents binding of 2′-C-methyl modified nucleotides. (B) The loss of a hydrogen bond between G283 and a DNA template increases the flexibility at S282T and facilitates binding the inhibitor. (C) The loss of a hydrogen bond between G283 and a DNA template prevents binding of ribavirin (R) that shows per se weak base pairing. (D) S282T can partially compensate for this deficiency and facilitates the binding and/or incorporation of ribavirin.
The model is derived from Fig. 1 and depicts the relevant interactions between the three components that form the ternary NS5B-primer/template/NTP complex (Fig. 7). The structural data suggest that the covalently linked residues S282 and G283 facilitate binding and proper positioning of the NTP substrate and template (Fig. 1). The loss of the hydrogen bond between G283 and a DNA template causes in some cases enzyme pausing, which supports this notion. Pausing is evident with dT, dU, and 2′-F-U modifications in the template strand, while pausing is not seen with the dG modification. The additional hydrogen bond in GC versus AT or AU base pairs may at least partially compensate for the loss of the hydrogen bond between G283 and the 2′-deoxy residue in the template.
S282T diminishes binding and/or incorporation of 2′C-methylated compounds. The putative steric clash between S282T and the 2′-C-methyl group of the inhibitor can diminish the interaction between the side chain of residue 282 and the sugar moiety of the nucleotide analogue. The altered interaction may in turn affect proper base pairing (Fig. 7A). As a result, the compound is not in a favorable position for incorporation. However, a DNA residue in the template appears to neutralize this effect. A possible explanation is that the lack of the hydrogen bond between G283 and the 2′-OH of the template causes an increase in flexibility at the nucleotide binding site that permits incorporation of the analogue. It is therefore conceivable that the postulated constraints introduced by the S282T mutation are relieved through a more favorable positioning of the template for base pairing with the inhibitor (Fig. 7B). The total available space for productive inhibitor binding is likely increased. This model is consistent with an increase in the rates of incorporation of ribavirin-TP in the context of S282T. Ribavirin-TP is a poor substrate for NS5B, which is likely due to weak base pairing between the pseudo base of ribavirin and the template. The DNA residue of the template further enhances this effect, which helps to explain the lack of activity in this context (Fig. 7C). The ability to incorporate ribavirin-TP is partially restored with the S282T mutant. This suggests an improvement in inhibitor binding, although the nature of the altered interaction remains to be characterized (Fig. 7D).
Taken together, our findings provide strong evidence demonstrating that susceptibility to nucleotides and nucleotide analogs depends crucially on a network of interdependent hydrogen bonds that involve the adjacent residues S282 and G283 and their interactions with the incoming nucleotide and complementary template residue, respectively. Changes in this ring-like structure can affect substrate binding and/or the precise alignment of substrate and primer for catalysis. However, the inherently flexible nature of this hydrogen bonding network may also be exploited in drug development efforts. The data provide a rationale and the experimental tools for the design of inhibitors that are simultaneously modified at their base and sugar moieties in attempts to maximize inhibitor binding and rates of incorporation.
ACKNOWLEDGMENTS
We thank the Genome Quebec Innovation Center for sequencing our mutagenesis samples.
Funding Statement
A.S.K. received a graduate student fellowship from the National CIHR Research Training Program in Hepatitis C (NCRTP). This study was sponsored by the Canadian Institutes of Health Research (CIHR), the National Sciences and Engineering Research Council of Canada (NSERC), and Gilead Sciences, Inc. R.F.S. is supported by NIH CFAR grant 2P30AI050409 and by the Department of Veterans Affairs.
REFERENCES
- 1.Major ME, Feinstone SM. 1997. The molecular virology of hepatitis C. Hepatology 25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson IM, Davis GL, El-Serag H, Negro F, Trepo C. 2010. Prevalence and challenges of liver diseases in patients with chronic hepatitis C virus infection. Clin Gastroenterol Hepatol 8:924–933. doi: 10.1016/j.cgh.2010.06.032. [DOI] [PubMed] [Google Scholar]
- 3.McHutchison JG, Lawitz EJ, Shiffman ML, Muir AJ, Galler GW, McCone J, Nyberg LM, Lee WM, Ghalib RH, Schiff ER, Galati JS, Bacon BR, Davis MN, Mukhopadhyay P, Koury K, Noviello S, Pedicone LD, Brass CA, Albrecht JK, Sulkowski MS, Team IS. 2009. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N Engl J Med 361:580–593. doi: 10.1056/NEJMoa0808010. [DOI] [PubMed] [Google Scholar]
- 4.Feld JJ, Hoofnagle JH. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967–972. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- 5.Sulkowski MS, Sherman KE, Dieterich DT, Bsharat M, Mahnke L, Rockstroh JK, Gharakhanian S, McCallister S, Henshaw J, Girard PM, Adiwijaya B, Garg V, Rubin RA, Adda N, Soriano V. 2013. Combination therapy with telaprevir for chronic hepatitis C virus genotype 1 infection in patients with HIV: a randomized trial. Ann Intern Med 159:86–96. doi: 10.7326/0003-4819-159-2-201307160-00654. [DOI] [PubMed] [Google Scholar]
- 6.Soriano V, Vispo E, Poveda E, Labarga P, Barreiro P. 2012. Treatment failure with new hepatitis C drugs. Expert Opin Pharmacother 13:313–323. doi: 10.1517/14656566.2012.653341. [DOI] [PubMed] [Google Scholar]
- 7.Zeng QL, Zhang JY, Zhang Z, Wang LF, Wang FS. 2013. Sofosbuvir and ABT-450: terminator of hepatitis C virus? World J Gastroenterol 19:3199–3206. doi: 10.3748/wjg.v19.i21.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. 2013. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med 368:1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 9.Zeuzem S, Dusheiko GM, Salupere R, Mangia A, Flisiak R, Hyland RH, Illeperuma A, Svarovskaia E, Brainard DM, Symonds WT, Subramanian GM, McHutchison JG, Weiland O, Reesink HW, Ferenci P, Hezode C, Esteban R, Investigators V. 2014. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med 370:1993–2001. doi: 10.1056/NEJMoa1316145. [DOI] [PubMed] [Google Scholar]
- 10.Ma H, Leveque V, De Witte A, Li W, Hendricks T, Clausen SM, Cammack N, Klumpp K. 2005. Inhibition of native hepatitis C virus replicase by nucleotide and non-nucleoside inhibitors. Virology 332:8–15. doi: 10.1016/j.virol.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 11.Sofia MJ. 2013. Nucleotide prodrugs for the treatment of HCV infection. Adv Pharmacol 67:39–73. doi: 10.1016/B978-0-12-405880-4.00002-0. [DOI] [PubMed] [Google Scholar]
- 12.Luo G, Hamatake RK, Mathis DM, Racela J, Rigat KL, Lemm J, Colonno RJ. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J Virol 74:851–863. doi: 10.1128/JVI.74.2.851-863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong W, Ferrari E, Lesburg CA, Maag D, Ghosh SK, Cameron CE, Lau JY, Hong Z. 2000. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74:9134–9143. doi: 10.1128/JVI.74.19.9134-9143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohmann V, Korner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol 71:8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powdrill MH, Tchesnokov EP, Kozak RA, Russell RS, Martin R, Svarovskaia ES, Mo H, Kouyos RD, Gotte M. 2011. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc Natl Acad Sci U S A 108:20509–20513. doi: 10.1073/pnas.1105797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Appleby TC, Perry JK, Murakami E, Barauskas O, Feng J, Cho A, Fox D III, Wetmore DR, McGrath ME, Ray AS, Sofia MJ, Swaminathan S, Edwards TE. 2015. Viral replication: structural basis for RNA replication by the hepatitis C virus polymerase. Science 347:771–775. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 17.Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol 6:937–943. doi: 10.1038/13305. [DOI] [PubMed] [Google Scholar]
- 18.Mosley RT, Edwards TE, Murakami E, Lam AM, Grice RL, Du J, Sofia MJ, Furman PA, Otto MJ. 2012. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J Virol 86:6503–6511. doi: 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joyce CM, Steitz TA. 1995. Polymerase structures and function: variations on a theme? J Bacteriol 177:6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Z, Cameron CE, Walker MP, Castro C, Yao N, Lau JY, Zhong W. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6–11. doi: 10.1006/viro.2001.0948. [DOI] [PubMed] [Google Scholar]
- 21.Feng JY, Cheng G, Perry J, Barauskas O, Xu Y, Fenaux M, Eng Tirunagari SN, Peng B, Yu M, Tian Y, Lee YJ, Stepan G, Lagpacan LL, Jin D, Hung M, Ku KS, Han B, Kitrinos K, Perron M, Birkus G, Wong KA, Zhong W, Kim CU, Carey A, Cho A, Ray AS. 2014. Inhibition of hepatitis C virus replication by GS-6620, a potent C-nucleoside monophosphate prodrug. Antimicrob Agents Chemother 58:1930–1942. doi: 10.1128/AAC.02351-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedskog C, Chodavarapu K, Ku KS, Xu S, Martin R, Miller MD, Mo H, Svarovskaia E. 2015. Genotype- and subtype-independent full-genome sequencing assay for hepatitis C virus. J Clin Microbiol 53:2049–2059. doi: 10.1128/JCM.02624-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutartre H, Bussetta C, Boretto J, Canard B. 2006. General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob Agents Chemother 50:4161–4169. doi: 10.1128/AAC.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deval J, Powdrill MH, D'Abramo CM, Cellai L, Gotte M. 2007. Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob Agents Chemother 51:2920–2928. doi: 10.1128/AAC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao CC, Yang X, Kline A, Wang QM, Barket D, Heinz BA. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J Virol 74:11121–11128. doi: 10.1128/JVI.74.23.11121-11128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams AA, Darwanto A, Theruvathu JA, Burdzy A, Neidigh JW, Sowers LC. 2009. Impact of sugar pucker on base pair and mispair stability. Biochemistry 48:11994–12004. doi: 10.1021/bi9014133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curry MP, Forns X, Chung RT, Terrault NA, Brown R Jr, Fenkel JM, Gordon F, O'Leary J, Kuo A, Schiano T, Everson G, Schiff E, Befeler A, Gane E, Saab S, McHutchison JG, Subramanian GM, Symonds WT, Denning J, McNair L, Arterburn S, Svarovskaia E, Moonka D, Afdhal N. 2015. Sofosbuvir and ribavirin prevent recurrence of HCV infection after liver transplantation: an open-label study. Gastroenterology 148:100–107 e101. doi: 10.1053/j.gastro.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Osinusi A, Meissner EG, Lee YJ, Bon D, Heytens L, Nelson A, Sneller M, Kohli A, Barrett L, Proschan M, Herrmann E, Shivakumar B, Gu W, Kwan R, Teferi G, Talwani R, Silk R, Kotb C, Wroblewski S, Fishbein D, Dewar R, Highbarger H, Zhang X, Kleiner D, Wood BJ, Chavez J, Symonds WT, Subramanian M, McHutchison J, Polis MA, Fauci AS, Masur H, Kottilil S. 2013. Sofosbuvir and ribavirin for hepatitis C genotype 1 in patients with unfavorable treatment characteristics: a randomized clinical trial. JAMA 310:804–811. doi: 10.1001/jama.2013.109309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svarovskaia ES, Dvory-Sobol H, Parkin N, Hebner C, Gontcharova V, Martin R, Ouyang W, Han B, Xu S, Ku K, Chiu S, Gane E, Jacobson IM, Nelson DR, Lawitz E, Wyles DL, Bekele N, Brainard D, Symonds WT, McHutchison JG, Miller MD, Mo H. 2014. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis 59:1666–1674. doi: 10.1093/cid/ciu697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Migliaccio G, Tomassini JE, Carroll SS, Tomei L, Altamura S, Bhat B, Bartholomew L, Bosserman MR, Ceccacci A, Colwell LF, Cortese R, De Francesco R, Eldrup AB, Getty KL, Hou XS, LaFemina RL, Ludmerer SW, MacCoss M, McMasters DR, Stahlhut MW, Olsen DB, Hazuda DJ, Flores OA. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J Biol Chem 278:49164–49170. doi: 10.1074/jbc.M305041200. [DOI] [PubMed] [Google Scholar]
- 31.Murakami E, Tolstykh T, Bao H, Niu C, Steuer HM, Bao D, Chang W, Espiritu C, Bansal S, Lam AM, Otto MJ, Sofia MJ, Furman PA. 2010. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J Biol Chem 285:34337–34347. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sofia MJ, Bao D, Chang W, Du J, Nagarathnam D, Rachakonda S, Reddy PG, Ross BS, Wang P, Zhang HR, Bansal S, Espiritu C, Keilman M, Lam AM, Steuer HM, Niu C, Otto MJ, Furman PA. 2010. Discovery of a β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem 53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]