Abstract
Bacterial persisters are a small fraction of quiescent cells that survive in the presence of lethal concentrations of antibiotics. They can regrow to give rise to a new population that has the same vulnerability to the antibiotics as did the parental population. Although formation of bacterial persisters in the presence of various antibiotics has been documented, the molecular mechanisms by which these persisters tolerate the antibiotics are still controversial. We found that amplification of the fumarate reductase operon (FRD) in Escherichia coli led to a higher frequency of persister formation. The persister frequency of E. coli was increased when the cells contained elevated levels of intracellular fumarate. Genetic perturbations of the electron transport chain (ETC), a metabolite supplementation assay, and even the toxin-antitoxin-related hipA7 mutation indicated that surplus fumarate markedly elevated the E. coli persister frequency. An E. coli strain lacking succinate dehydrogenase (SDH), thereby showing a lower intracellular fumarate concentration, was killed ∼1,000-fold more effectively than the wild-type strain in the stationary phase. It appears that SDH and FRD represent a paired system that gives rise to and maintains E. coli persisters by producing and utilizing fumarate, respectively.
INTRODUCTION
Bacterial persisters are phenotypic variants that are tolerant even to supralethal concentrations of multiple antibiotics (1–3). Reseeding of the persisters yields a bacterial population with a frequency of antibiotic-tolerant cells that is similar to that of the parental population (4, 5). Persisters are distinct from antibiotic-resistant cells because the ability to tolerate antibiotics is neither genetically determined nor inherited. Persisters showing tolerance of different classes of antibiotics are observed in most microbial species and have been implicated in chronic and recurrent infections (1). Furthermore, it is highly probable that persisters are a potential reservoir for the development of drug resistance in pathogenic bacteria (6, 7).
Despite the discovery of bacterial persisters more than 70 years ago (4), the mechanisms that underlie noninheritable persistence phenotypes remain unclear. Various researchers recently identified a number of genes and pathways that lead to persister formation or survival upon antibiotic treatments. These include toxin-antitoxin (TA) modules, a stringent response, phosphate metabolism, alternative energy production, and antioxidative defense (8–13). Because nongrowing or slow-growing bacteria are less sensitive to antibiotics, dormancy has been proposed to be the mechanism of last resort in many of these persistence studies. Thus, many recent mechanistic studies have focused on how bacterial cells reach the dormant state (8–15). Nonetheless, the prevailing hypothesis that persisters might survive solely because of dormancy is being challenged. A lack of significant growth or metabolic activity does not guarantee persistence, and dormancy is neither necessary nor sufficient for bacterial persistence (16, 17). Although the mechanisms behind dormancy are highly redundant, whether dormancy is the cause or result of persistence is a controversial topic.
Most insights into bacterial persistence were obtained by means of screening techniques allowing the isolation of mutants exhibiting higher frequencies of persister formation. The first mutant that was identified in such studies was a high-persister (hip) mutant of Escherichia coli isolated by Moyed and Broderick (18). The best-studied allele of hipA (hipA7) raises the persister frequency as much as 1,000-fold (19). Screening of a library of transposon insertion mutants produced a large set of candidate genes (20, 21). Because there was a chance that the transposon library screen missed some genes that are crucial for persistence, similar screening assays were performed several times (22, 23), and a complete E. coli gene knockout library called the Keio collection (24) was screened again. These screening experiments produced additional interesting candidate genes. Screening of an expression library (20) and gene expression analysis of persister-enriched samples (25) yielded additional persistence genes, suggesting that multiple metabolic pathways can contribute to persistence in bacteria. We also attempted to use an overexpression library to identify and enrich for E. coli mutants capable of generating persisters at a high frequency. Rather than using the existing single-gene overexpression libraries, we constructed an E. coli genomic library with large inserts to ensure overexpression of not only a single gene but also entire operons of genes. This library made gain-of-function screening possible and helped to identify a new persistence gene: the fumarate reductase gene (FRD). After testing of various properties of this candidate gene, we propose a new persistence model that does not necessarily rely on either slow growth or dormancy.
MATERIALS AND METHODS
Reagents and bacterial strains.
Escherichia coli K-12 BW25113 was purchased from Open Biosystems (Thermo Fisher Scientific, Waltham, MA). For evaluations of persister frequency, antibiotics were administered at the following concentrations: 100 μg/ml ampicillin (Amp), 50 μg/ml kanamycin (Kan), or 5 μg/ml norfloxacin (Nor). Hydroxyphenyl fluorescein (HPF) was employed for detection of hydroxyl radicals inside the cell (26).
Construction of the overexpression library and screening for persister-rich mutants.
E. coli K-12 genomic fragments were inserted into the plasmid pZE21, which enables constitutive overexpression of multiple genes present in a specific genomic fragment. Genomic DNA of E. coli K-12 MG1655 was partially digested by use of Tsp509I (New England BioLabs, Ipswich, MA). Approximately 2- to 6-kbp fragments were isolated from an agarose gel and ligated into the EcoRI site of the pZE21 plasmid. The ligation mixture was introduced into the E. coli DH10 strain for creation of an amplified genomic library. More than 50,000 separate colonies were collected for construction of the library. The library was then introduced into the E. coli DH5α strain for serial subculture experiments. After isolation of the plasmid-harboring cells by Kan selection, the E. coli DH5α strain (an equivalent of 107 cells) was transformed with the genomic library and spread on an LB agar plate containing 50 μg/ml Amp. Plasmids containing genomic fragments were isolated from each colony that survived the Amp selection and were sequenced (see Fig. S1 in the supplemental material).
Construction of E. coli knockout mutants.
E. coli BW25113 carrying the Red helper plasmid (pKD46) was grown in LB broth supplemented with 50 μg/ml Amp and 1 mM l-arabinose at 30°C until the optical density at 600 nm reached ∼0.8 to attain Red recombinase expression. The cells were made electrocompetent via 100-fold concentration and washing with chilled 10% glycerol. PCR products were generated using several pairs of ∼70-mer primers (see Table S1 in the supplemental material) that included an ∼50-base homology extension and an ∼20-base priming sequence for pKD3 (for a chloramphenicol selection marker) or pKD4 (for a kanamycin selection marker) as the template. The amplicons were gel purified, digested with DpnI, and repurified. Electroporation was performed using 50 μl of electrocompetent cells and 100 ng of PCR product on a Gene Pulser system (Bio-Rad Laboratories, Hercules, CA). The electroshocked cells were added to 1 ml of LB broth and incubated at 37°C for 2 h. The cells were then seeded into LB agar plates containing 30 μg/ml kanamycin or 24 μg/ml chloramphenicol. After primary selection, the resulting knockout mutants were cultured at 43°C to achieve pKD46 plasmid curing and were tested for the loss of the helper plasmid. To prepare a double-knockout mutant, we utilized the modified P1 phage transduction method (27), as follows. A P1 lysate was prepared from the wild-type (WT) strain of E. coli BW25113. Each single-knockout mutant was further mutated using different antibiotic selection markers, e.g., chloramphenicol or kanamycin. The donor mutant for P1 transduction was precultured overnight in LB broth, and 200 μl of the culture was removed and supplemented with 5 mM CaCl2 and 50 μl of the P1 lysate. The mixture was seeded into fresh LB broth containing 5 mM CaCl2, and this culture was incubated at 37°C until the cells were lysed. The lysate was incubated at room temperature for 20 min with 1 or 2 drops of chloroform. Next, the supernatant was removed and stored at 4°C after centrifugation. An acceptor mutant was precultured overnight, and 5 mM CaCl2 was added; after that, 200 μl of culture and 5 μl of P1 lysate, which contained a genomic fragment of the donor mutant, were mixed and incubated at room temperature for 15 min. The mixture was incubated at 37°C for 1 h with 1 drop of 1 M sodium citrate and then seeded onto an LB agar plate containing antibiotics. Each knockout mutant was verified by colony PCR using common testing primers for an antibiotic resistance gene (see Tables S1 and S2).
Measurement of persister frequency.
The number of persisters was determined by counting CFU after exposure to antibiotics. For preparation of exponential-phase cells, 30 μl of a frozen cell stock was diluted in 3 ml LB broth and grown at 37°C and 250 rpm for 16 h. The precultured cells (0.2 ml) were seeded into 20 ml of LB broth containing 100 mM fumarate or other compounds of interest and then cultured to ∼108 CFU/ml, which took 2 to 3 h after the inoculation. Severe cell growth inhibition was not observed in the presence of 100 mM sodium fumarate and sodium citrate (see Fig. S2A in the supplemental material). Three milliliters of the culture was centrifuged at 10,000 × g for 3 min to collect cells. The cells were resuspended with 3 ml fresh LB broth containing antibiotics, transferred to a 14-ml round-bottom tube, and incubated at 37°C for 9 h. Note that fumarate was not contained in the culture broth during the time of antibiotic exposure. Time-dependent killing curves (see Fig. S3) (3) showed that the cell death rate was decreased after 3 h of antibiotic treatment, and a second slow death phase continued after that time. After incubation for 9 h, the cells were collected by centrifugation at 10,000 × g for 3 min, washed, and serially diluted with phosphate-buffered saline (PBS). Each 100-μl aliquot was spread on an LB agar plate. Visible viable colonies were counted to determine the number of CFU after overnight incubation of the plates. Only dilutions that yielded 50 to 250 colonies were subjected to counting. The persister frequency was obtained by dividing the number of CFU after antibiotic treatment by that before treatment. For preparation of stationary-phase cells, 30 μl of a frozen cell stock was diluted in 3 ml LB broth and grown at 37°C and 250 rpm for 16 h. The precultured cells (0.2 ml) were seeded into 20 ml LB broth containing compounds of interest and again grown for 16 h. This resulted in ∼109 cells/ml. Three milliliters of the preculture at stationary phase was centrifuged, and the pelleted cells were resuspended with 3 ml of prewarmed fresh LB broth containing antibiotics in a 14-ml polypropylene round-bottom tube to prevent the influence of remaining intermediates and chemicals contained in the preculture. The persister frequency was measured in the same way as that for the exponential-phase cells. To measure the persister frequency under anaerobic conditions, E. coli was cultured until the cell number reached ∼108 CFU/ml (exponential phase) or ∼109 cells/ml (stationary phase), and the cultures were put into a GasPak jar with a GasPak pouch (BD Biosciences) to remove oxygen inside the jar. The LB medium and agar plates were incubated in a GasPak jar with a pouch before use to remove dissolved oxygen.
Analysis of extra- and intracellular fumarate and other TCA cycle intermediates.
Cells were precultured in 5 ml of M9 broth with 0.4% (wt/vol) glucose overnight. Next, 0.5 ml of the preculture was seeded into 50 ml of fresh M9 broth with 50 mM glucose in a 250-ml flask and cultured at 37°C and 250 rpm for 16 h. One of the tricarboxylic acid (TCA) cycle intermediates (100 mM)—sodium fumarate, sodium succinate, sodium malate, sodium citrate, sodium isocitrate, or α-ketoglutarate—was added to the medium instead of glucose. Samples were taken 16 h after batch culture initiation. The culture sample vials (30 ml) were rapidly plunged into an equal volume of a 60% aqueous methanol solution (−40°C). The quenched biomass was centrifuged at 5,000 × g and 4°C for 10 min. The supernatant was removed rapidly. The pellets and supernatants were frozen in liquid nitrogen and stored at −80°C until analysis. After storage, the pellets were resuspended in 1 ml of 100% methanol (−40°C) and sonicated (35% amplitude, 1 min). The pellets were frozen in liquid nitrogen and thawed again; there were three freeze-thaw cycles in total. The suspensions were then centrifuged at 13,000 × g and 4°C for 30 min. The supernatant was retained and stored at −80°C. Metabolites were analyzed by high-performance liquid chromatography (HPLC) and gas chromatography with time of flight mass spectrometry (GC-TOF-MS). Samples of the supernatant (20 μl) were analyzed using HPLC (Waters 2487 dual λ absorbance detector; Waters, Milford, MA) on an Aminex HPX-87H column (300 mm × 7.8 mm; Bio-Rad), using 0.01 N H2SO4 as the mobile phase at 30°C and a flow rate of 0.6 ml/min. HPLC chromatograms were normalized by using the area under the curve.
For analysis of metabolites by GC-TOF-MS, 800 μl of the supernatant was dried using a SpeedVac machine (Biotron, South Korea). The dried samples were methoximated with 100 μl of methoxyamine hydrochloride in pyridine (20 mg/ml) at 30°C for 90 min. Derivatization (silylation) was performed by adding 100 μl of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) containing 1% trimethylchlorosilane (TMCS), and the samples were incubated at 37°C for 30 min. An Agilent 7890A gas chromatograph coupled with a Pegasus HT TOF-MS instrument (Leco Corporation, St. Joseph, MI) was used for the analysis. The metabolites were separated on an RTx-5MS column (30 m × 0.25 mm; film thickness, 0.25 μm), and helium served as the carrier gas at a constant flow rate of 1.5 ml/min. The injector temperature was 250°C, and the transfer line and ion source temperatures were 250°C and 230°C, respectively. The initial temperature was held at 75°C for 2 min, raised to 300°C at a rate of 15°C/min, and held there for 3 min. The injection volume was 1 μl in split mode (10:1 [vol/vol]), and the mass spectra were recorded across the mass range of 45 to 1,000 m/z. Identification and semiquantification of metabolites were carried out based on ChromaTOF software (Leco). This software automatically detected all peaks and deconvoluted the mass spectra for searches in libraries, such as the NIST05 MS library and the Wiley mass spectral database. The identification of each metabolite was confirmed by comparison of retention times and mass spectra with those of authentic standard compounds, and semiquantification was represented by the relative peak area, which was automatically calculated by the software. Peak areas for intracellular TCA cycle intermediates obtained after incubation of cells in the presence of the TCA cycle intermediates (100 mM sodium salts of citrate, succinate, fumarate, α-ketoglutarate, and malate) in M9 medium were divided by those obtained after incubating the cells with 50 mM glucose in M9 medium (see Fig. S6 in the supplemental material).
Hydroxyl radical generation assay.
Precultured cells were diluted 100 times with LB broth and cultured until the number of CFU reached ∼1 × 108/ml. Each antibiotic (10 μg/ml Amp, 10 μg/ml Kan, and 0.5 μg/ml Nor) and HPF (5 μM) were administered together at 37°C and 250 rpm for 3 h. Cells were collected by centrifugation (14,500 rpm, 3 min), washed, and serially diluted in PBS. HPF signals were measured by use of a FACSCalibur flow cytometer (BD Biosciences). A total of 30,000 cells were collected for each sample. The following photomultiplier tube (PMT) voltage settings were used: E01 (forward scatter [FSC]), 360 (side scatter [SSC]), 720 (FL1), and 680 (FL3). All flow data were processed and analyzed with FCS Express 4 software (De Novo Software).
Probability of concurrence of FRD and antibiotic resistance genes.
According to the orthology data available in the Kyoto Encyclopedia of Genes and Genomes (KEGG), we analyzed the presence or absence of orthologs of FRD, ampC, and emrB across 588 bacterial species. To prevent any possible biases arising from redundant genomes, we considered only one subspecies of each species that comprised multiple subspecies. Furthermore, we excluded a few species because of suspect annotation of the genome. For a given set of genes of interest, we obtained the number of species (N) containing all the genes and compared it with the number (Nnull) expected from randomly shuffled phylogenetic profiles. Application of the central limit theorem ensured that the null distribution converged well with the Gaussian distribution, which yielded a P value indicating how frequently Nnull exceeded N.
Statistical analysis.
All experiments were performed at least in triplicate. Data are presented as means and standard deviations. Statistical significance was determined by unpaired Student's t test and unpaired Student's t test with Welch's correction. P values of <0.05 were considered significant.
RESULTS
Persistence is related to fumarate utilization.
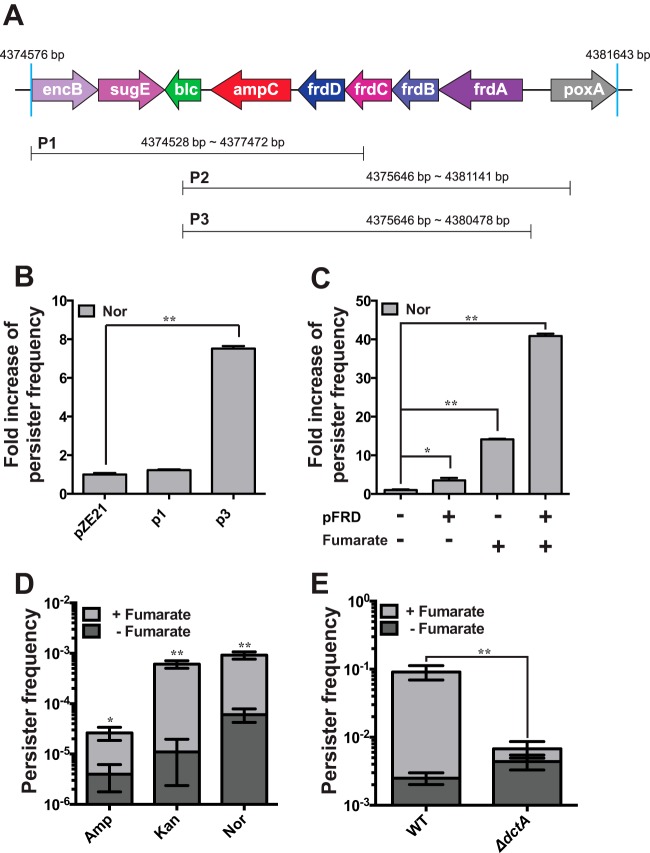
To construct a genomic library that may cover entire operons of genes, 2- to 6-kbp fragments of E. coli K-12 genomic DNA were cloned into a multicopy plasmid (28). To identify the genes enhancing persister formation, we screened the E. coli DH5α clones that were transformed with the genomic library against lethal concentrations of ampicillin (Amp). After a series of screening procedures (see Fig. S1A in the supplemental material), we isolated surviving colonies during Amp exposure and purified 19 plasmids conferring enhanced survival in the presence of Amp. All plasmids could be classified as one of three plasmids (P1, P2, and P3) and commonly contained ampC, coding for β-lactamase (Fig. 1A). Identification of ampC-containing fragments appeared to be confirmatory because Amp could work as a selection pressure. However, there was the possibility that the resistance to Amp was derived from another part of the DNA fragments, because translation of ampC mRNA is strongly attenuated in E. coli (29) and ampC itself is insufficient for conferring resistance when it is induced in multiple copy numbers (30). We found that the P3 plasmids containing both ampC and the fumarate reductase operon (frdABCD; called FRD here) could confer higher tolerance to the distinct antibiotic Nor, a DNA gyrase inhibitor, but that the P1 plasmid containing only ampC could not (Fig. 1B). When the ampC gene was deleted from a P3 plasmid, the resulting plasmid, pFRD (which contains only FRD), enhanced the tolerance of E. coli toward both Amp and Nor (see Fig. S1B and C). The Nor MIC for E. coli containing the P3 plasmid was 0.125 μg/ml and was the same as that for cells without the plasmid.
FIG 1.
Identification of FRD as a gene responsible for persistence. (A) Plasmids screened by means of the procedure shown in Fig. S1 in the supplemental material and which conferred tolerance against Amp. (B) The P3 plasmid increased the persister frequency in the presence of Nor. (C) Fumarate and FRD elevated the persister frequency synergistically. (D) Preexposure of cells to fumarate increased the frequency of persisters in the presence of all three major classes of antibiotics (Amp, Kan, and Nor). (E) Deletion of dctA (dicarboxylate and fumarate transporter) strongly attenuated the effect of fumarate preexposure on the persister frequency in the presence of Nor. WT, wild type. *, P < 0.05; **, P < 0.01. Experiments were performed in triplicate.
To identify the putative mechanism by which antibiotic tolerance was elicited by FRD, sodium fumarate, the substrate of FRD, was added to the culture medium of E. coli, i.e., Luria-Bertani (LB) broth. The cells were harvested and resuspended in LB broth containing antibiotics, and the number of CFU was counted after 9 h of incubation. The cells preexposed to fumarate before antibiotic treatment showed an ∼15-fold increase in the persister frequency in the presence of Nor (Fig. 1C), in line with a recent report that fumarate yields the strongest persister formation response during a diauxic growth switch from glucose (10). Introduction of pFRD into E. coli further enhanced the effect of preexposure to fumarate on persister formation (Fig. 1C). Moreover, the preexposure of cells to fumarate conferred multidrug tolerance on E. coli toward three distinct antibiotics (Amp, Nor, and Kan) with different modes of action (Fig. 1D) when the harvested cells were tested for their persister frequency. When a deletion mutant of a dicarboxylate transporter (ΔdctA) (31) was tested, the fumarate-mediated increase in the tolerance of antibiotics was strongly attenuated (Fig. 1E) due to impaired fumarate influx into the cell. Cell death kinetics of exponential-phase cells were biphasic (see Fig. S3A in the supplemental material), while those of stationary-phase cells were almost linear (see Fig. S3B). Taxonomic and genomic signatures associated with FRD in bacterial genomes also supported a putative role of the FRD complex in the antibiotic tolerance by showing the cooccurrence of FRD and antibiotic resistance genes, such as ampC and emrB (see Tables S3 and S4), throughout the lineages of two phyla: Actinobacteria and Proteobacteria (see Fig. S4 and S5). These observations provided evidence that intracellular fumarate and FRD are involved in persistence in the presence of antibiotics.
Persister frequency is proportional to the intracellular fumarate concentration.
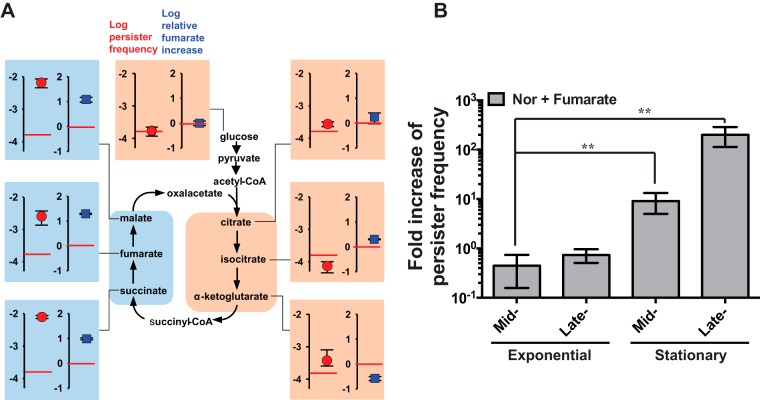
To establish the relationship between the intracellular fumarate concentration and persistence, we changed the intracellular fumarate concentration by supplementation with TCA cycle intermediates. As expected, preexposure of cells to TCA cycle intermediates had profound effects on persister frequency, in parallel with the changes in the intracellular fumarate concentration. Preexposure of cells to fumarate, succinate, or malate commonly resulted in 9- to 18-fold higher intracellular fumarate accumulations (relative to that in cells grown on glucose). In contrast, preexposure of cells to the other intermediates (citrate, isocitrate, or α-ketoglutarate) did not substantially alter the intracellular fumarate concentration (relative to that in cells grown on glucose). Preexposure of cells to one of three intermediates (citrate, isocitrate, or α-ketoglutarate) located on the right side of the TCA cycle (Fig. 2A) slightly affected the frequency of persisters in the presence of antibiotics, but fumarate, succinate, and malate—which are located on the left side of the TCA cycle (Fig. 2A)—dramatically increased the persister frequency in the presence of Nor. This supplementation assay clearly showed that the persister frequency was tightly linked to the intracellular fumarate concentration (Fig. 2A; see Fig. S6 in the supplemental material). We note that conversions of succinyl-coenzyme A (CoA)–succinate–fumarate–malate–oxaloacetate in the TCA cycle are reversible, while addition of acetyl-CoA to oxaloacetate and conversions of citrate–isocitrate–α-ketoglutarate are irreversible.
FIG 2.
Correlation between the fumarate concentration and persister frequency. (A) Colligative properties of fumarate content and persister frequency. Perturbation of the TCA cycle by preexposure of cells to one of its intermediates differentially altered the intracellular fumarate concentration (squares) and the persister frequency (circles). The fumarate content of cells preexposed to each intermediate was compared to that of cells grown on glucose (red horizontal lines). The persister frequency of the cells grown on glucose is shown as a reference (red horizontal lines). Cells preexposed to each intermediate were harvested, resuspended in fresh LB broth containing Nor, and incubated for 9 h before counting of CFU on an LB plate. The cell growth profiles in LB broth during preexposure to TCA cycle intermediates are shown in Fig. S2A in the supplemental material. When preexposed cells were resuspended in M9 broth containing Nor, fumarate and succinate increased the persister frequency, while citrate did not, similar to the results for LB broth (see Fig. S2B), but the degrees of persister frequency elevation were smaller than those in LB broth. When fumarate, succinate, and citrate were used at 6.75 mM to supplement the culture with Nor, they similarly affect the persister frequency (see Fig. S2C). (B) Growth-stage-dependent effects of fumarate on persister frequency. The persister frequency obtained in the presence of 100 mM fumarate was divided by that without fumarate. *, P < 0.05; **, P < 0.01. Experiments were performed in triplicate.
The persister frequency of stationary-phase cells is known to be 10- to 1,000-fold higher than that of exponential-phase cells (1, 2). The enhanced persister formation under the influence of fumarate preexposure varied depending on the growth stage of the cells (Fig. 2B). When the bacterial cells were incubated with antibiotics during the exponential growth phase, the effect of fumarate preexposure was weak. In contrast, stationary-phase cells responded strongly to fumarate preexposure. These variations suggested that the fumarate-mediated persistence may require growth-phase-dependent regulation. According to gene expression profiling studies of E. coli under various culture conditions, FRD was upregulated in the stationary phase (see Fig. S7 in the supplemental material). Fumarate was one of the major metabolites that was accumulated during the switch from the growth stage to the stationary phase (see Fig. S7). Thus, the intracellular fumarate concentration was also linked to the growth-stage-related change in persister frequency.
A paired FRD/SDH system underlies persistence.
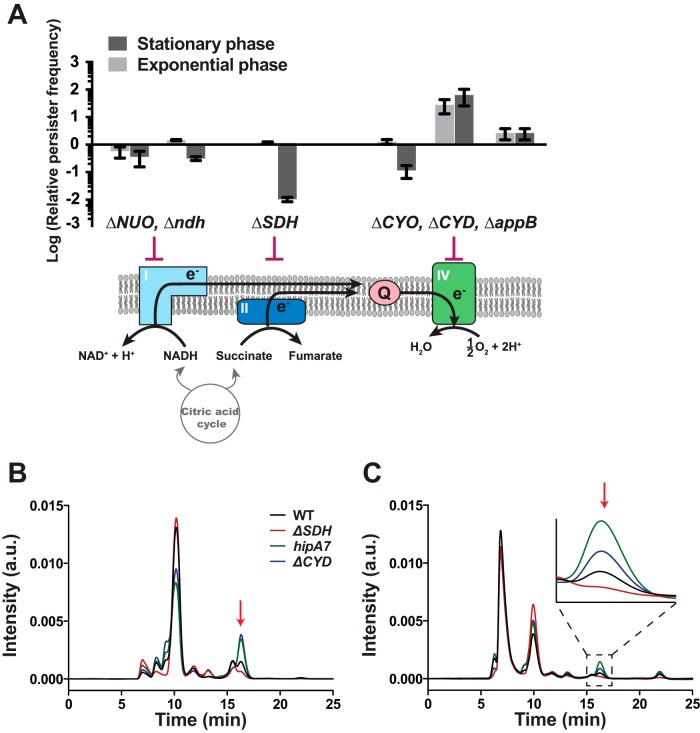
Aside from the identification of FRD as a putative persistence mediator, we examined the effects of deletion of genes involved in the electron transport chain (ETC) on persister frequency due to the evoked role of the ETC in antibiotic-affected bacterial cells (26, 32, 33). Each step of the ETC was disrupted by a gene knockout, and the resulting persister frequency was measured. Deletions affecting complex I (ΔNUO or Δndh) did not substantially affect the persister frequency, regardless of the growth phase (Fig. 3A). The deletion of a gene in complex II (succinate dehydrogenase complex; SDH) reduced the persister frequency only in stationary-phase cells, whereas deletions of complex IV genes mostly increased the persister frequency (Fig. 3A).
FIG 3.
Effects of perturbations of the electron transport chain (ETC) on persister frequency. (A) Operons (in uppercase letters) or specific genes (in lowercase letters) that participate in the ETC were knocked out, and the persister frequency of each knockout mutant was measured in the presence of 5 μg/ml Nor. The persister frequency of each deletion mutant was divided by that of the WT strain to obtain the relative value. Abbreviations: NUO, NADH dehydrogenase I (nuoABCDEFGHIJKLMN); ndh, NADH dehydrogenase II (Ndh-2); SDH, succinate dehydrogenase (sdhCDAB); CYO, cytochrome bo oxidase (cyoABCD); CYD, cytochrome bd-I oxidase (cydAB); appB, cytochrome bd-II oxidase (appBC); Q, ubiquinone. (B) Comparison of intracellular fumarate levels among the WT, highly persistent mutants (hipA7 and ΔCYD), and a less persistent mutant (ΔSDH). a.u., arbitrary units. (C) Comparison of extracellular fumarate levels among the mutants. The arrows indicate a fumarate peak.
The ETC deletion mutants were then examined to determine their intracellular fumarate concentrations. We used two ETC mutants: the ΔCYD mutant for highly persistent disruption of the ETC and the ΔSDH mutant for less persistent disruption. We also determined the intracellular fumarate concentration of the hipA7 mutant, the first highly persistent mutant identified, which has long been accepted as the gold standard of a persister strain; this mutant exhibits persister phenotypes mediated by a toxin-antitoxin module (34). Both highly persistent mutants (ΔCYD and hipA7 strains) contained ∼3-fold larger amounts of fumarate than did the wild-type (WT) cells, whereas the intracellular fumarate concentration of the ΔSDH strain was substantially lower than that of the WT (∼30% of the WT level) (Fig. 3B). Extracellular levels of fumarate changed in the same way as the intracellular concentrations (Fig. 3C).
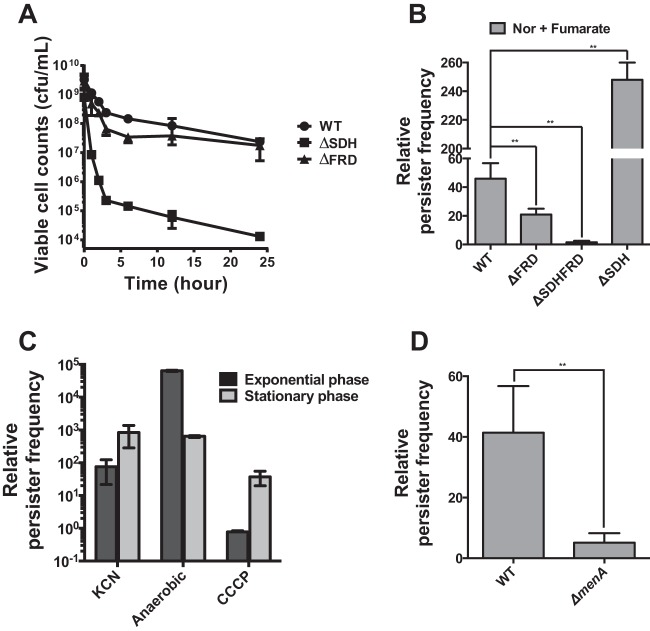
The Nor-induced death rate for the ΔSDH strain in the stationary phase was a few orders of magnitude higher than that for WT cells (Fig. 4A; see Fig. S3 in the supplemental material). Deletion of SDH reduced the decimal reduction time for Nor-treated stationary-phase cells to 3 h, from 19 h for the WT strain (see Fig. S3B). The roles of FRD and SDH in persistence were sometimes complementary in our experiments. The ΔFRD strain in the stationary phase did not show a reduction of persistence in comparison with the WT strain (Fig. 4A); however, overexpression of FRD and SDH by use of a strong arabinose-inducible promoter increased the persister frequency (Fig. 1; see Fig. S8). This phenomenon may happen when SDH can compensate for the absence of FRD, and it is known that both FRD and SDH can reduce fumarate and oxidize succinate, with a preference for one substrate (35, 36). To better understand the role of the paired SDH/FRD system in persistence, a fumarate supplementation assay was again performed by using the knockout strains (Fig. 4B). The ΔFRD strain responded to fumarate supplementation weakly in comparison with the WT strain, whereas the lack of both enzymes (ΔSDH ΔFRD) almost completely abrogated the fumarate-mediated increase in persister frequency (Fig. 4B). In contrast, the ΔSDH strain, which did not accumulate fumarate but retained FRD, showed the strongest increase in fumarate-mediated persistence. These results suggested that SDH contributed to persister formation by producing fumarate, whereas FRD used fumarate to drive the persistence mechanism. The lack of FRD was complemented by SDH during operation of the fumarate-mediated persistence mechanism, whereas SDH was indispensable for persister formation.
FIG 4.
FRD and SDH as a paired-enzyme system underlying persistence. (A) Cell death kinetics of E. coli mutants lacking either SDH or FRD in the presence of 5 μg/ml Nor. Stationary-phase cells were used. (B) Effects of SDH and/or FRD deletion on the fumarate-mediated change in persister frequency in the presence of 5 μg/ml Nor. Stationary-phase cells were used. (C) Increased persister frequency under anaerobic conditions. The persister frequency obtained under each condition was divided by that under aerobic conditions in the presence of 5 μg/ml Nor. KCN and CCCP are an inhibitor of complex IV and an uncoupler of the electron transport chain, respectively. Anaerobic conditions were created as described in Materials and Methods. KCN and CCCP, at 1 mM and 20 μM, respectively, were provided with Nor under aerobic conditions. (D) Deletion of menA (encoding a menaquinone biosynthetic enzyme) strongly attenuated the effect of fumarate preexposure on the persister frequency in the presence of 5 μg/ml Nor. Stationary-phase cells were used. *, P < 0.05; **, P < 0.01. Experiments were performed in triplicate.
DISCUSSION
We observed that increased fumarate content elevated the persister frequency. Preexposure of cells to TCA cycle intermediates (Fig. 1 and 2) and perturbations of the ETC (Fig. 3) indicated that the persister frequency might change proportionally to changes in the intracellular fumarate concentration. Furthermore, growth-stage-dependent changes in persister frequency (Fig. 2B) and hipA7-mediated persistence (Fig. 3B and C), which apparently are not linked directly to the fumarate concentration, pointed to the relationship between persister frequency and the intracellular fumarate concentration.
Our experiments simultaneously identified fumarate as a common mediator of bacterial persistence, as a substrate of FRD (Fig. 1) and as the product of SDH (Fig. 3). Fumarate is an intermediate of the TCA cycle and is produced via succinate oxidation, but there is evidence that antibiotic-induced cell death is not simply affected by the overall activity of the TCA cycle (37, 38). Because the cells were tolerant of antibiotics when their respiration was blocked either by KCN (an inhibitor of complex IV of the ETC) or by anaerobic incubation (Fig. 4C), in agreement with other studies (32, 33, 39, 40), we speculate that E. coli persisters survive in the presence of antibiotics by using fumarate as the terminal electron acceptor, as in the case of anaerobic respiration. SDH and FRD represent a paired system involved in the anaerobic respiration of E. coli (35, 41), and fumarate is the terminal electron acceptor in the anaerobic respiration pathway of E. coli. Furthermore, the ΔmenA strain, which lacks a menaquinone biosynthetic enzyme (42, 43), showed a fumarate-mediated increase of persister frequency that was much smaller than that of the WT strain (Fig. 4D). This result supports the role of fumarate as the electron acceptor in the persistence mechanism considering that fumarate is reduced by FRD with electrons from menaquinone during anaerobic respiration. Accordingly, it is likely that SDH contributes to persistence by producing fumarate, whereas FRD drives the anaerobic respiration-like persistence mechanism by using the accumulated fumarate as a terminal electron acceptor. Our results also suggest that the formation of persisters may be prevented efficiently by blocking the intracellular accumulation of fumarate.
Additionally, we tested whether other electron acceptors, such as nitrate, dimethyl sulfoxide (DMSO), and trimethylamine N-oxide (TMAO), could affect the persister frequency as fumarate did (see Fig. S9B in the supplemental material), but none of them did so. When the terminal oxidoreductases of these alternative electron acceptors were deleted, none of the deletion mutations reduced the persister frequency to the extent that the SDH deletion did (see Fig. S9A and C). Fumarate is not only an anaerobic terminal electron acceptor but also an energy source, while DMSO, TMAO, and nitrate are not. We speculate that E. coli persisters use fumarate among alternative electron acceptors, perhaps because the cells can make their own supply.
Fumarate accumulated inside the cell might have induced persister formation in several ways. First, a ppGpp-dependent mechanism might have elicited persister formation during carbon source transition (10). Diauxic growth in glucose-fumarate strongly elicits a persister formation response, and the increase in persisters is a result of carbon source transition rather than slower growth on fumarate (10, 44). Because we used LB broth for cell culture, there is a possibility that carbon sources enriched in LB broth were used as the first carbon source and that the intracellularly accumulated fumarate was used as a second carbon source. This sequential carbon source utilization might have resulted in a carbon source transition-like effect on cells, leading to ppGpp-dependent persister elicitation. However, because of the strong relationship between intracellular fumarate content and persister frequency, it is tempting to speculate that fumarate did not merely play a role as a second carbon source during persister formation but actively participated in the persistence mechanism. Second, accumulated fumarate inside cells might have affected the electron transport chain, leading to a decreased proton motive force (PMF). PMF is required for aminoglycoside uptake (45), and certain metabolites can elevate PMF, facilitating the uptake of aminoglycosides and persister death (38). Amino acid biosynthesis and motility may play important roles in tolerance toward aminoglycosides in the stationary growth phase, through alteration of central metabolism and PMF (46), but fumarate-mediated persistence is not well explained by PMF because fumarate increased the persister frequency against all 3 kinds of antibiotics. Third, intracellular fumarate might have reduced the generation of hydroxyl radicals (·OH). Hydroxyl radical generation has been suspected to be a common mechanism of cell death induced by antibiotics for years (26, 32, 33). Fluorescence-activated cell sorting (FACS) analysis using HPF indicated that ·OH generation was reduced in the absence of O2, in the presence of the complex IV inhibitor KCN, and in the absence of the complex IV protein CYD, suggesting that ·OH is generated during electron transport to oxygen (see Fig. S10 in the supplemental material). Even though the cells were grown under aerobic conditions, fumarate preexposure efficiently reduced ·OH generation as if the cells were not respiring oxygen (see Fig. S10). The hipA7 mutant also showed reduced ·OH formation. Thus, it is also probable that persisters avoid ·OH generation by detouring electrons from the ETC to fumarate as if the cells are respiring anaerobically. Interpretation of FACS results should be cautious, though, as the role of reactive oxygen species (ROS) in antibiotic-mediated bacterial killing and the use of HPF in measuring ·OH have recently become matters of debate (47–50). Nevertheless, we have established a relationship between fumarate-mediated persisters and ·OH generation, because substantial amounts of evidence still support the use of HPF and the role of ·OH in antibiotic-induced cell death (26, 32, 33, 40, 51).
Supplementary Material
ACKNOWLEDGMENT
We thank Chris Rao for comments on the manuscript.
Funding Statement
This work was supported by the Basic Science Research Program through a National Research Foundation of Korea grant funded by the Ministry of Science, ICT & Future Planning (2009-0083540 and NRF-2013R1A1A2010775) to D.-H.K.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01794-15.
REFERENCES
- 1.Grant SS, Hung DT. 2013. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4:273–283. doi: 10.4161/viru.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 3.Kim JS, Heo P, Yang TJ, Lee KS, Cho DH, Kim BT, Suh JH, Lim HJ, Shin D, Kim SK, Kweon DH. 2011. Selective killing of bacterial persisters by a single chemical compound without affecting normal antibiotic-sensitive cells. Antimicrob Agents Chemother 55:5380–5383. doi: 10.1128/AAC.00708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230:13–18. doi: 10.1016/S0378-1097(03)00856-5. [DOI] [PubMed] [Google Scholar]
- 5.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet ii:497–500. [Google Scholar]
- 6.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin BR. 2004. Microbiology. Noninherited resistance to antibiotics. Science 305:1578–1579. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes K, Maisonneuve E. 2012. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol 66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- 10.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 11.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y. 2014. Persisters, persistent infections and the Yin-Yang model. Emerg Microbes Infect 3:e3. doi: 10.1038/emi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat Rev Microbiol 9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- 14.Luidalepp H, Joers A, Kaldalu N, Tenson T. 2011. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. J Bacteriol 193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 16.Orman MA, Brynildsen MP. 2013. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother 57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. 2013. Dynamic persistence of antibiotic-stressed mycobacteria. Science 339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 18.Moyed HS, Broderick SH. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 166:399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korch SB, Henderson TA, Hill TM. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol 50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- 20.Spoering AL, Vulic M, Lewis K. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J Bacteriol 188:5136–5144. doi: 10.1128/JB.00369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu YM, Coates ARM. 2005. Transposon mutagenesis identifies genes which control antimicrobial drug tolerance in stationary-phase Escherichia coli. FEMS Microbiol Lett 243:117–124. doi: 10.1016/j.femsle.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 22.Ma C, Sim S, Shi W, Du L, Xing D, Zhang Y. 2010. Energy production genes sucB and ubiF are involved in persister survival and tolerance to multiple antibiotics and stresses in Escherichia coli. FEMS Microbiol Lett 303:33–40. doi: 10.1111/j.1574-6968.2009.01857.x. [DOI] [PubMed] [Google Scholar]
- 23.Hansen S, Lewis K, Vulic M. 2008. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother 52:2718–2726. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 27.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol 2007:Unit 1.17. doi: 10.1002/0471142727.mb0117s79. [DOI] [PubMed] [Google Scholar]
- 28.Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaurin B, Grundstrom T, Edlund T, Normark S. 1981. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature 290:221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- 30.Soo VW, Hanson-Manful P, Patrick WM. 2011. Artificial gene amplification reveals an abundance of promiscuous resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A 108:1484–1489. doi: 10.1073/pnas.1012108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies SJ, Golby P, Omrani D, Broad SA, Harrington VL, Guest JR, Kelly DJ, Andrews SC. 1999. Inactivation and regulation of the aerobic C(4)-dicarboxylate transport (dctA) gene of Escherichia coli. J Bacteriol 181:5624–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. 2012. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci U S A 109:12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu YL, Liu XH, Qu YL, Wang XH, Li LP, Zhao XL. 2012. Inhibitors of reactive oxygen species accumulation delay and/or reduce the lethality of several antistaphylococcal agents. Antimicrob Agents Chemother 56:6048–6050. doi: 10.1128/AAC.00754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moyed HS, Bertrand KP. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320:217–234. doi: 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 36.Cecchini G, Schroder I, Gunsalus RP, Maklashina E. 2002. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta 1553:140–157. doi: 10.1016/S0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 37.Prax M, Bertram R. 2014. Metabolic aspects of bacterial persisters. Front Cell Infect Microbiol 4:148. doi: 10.3389/fcimb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allison KR, Brynildsen MP, Collins JJ. 2011. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verklin RM Jr, Mandell GL. 1977. Alteration of effectiveness of antibiotics by anaerobiosis. J Lab Clin Med 89:65–71. [PubMed] [Google Scholar]
- 40.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CTY, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iverson TM, Luna-Chavez C, Cecchini G, Rees DC. 1999. Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284:1961–1966. doi: 10.1126/science.284.5422.1961. [DOI] [PubMed] [Google Scholar]
- 42.Korshunov S, Imlay JA. 2006. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol 188:6326–6334. doi: 10.1128/JB.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suvarna K, Stevenson D, Meganathan R, Hudspeth ME. 1998. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menA gene from Escherichia coli. J Bacteriol 180:2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amato SM, Brynildsen MP. 2015. Persister heterogeneity arising from a single metabolic stress. Curr Biol 25:2090–2098. doi: 10.1016/j.cub.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 45.Taber HW, Mueller JP, Miller PF, Arrow AS. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51:439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. 2015. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6:e00078-15. doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulander W, Wang Y, Folkesson A, Charbon G, Lobner-Olesen A, Ingmer H. 2014. Bactericidal antibiotics increase hydroxyphenyl fluorescein signal by altering cell morphology. PLoS One 9:e92231. doi: 10.1371/journal.pone.0092231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu YY, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 50.Renggli S, Keck W, Jenal U, Ritz D. 2013. Role of autofluorescence in flow cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J Bacteriol 195:4067–4073. doi: 10.1128/JB.00393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Halfawy OM, Valvano MA. 2014. Putrescine reduces antibiotic-induced oxidative stress as a mechanism of modulation of antibiotic resistance in Burkholderia cenocepacia. Antimicrob Agents Chemother 58:4162–4171. doi: 10.1128/AAC.02649-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.