Abstract
The human immunodeficiency virus type 1 (HIV-1) capsid plays crucial roles in HIV-1 replication and thus represents an excellent drug target. We developed a high-throughput screening method based on a time-resolved fluorescence resonance energy transfer (HTS-TR-FRET) assay, using the C-terminal domain (CTD) of HIV-1 capsid to identify inhibitors of capsid dimerization. This assay was used to screen a library of pharmacologically active compounds, composed of 1,280 in vivo-active drugs, and identified ebselen [2-phenyl-1,2-benzisoselenazol-3(2H)-one], an organoselenium compound, as an inhibitor of HIV-1 capsid CTD dimerization. Nuclear magnetic resonance (NMR) spectroscopic analysis confirmed the direct interaction of ebselen with the HIV-1 capsid CTD and dimer dissociation when ebselen is in 2-fold molar excess. Electrospray ionization mass spectrometry revealed that ebselen covalently binds the HIV-1 capsid CTD, likely via a selenylsulfide linkage with Cys198 and Cys218. This compound presents anti-HIV activity in single and multiple rounds of infection in permissive cell lines as well as in primary peripheral blood mononuclear cells. Ebselen inhibits early viral postentry events of the HIV-1 life cycle by impairing the incoming capsid uncoating process. This compound also blocks infection of other retroviruses, such as Moloney murine leukemia virus and simian immunodeficiency virus, but displays no inhibitory activity against hepatitis C and influenza viruses. This study reports the use of TR-FRET screening to successfully identify a novel capsid inhibitor, ebselen, validating HIV-1 capsid as a promising target for drug development.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) capsid plays essential roles in the viral life cycle; e.g., the precise uncoating process (or disassembly) of capsid is tightly associated with reverse transcription (RT) (1), and capsid assembly and maturation are essential for viral particle integrity and infectivity (2). The mature HIV-1 conical core consists of 1,500 capsid (CA, or p24) monomers assembled into 250 hexamers and 12 pentamers. Antiretroviral therapy (ART) currently administered for HIV treatment does not directly target the uncoating or assembly of the viral capsid protein; instead, it consists of a cocktail of at least three drugs that include a combination of nonnucleoside reverse transcriptase inhibitors (NNRTIs), nucleoside reverse transcriptase inhibitors (NRTIs), protease inhibitors (PIs), integrase inhibitors (INIs), fusion inhibitors (FIs), and entry inhibitors. Given the importance of the capsid for HIV-1 replication, it represents an attractive target for drug development.
HIV-1 capsid originates from a 55-kDa Gag precursor proteolyzed into three folded proteins (matrix [MA], capsid [CA], and nucleocapsid [NC]) and 3 small peptides (spacer peptides 1 and 2 [SP1 and SP2, respectively] and p6). The Gag polyprotein plays an important role in cell membrane binding and in Gag-Gag lattice interaction in immature virions. Upon budding and release of the immature viral particles from the host cell, HIV maturation is initiated by proteolysis of Gag by the viral protease, resulting in the assembly of mature fullerene-like conical capsids. Capsid is critical for both early and late events in the HIV-1 life cycle. Early on, the fusion of the virus with target cell membranes triggers disassembly or uncoating of CA, which promotes completion of reverse transcription and synthesis of viral cDNA (1, 3). Even though viral uncoating is not completely understood, the capsid's stability and integrity during the early stages of infection are essential for efficient reverse transcription and infectivity. Mutations in CA that alter its stability compromise HIV-1 uncoating and infection (4, 5). Furthermore, two cellular restriction factors target the viral capsid during the uncoating process. Tripartite motif 5-alpha (TRIM5α) promotes rapid and premature disassembly of viral capsids, abrogating productive reverse transcription (6, 7), while the interferon-inducible MxB protein prevents uncoating by stabilizing the HIV-1 core during infection, limiting integration of viral DNA (8, 9). During the late stages of the viral life cycle, capsid assembly and maturation are essential for the formation of infectious viral particles, and mutations in capsid are detrimental for HIV-1 assembly and particle release (10, 11).
The CA protein consists of an independently folded N-terminal domain (NTD; residues 1 to 145) and C-terminal domain (CTD; residues 151 to 231) connected by a 5-residue flexible linker. Structures of the full-length capsid, NTD, and CTD have been studied by crystallography, cryo-electron microscopy, and nuclear magnetic resonance (NMR) (12–16). The CA-NTD is composed of seven α-helices (CA helices 1 to 7), while the smaller CA-CTD is composed of a short 310-helix followed by an extended strand and four α-helices (CA helices 8 to 11). In solution, CA dimerization (dissociation constant [Kd] of 18 μM) is mainly dependent on Trp184 and Met185 residues in helix 9 of the CTD (13). Mutation of these residues interferes with CA assembly in vitro and abolishes viral infectivity (15, 17). This interface is therefore required for efficient assembly of both the mature and immature capsid lattice.
The HIV capsid protein is thus emerging as an important target for development of small-molecule compounds (reviewed in reference 18). Unfortunately, so far, none of the most promising candidates is used in the clinic. Bevirimat, which blocks HIV-1 replication by interfering with the Gag CA-SP1 cleavage site, was tested in clinical trials (19), but a high-baseline drug resistance was observed due to natural viral polymorphisms (20).
Here, we describe a time resolved-fluorescence energy transfer high-throughput screening (HTS-TR-FRET) assay to identify inhibitors of CTD capsid dimerization. A library of pharmacologically active compounds (LOPAC) composed of 1,280 compounds was screened, and ebselen was identified as an HIV-1 inhibitor. Ebselen was shown to directly bind the CTD and to inhibit early events of the HIV-1 life cycle. Our efforts not only resulted in the identification of a novel class of compounds with antiretroviral activity but also validated the use of HTS-TR-FRET for the identification of inhibitors of HIV-1 capsid dimerization, thereby confirming HIV-1 capsid as a promising antiviral target.
MATERIALS AND METHODS
Compounds: LOPAC library, ebselen and analogs of ebselen, and cysteine binding compounds.
A library of pharmacologically active compounds (LOPAC) containing 1,280 compounds representing all major target classes was purchased from Sigma-Aldrich (St. Louis, MO). The compounds were screened at a final concentration of 10 μM. Ebselen was purchased from Adipogen Corporation (catalog number AG-CR1-0031), and ebselen oxide was from Cayman Chemical (catalog number 10012298). Ebselen analogs with the phenyl ring converted to pyridine (2pyr- and 3pyr-ebselen) were synthesized at Wroclaw University of Technology, Poland, with purity and stability assessed at St. John's University, NY. Other ebselen analogs with selenium replaced by sulfur (S-ebselen), oxygen (O-ebselen), and carbon (C-ebselen) were provided by Barbara Slusher, Johns Hopkins University School of Medicine, Baltimore, MD. The cysteine binding compounds, omeprazole (AAJ62860-03; Afla-Aesar), methyl-3,4-dephostatin (M9440; Sigma), and 3-bromo-1,1,1-trifluoroacetone (BTFA) (18545; Sigma) were purchased.
TR-FRET.
A time-resolved fluorescence resonance energy transfer (TR-FRET) assay was used to monitor CTD dimerization and to screen for inhibitors of this interaction. This assay is based on the transfer of energy between two fluorophores, a donor and an acceptor labeled with europium cryptate (EU) and allophycocyanin (XL665), respectively (Cisbio Bioassays). The proteins, as well as the antibody-tagged fluorophores, were diluted in assay buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol, 0.05% Triton X-100, and 100 mM potassium fluoride). In a 384-well plate (total volume = 20 μl), glutathione S-transferase (GST)-CTD and CTD-Flag (200 nM each) were mixed together with EU-conjugated anti-GST antibody (1.8 ng/well) (catalog number 61GSTKLB; Cisbio) and XL665-conjugated anti-Flag antibody (20 ng/well) (61FG2XLB; Cisbio) and incubated overnight at room temperature. Interaction between the two proteins and illumination with a 320-nm beam resulted in transfer of fluorescence energy and emission at 665 nm. Untagged CTD was used as a control of inhibition of GST-CTD/CTD-Flag dimerization.
For HTS, the assay was further miniaturized to a 1,536-well plate format. All concentrations were retained, yet the volume was scaled down to 10 μl total. The proteins and the test compounds (10 μM) were mixed together and incubated for 20 min at room temperature; then the two antibody-tagged fluorophores were added. The mixture was incubated for 5 h at room temperature before the emission was read at 665 nm. The TR-FRET signal was calculated as a ratio of emission: (665 nm/620 nm) × 10,000. Hepatitis C virus (HCV) core protein was used as a counterscreen (23). All plates were analyzed and passed quality control (QC) if their Z′ score was greater than 0.5. All data were archived into the Scripps drug discovery database, and activity was determined based on that of high and low controls. The inhibitory activities of ebselen and its analogs were further evaluated by TR-FRET in a 384-well plate format at a 10 μM final concentration. The results represent the mean of at least three independent experiments with triplicate data points per experiment.
Multiple-round infection.
HeLa-CD4-LTR-LacZ (where LTR is long terminal repeat) cells were infected with NL4.3 replicative virus in the presence of increasing concentrations of ebselen for 72 h. HIV-1 replication was assessed by measuring the β-galactosidase (β-Gal) production in cells by chlorophenol red-β-d-galactopyranoside (CPRG) assay or by quantifying the viral capsid in the supernatant by p24 enzyme-linked immunosorbent assay (ELISA).
Quantification of early and late reverse transcription product and provirus integration.
HeLa-CD4 cells were plated at 2 × 105 cells per well in a six-well plate. Twenty-four hours later, cells were infected with vesicular stomatitis virus G protein (VSV-G)-NL4.3 pseudotyped viruses in the presence of ebselen, efavirenz, raltegravir, lopinavir, or dimethyl sulfoxide (DMSO). Virus that was heat inactivated for 10 min at 95°C was used as a control. At 18 h postinfection, genomic DNAs (gDNA) were prepared using a DNeasy blood and tissue kit (Qiagen), according to the manufacturer's protocol, and subjected to quantitative PCR (qPCR) using Sensifast Sybr mix (Bioline). The early viral DNA products were amplified using the primer pair 5′-GGC TAA CTA GGG AAC CCA CTG-3′ (P1) and 5′-CTG CTA GAG ATT TTC CAC ACT GAC-3′ (P2), and the late viral DNA products were amplified using the primer pair 5′-TGT GTG CCC GTC TGT TGT G-3′ (P3) and 5′-GAG TCC TGC GTC GAG G-3′ (P4) (9). The number of integrated proviruses was quantified by Alu-Gag PCR, followed by a nested qPCR (50). The Alu-PCR was run with the following conditions: initial denaturation of 2 min at 94°C, then 20 cycles of amplification of 20 s at 94°C, 10 s at 50°C, and 3.5 min at 65°C, and a final elongation step of 7 min at 65°C. Primers sequences were as follow: for Alu, 5′-TCC CAG CTA CTG GGG AGG CTG AGG-3′; for Gag, 5′-CCT GTG TCA GCT GCT TG-3′. Nested qPCR was done with primers P1 and P2. The qPCR amplification was run as follows: 3 min at 95°C and then 40 cycles of 5 s at 95°C, 20 s at 60°C, and 8 s at 68°C. Results correspond to the mean of three independent experiments with triplicate (RT products) or duplicate (Alu-qPCR) data points per experiment.
Fate of capsid in cells.
HeLa-CD4 cells were plated at 2 × 106 cells in a 10-cm plate. Twenty-four hours later, cells were infected with NL4.3-eGFP (where eGFP is enhanced green fluorescent protein) virus for 30 min at 4°C to allow viral attachment to cells and then moved to 37°C for 16 h in the presence of 20 μM ebselen, 6 μM PF74, or DMSO. Cells were washed three times with ice-cold phosphate-buffered saline (PBS) and then detached with 1 ml of Pronase (7.0 mg/ml prepared in Dulbecco's modified Eagle's medium [DMEM]) for 5 min at room temperature. Cell pellets were washed three times in PBS and resuspended in 2.5 ml of hypotonic buffer (10 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM EDTA, and protease inhibitors) for 15 min on ice. Cells were lysed with 15 strokes in a 7-ml Dounce homogenizer with pestle B, and cellular debris was cleared by centrifugation for 3 min at 2,000 × g. Cleared lysate (100 μl) was harvested and used as input for HIV-1 p24 assay. Two milliliters of cleared lysate was ultracentrifuged onto a 50% (wt/vol, in PBS) sucrose cushion at 125,000 × g for 2 h at 4°C (SW 41 Beckman rotor). The top-most part (100 μl) of the supernatant containing soluble CA was harvested. The pellet, containing fullerene CA, was resuspended in 100 μl of SDS sample buffer. The input, supernatant, and pellet fractions were run on SDS-PAGE gels followed by Western blotting with anti-p24 antibody. The bands corresponding to input and pellet were quantified, and the ratio of the value for the pellet fraction to that of the control was calculated. The error bars are representative of two independent experiments, normalized to the values of the DMSO control.
In vitro CA assembly assay.
The effect of ebselen on CA assembly was measured by monitoring the turbidity at 350 nm, as previously described (51). Briefly, 75 μl of NaCl solution (2 ml of 5 M NaCl mixed with 1 ml of 200 mM sodium phosphate, pH 8) containing increasing concentrations of ebselen, PF74, raltegravir (used as a negative control), or DMSO was placed into a 96-well plate. To initiate the assembly reaction, 25 μl of purified p24-His protein (100 μM; 25 μM final concentration) was added. Turbidity was monitored every 30 s for 20 min.
NMR.
Nuclear magnetic resonance (NMR) data were collected on a 700-MHz Bruker NMR instrument equipped with a quadruple resonance inverse (QCI) cryo-probe. The CA-CTD and the capsid mutant CA-CTD W184A/M185A (WAMA) were prepared in 25 mM phosphate (pH 6.5), 100 mM NaCl, 0.02% NaN3, and 10% D2O, with or without 5 mM dithiothreitol (DTT) to a final concentration of 1 mM and 330 μM, respectively. Two-dimensional (2D) [1H,15N] heteronuclear single-quantum coherence (HSQC) spectra were acquired at 298 K, with or without a 1.2-fold or 2.4-fold molar excess of ebselen prepared as a 50 or 100 mM stock in hexadeutero-DMSO (DMSO-d6). All samples contained the same amount of DMSO-d6 (2%). Chemical shifts were assigned using previously published CA-CTD (24) and WAMA HSQC peak assignments (25). Data were processed using Bruker Topspin, version 3.0, and analyzed with NMRViewJ (OneMoon Scientific, Inc.).
Tryptophan fluorescence assay.
CA-CTD and a negative-control protein, lysozyme, were prepared at 5 μM and 2.5 μM, respectively, in the same buffer as for NMR. Similarly, l-Trp was prepared at 25 μM. Serial dilutions (1:1) of ebselen and a negative-control ligand, raltegravir, were prepared in DMSO and added to the protein or l-Trp samples to final concentrations ranging from 25 nM to 50 μM. Samples were plated in duplicate at 12 concentration points in a 96-well black quartz microplate (Hellma) and read using a SpectraMax M5e at excitation and emission wavelengths of 280 nm and 335 nm, respectively. The reversible effect of ebselen to cysteine was investigated by postincubation with 5 mM DTT for 30 min at room temperature. The effect of the ligand on l-Trp fluorescence is subtracted from the ligand quenching to the intrinsic Trp fluorescence of CA-CTD or lysozyme and converted to percentage with respect to the fluorescence of vehicle control samples. Data were analyzed in GraphPad Prism.
CD.
CA-CTD (14 μM) was buffer exchanged in 10 mM potassium phosphate–100 mM KF, pH 6.6. For ligand-bound samples, ebselen (in DMSO) was added at a 2.4:1 molar ratio with respect to the protein, and after a 1-h incubation, was buffer exchanged anew to remove traces of DMSO. Experiments were performed in triplicate for each sample in a circular dichroism (CD) quartz cuvette (Hellma) with a path length of 1 mm. Wavelength scans from 190 to 250 nm at 1-nm increments were acquired at 25°C in a JASCO J-815 CD spectrometer.
Liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS).
The molecular mass of CA-CTD was measured on a LTQ XL linear ion trap mass spectrometer (Thermo-Fisher Scientific) connected to a liquid chromatography system. The CA-CTD was reacted with different molar ratios of ebselen and subsequently desalted and diluted to 4 μM at room temperature. The reversibility of ebselen modification was investigated by postincubation treatment with 5 mM DTT for 30 min at room temperature. The resulting protein was acidified using 0.1% formic acid (FA) and separated onto a C8 column employing a 0% to 100% acetonitrile gradient. The mass spectra were acquired in the positive-ion mode. Mass-to-charge ratios were extracted from the raw data, deconvoluted, and deisotoped with the MaqTran software.
For more details, see the supplemental material.
RESULTS AND DISCUSSION
The CTD of CA is responsible for capsid dimerization, and Trp184 and Met185 residues in helix 9 of the CTD mediate this interaction (13). A peptide mimicking the helix 9 sequence, CAC-1, was shown to inhibit CTD dimerization (21), and a stabilized version of this peptide, NYAD-201, inhibited mature-like virus particle formation (22). These results suggested that targeting CTD dimerization would be a good strategy to identify HIV inhibitors, and we initiated the development of an HTS-TR-FRET assay using the CTD of CA tagged with either GST (GST-CTD) or Flag (CTD-Flag) (Fig. 1A). The proteins were purified, and their identity and homogeneity were verified by SDS-PAGE, followed by Coomassie blue staining and Western blotting, revealing major expected bands at 37 kDa for GST-CTD, 12 kDa for CTD-Flag, and 11 kDa for untagged CTD (see Fig. S1A and B in the supplemental material).
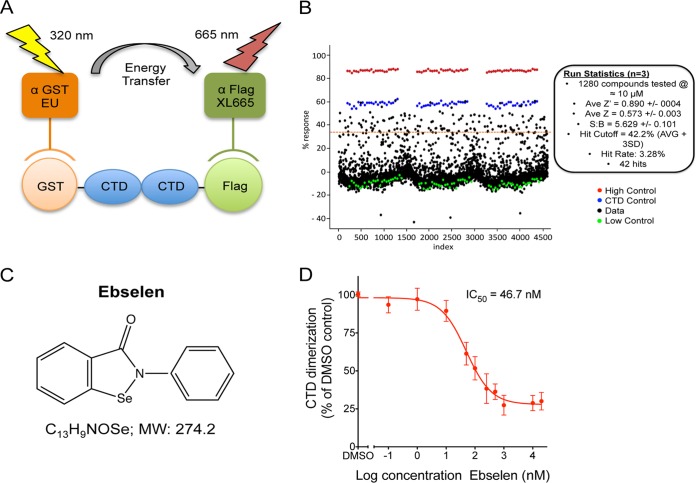
FIG 1.
LOPAC library screen. (A) Schematic representation of the TR-FRET assay. (B) Primary results screen. Each black dot represents a single data point, red dots correspond to GST-CTD alone and represent the “no CTD dimerization” baseline, green dots correspond to GST-CTD and CTD-Flag interaction and represent the “100% dimerization” baseline, and blue dots correspond to CTD dimerization inhibited by 3 μM untagged CTD, which results in 75% inhibition of CTD dimerization. The HTS assay was performed in triplicate. Statistics of the screen are indicated. Ave/AVG, average; SD, standard deviation. (C) Structure of ebselen. MW, molecular weight. (D) Inhibition of CTD dimerization by Ebselen in TR-FRET assay.
HTS-TR-FRET for the identification of inhibitors of CTD dimerization.
We first performed an ELISA to validate the GST-CTD/CTD-Flag dimerization. Using a range of CTD-Flag concentrations, a dose-dependent signal was observed, revealing successful dimerization of CTD capsid (see Fig. S2A in the supplemental material). When increasing concentrations of untagged CTD were included, CTD dimerization was inhibited, with a 50% effective concentration (EC50) of 1.9 ± 0.12 μM (see Fig. S2B). To screen a small-molecule compound library, we developed a HTS-TR-FRET method assessing CTD dimerization. A strong and specific absorbance was observed between GST-CTD and CTD-Flag, while interaction between free GST and CTD-Flag gave a background signal (see Fig. S2C). The CTD dimerization was inhibited by untagged CTD concentrations, with an EC50 of 1.07 μM (see Fig. S2D). This assay presented excellent statistics, with a Z′ score of >0.7 and a signal-to-background ratio (S/B) of 3.5, allowing the screen of the LOPAC library. The results were summarized as a scatterplot (Fig. 1B). The LOPAC screen was completed, with excellent assay statistics between triplicates with an average Z′ score of 0.890 ± 0.004, an average Z score of 0.573 ± 0.003, and an S/B value of 5.629 ± 0.101. The hit cutoff was set at 42% (average + 3 standard deviations [SD]), and the hit rate was 3.28%, corresponding to 42 compounds. The top 10 hit compounds inhibited CTD-CTD interaction by 59% to 71%. The LOPAC library was cross-referenced for HCV core inhibitors (23), and out of the 42 hits identified in our screen, 9 displayed inhibitory activity against HCV core dimerization (15.7% to 51.6%). We tested 20 of the top hits in multiple rounds of HIV-1 infection in HeLa-CD4 cells and identified ebselen, an organoselenium compound (Fig. 1C), as the most active inhibitor of HIV-1 replication. The inhibitory effect of ebselen on CTD dimerization was confirmed by TR-FRET, with ebselen displaying a 50% inhibitory concentration (IC50) of 46.1 nM (Fig. 1D). In comparison, ebselen presented no activity on HCV core C106 dimerization by TR-FRET assays (23).
Ebselen inhibits HIV-1 replication.
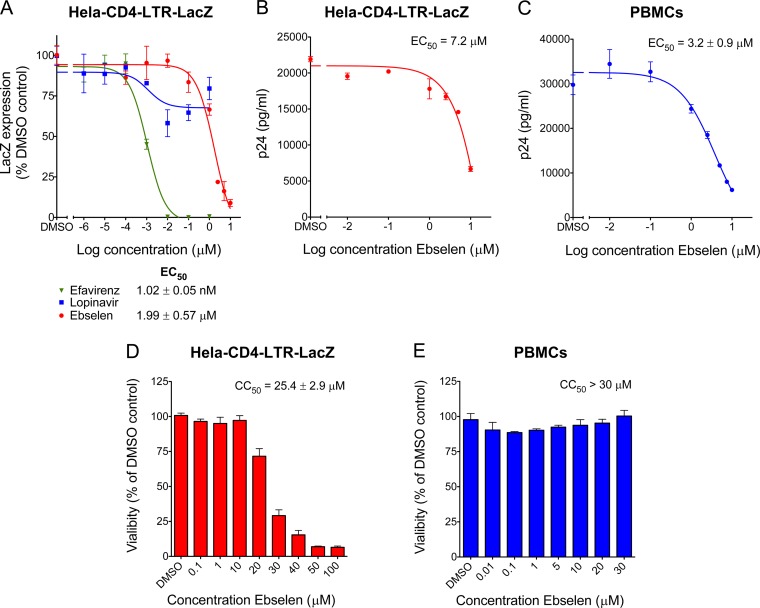
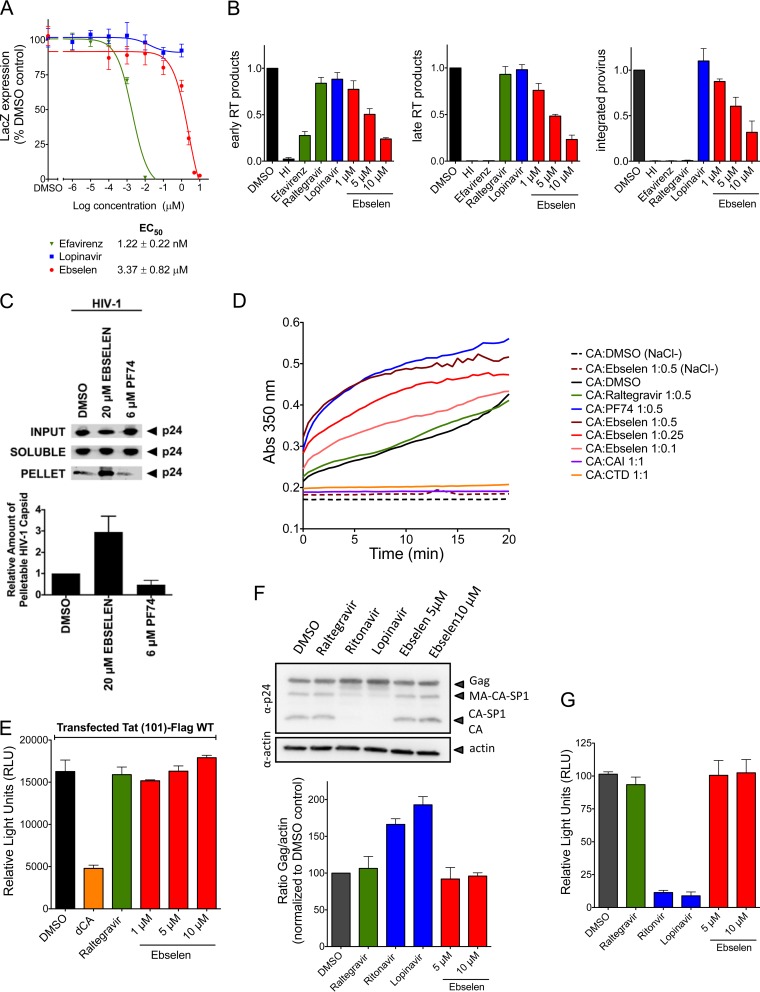
We evaluated HIV-1 susceptibility to ebselen using a reporter cell line that stably expresses the β-galactosidase (LacZ) gene driven by the HIV-1 5′ long terminal repeat (LTR) and responds to the viral protein Tat expressed by an incoming virus. Thus, HeLa-CD4-LTR-LacZ cells were infected with the HIV isolate NL4.3 in the presence of increasing concentrations of ebselen, and the measured β-Gal activity was directly correlated with viral replication. Ebselen inhibited the infection in a dose-dependent manner, with an EC50 of 1.99 ± 0.57 μM (Fig. 2A), while efavirenz, a nonnucleoside reverse transcriptase inhibitor (NNRTI) presented an EC50 of 1.02 ± 0.05 nM; lopinavir, a protease inhibitor (PI), showed maximal inhibition of only 30%. Given that PIs inhibit capsid maturation and only act upon spreading from the initial infection, it was not surprising to observe lopinavir's low inhibitory activity in this 72-h assay. The activity of ebselen on HIV-1 replication was also assessed by p24 ELISA in the supernatant, with an EC50 of 7.2 μM in HeLa-CD4-LTR-LacZ cells (Fig. 2B) and 3.2 ± 0.9 μM in peripheral blood mononuclear cells (PBMCs) (Fig. 2C). Given that ebselen is expected to bind to capsid, we made sure that ebselen was not interfering with p24 detection by the ELISA (see Fig. S3 in the supplemental material). The concentration of ebselen used in these assays did not impact cell viability as ebselen displayed a half-maximal cytotoxic concentration (CC50) of 25.4 ± 2.9 μM in HeLa-CD4-LTR-LacZ cells (Fig. 2D) and >30 μM in PBMCs (Fig. 2E). We then analyzed the antiviral activity of ebselen against 13 HIV-1 primary isolates belonging to seven different clades and two drug-resistant viruses in the presence of 10 μM ebselen in TZM-bl cells. TZM-bl cells are HeLa-CD4 cells that express the receptor (CD4) and the two coreceptors (CXCR4 and CCR5) of HIV-1 and express the β-galactosidase and luciferase genes under the control of the HIV-1 promoter. In this reporter system, ebselen displayed between 86.2 and 98.6% inhibitory activity against all viruses (see Table S1 in the supplemental material).
FIG 2.
Activity of ebselen on HIV-1 replication. (A) HeLa-CD4-LacZ cells were infected with NL4.3 replicative virus in the presence of increasing concentrations of ebselen for 72 h. β-Galactosidase activity was determined by quantitative CPRG assay. (B) Viral supernatants from HeLa-CD4 cell infections were assayed for their p24 antigen content using a sandwich ELISA kit. (C) Viral supernatants from PBMC infections were assayed for their p24 antigen content using a sandwich ELISA kit. (D and E) Cell viability MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assays on HeLa-CD4 cells and PBMCs, as indicated, treated with increasing concentrations of ebselen for 72 h (D) and for 4 days (E).
Ebselen covalently binds to Cys residues in the CTD of capsid.
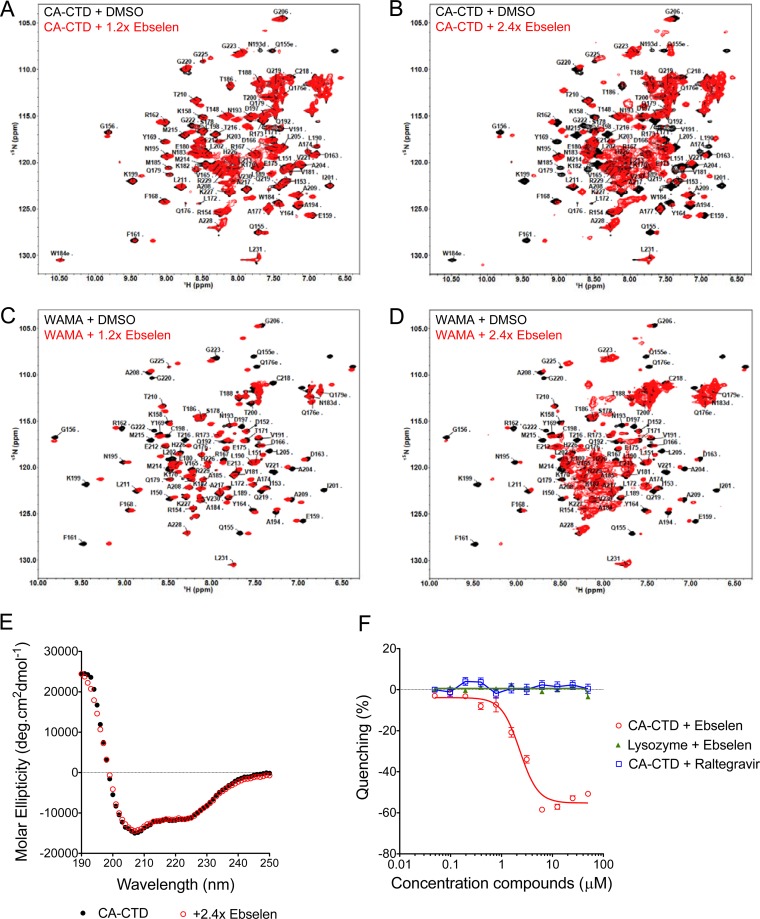
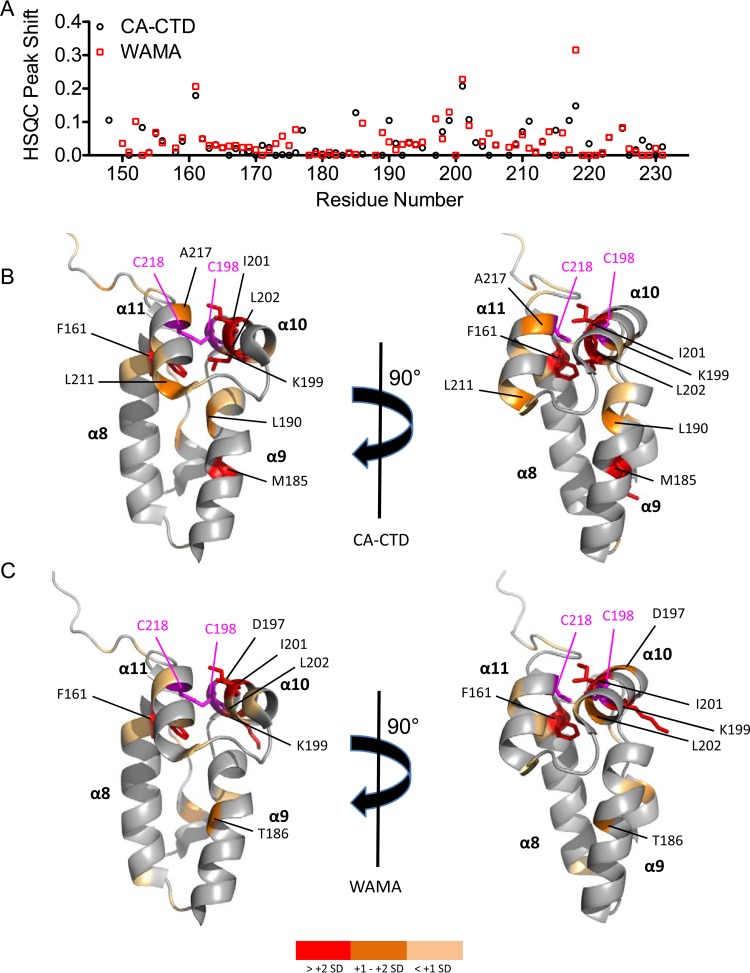
We monitored the binding of ebselen to CTD via [1H,15N] heteronuclear single-quantum coherence (HSQC) NMR on CA-CTD and on the capsid mutant CA-CTD W184A/M185A (WAMA) that bears amino acid substitutions that disrupt dimerization (13). Peak assignments were based on published HSQC chemical shifts for CA-CTD (24) and the WAMA mutant (25). The addition of 1.2- and 2.4-fold molar excesses of ebselen to CA-CTD and WAMA revealed peaks corresponding to free CA-CTD but with lower intensity than that of the apo form and the concomitant appearance of new resonances from ligand-bound protein shifted from initial peak positions, consistent with ligand binding in slow exchange on the NMR time scale (Fig. 3A to D). After the addition of 2.4-fold ebselen to either protein, high-intensity peaks clustered at the spectral center were observed, indicating protein aggregation (Fig. 3B and D). Circular dichroism (CD) spectrometry showed identical spectra for the free and ebselen-saturated protein (Fig. 3E), suggesting that the aggregated protein observed at 2.4-fold ebselen did not result from protein unfolding but from structural rearrangement to accommodate the bulky and hydrophobic ligand, resulting in a conformation susceptible to aggregation.
FIG 3.
Ebselen induces local structural changes in CA-CTD. (A and B) 2D [1H-15N] HSQC spectra of CA-CTD with a 1.2-fold (A) or 2.4-fold (B) molar excess of ebselen. (C and D) 2D [1H-15N] HSQC spectra of CA-CTD W184A/M185A (WAMA) with a 1.2-fold (C) or 2.4-fold (D) molar excess of ebselen. Peak assignments were based on published HSQC chemical shifts for CA-CTD (24) and the WAMA mutant (25). (E) Circular dichroism of CA-CTD with or without 2.4-fold ebselen. (F) Tryptophan fluorescence quenching assay of CA-CTD or lysozyme as a negative control, titrated with ebselen or raltegravir.
Binding of ebselen to CA-CTD was also confirmed by tryptophan fluorescence quenching, showing a decrease in fluorescence proportional to increased ebselen concentrations (Fig. 3F). No fluorescence quenching was observed in the presence of the integrase inhibitor (INI), raltegravir, or when ebselen was incubated with lysozyme, demonstrating the specificity of the ebselen–CA-CTD interaction. Furthermore, the addition of dithiothreitol (DTT) to ebselen–CA-CTD resulted in a loss of fluorescence quenching, suggesting that the interaction between ebselen and CA-CTD was reversible by a reducing agent (see Fig. S4A in the supplemental material).
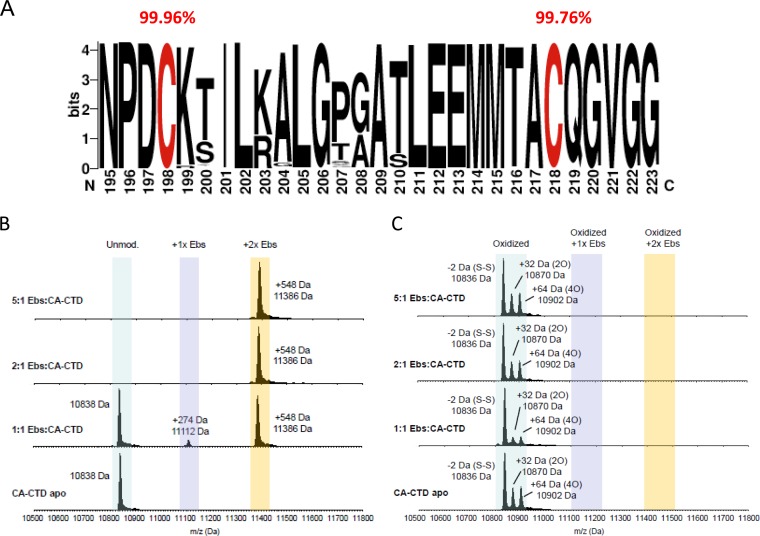
Ebselen is an organoselenium compound that mimics glutathione peroxidase activity (reviewed in reference 26). Previous studies show that this compound forms a covalent bond between its selenium atom and thiols in cysteine residues by forming a selenylsulfide (-Se-S-) linkage (27–29). HIV-1 capsid contains two cysteine residues, Cys198 and Cys218 (full-length CA numbering), both located in the CTD. These cysteine residues are highly conserved across HIV-1 subtypes, with 99.96% and 99.76% conservation for Cys198 and Cys218, respectively (Fig. 4A). These cysteines do not form a disulfide bond in the viral particle, and mutation of Cys198 interferes with viral particle disassembly, while mutation of Cys218 drastically reduces viral particle assembly (30). Liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) confirmed the covalent linkage of ebselen to CA-CTD (Fig. 4B). Incubation of CA-CTD with equimolar amounts of ebselen showed the presence of intact CA-CTD (∼10,838 Da) as well as ion masses that are 274 Da and 548 Da higher than the mass of CA-CTD, consistent with mass increases of one or two covalently linked ebselen molecules (∼274 Da). Notably, CA-CTD became preferentially double labeled with two molecules of ebselen even at a compound-to-protein molar ratio of 1:1. At higher ratios of ebselen to CA-CTD, only the doubly modified CA-CTD was detected. These observations highlight the high reactivity of both cysteine residues for ebselen modification. In accordance with the tryptophan fluorescence quenching results, we confirmed that addition of DTT reduced the selenylsulfide bond between ebselen and CA-CTD (see Fig. S4B in the supplemental material). Furthermore, small amounts of CA-CTD protein isoforms with intramolecular disulfide-linked (Cys-S-S-Cys) and oxidized Cys residues (Cys-SO2H) were also observed but not the ion species equivalent to the corresponding ebselen-linked oxidized CA-CTD (Fig. 4C). Collectively, these results suggest that ebselen covalently links to thiol groups (Cys-SH) in Cys198 and Cys218 of CA-CTD via a selenylsulfide linkage, forming S-[(2-carbamoylphenyl)selanyl]-l-cysteine, as illustrated in the structural model of CA-CTD bound to two ebselen molecules (see Fig. S5 in the supplemental material). The binding of ebselen to CTD is specific as omeprazole (31), methyl-3,4-dephostatin (27), and 3-bromo-1,1,1-trifluoroacetone (BTFA) (32), three cysteine-binding compounds, have no effect on HIV-1 replication and display less or no activity on CTD dimerization inhibition compared to the effect of ebselen (see Table S2 in the supplemental material).
FIG 4.
Ebselen covalently binds to both CA-CTD Cys residues. (A) Representation of frequencies of Cys198 and Cys218 residues from 2,890 sequences from eight different clades (A, B, C, D, F, G, CRF01_AE, and CRF01_AG) (HIV sequence database alignment) using the WebLogo application (http://weblogo.berkeley.edu). (B) ESI-MS analysis of CA-CTD incubated with different molar ratios of ebselen (Ebs). (C) ESI-MS analysis of oxidized CA-CTD species (intramolecular disulfide bridge, or higher-order cysteine oxidations) treated with different molar ratios of ebselen.
NMR chemical shift perturbations in CA-CTD and WAMA in the presence of a 1.2-fold molar excess of ebselen were quantitatively analyzed in a plot overlay that showed similar perturbation footprints (Fig. 5A). We mapped the residues that were affected by ebselen in CA-CTD and WAMA to the crystal structure of CA-CTD (33), and the most perturbed residues were found to colocalize in close proximity to Cys198 and Cys218. Some residues distal from these cysteine targets were also perturbed allosterically as a result of either structural rearrangements or conformational change after ebselen linkage (Fig. 5B and C). At a 2.4-fold molar excess of ebselen, additional residues in CA-CTD were perturbed that coincide with peaks that were substantially broadened in WAMA relative to those in CA-CTD (see Fig. S6 in the supplemental material). These peaks correspond to several critical residues within, or in close proximity to, the dimerization region, including W184 and M185. These results indicate that covalent attachment of two molar equivalents of ebselen promotes a conformational change that shifts the monomer-dimer equilibrium in favor of the ebselen-linked monomer that is susceptible to aggregation.
FIG 5.
Residues affected by ebselen in 15N–labeled CA-CTD. (A) HSQC peak shift with respect to CA-CTD (black) and WAMA (red) apo form after the addition of a 1.2-fold molar excess of ebselen. (B and C) HIV capsid CTD structure (33) (Protein Data Bank code 2BUO) rendered in PyMOL as a ribbon diagram highlighting residues affected by adding a 1.2-fold molar excess of ebselen to CA-CTD (B) and WAMA (C). Residue color scheme reflects peak shifts that are >+2 SD (red sticks), between +1 and +2 SD (orange), and within +1 SD (light orange) from the mean peak perturbation. Residues Cys198 and Cys218 are shown in magenta.
Mechanism of action of ebselen.
Capsid inhibitors as well as restriction factors targeting the viral capsid can inhibit HIV-1 replication at different steps of the viral life cycle, such as postentry events (uncoating), capsid assembly, or capsid maturation. We investigated in detail the step of the HIV-1 life cycle inhibited by ebselen, starting with the analysis of the activity of ebselen in early stages of HIV-1 replication (postentry events) in single-round infections using VSV-G pseudotyped virus (VSV-G-NL4-3). Ebselen was active in a single-round infection assay, with an EC50 of 3.37 ± 0.82 μM. Efavirenz displayed an EC50 of 1.22 ± 0.22 nM, while lopinavir, as expected, was not active in this assay (Fig. 6A). We subsequently analyzed the viral DNA species, such as the early and late reverse transcription (RT) products, via quantitative PCR (qPCR). In addition, we also analyzed proviral integration by Alu-PCR followed by qPCR (Fig. 6B). Ebselen decreased early and late RT products in a dose-dependent manner and, as a result, inhibited the levels of integrated proviral DNA. Raltegravir (INI) and lopinavir (PI) had no impact on RT activity, and as expected raltegravir inhibited proviral integration. The profile of ebselen was similar to that observed for the RT inhibitor efavirenz (NNRTI), which suggests that ebselen may affect the reverse transcription process by negatively impacting the uncoating process. To test this hypothesis, we investigated the fate of the HIV-1 capsid in the presence of ebselen. This assay monitors the uncoating process by measuring the relative level of soluble (disassembled CA) versus pelletable (fullerene core) capsid during HIV-1 infection (34). As shown in Fig. 6C, ebselen increased the amount of pelletable CA compared to that of the DMSO control, suggesting a stabilization of the mature capsid, similar to what is observed for the cellular restriction factor MxB (8). In contrast, the NTD inhibitor PF74 decreased the amount of pelletable CA protein during infection by destabilizing the mature capsid, as previously observed (35). The stabilization effect observed in the presence of ebselen was also shown in an in vitro capsid multimerization assay. While the addition of CTD and capsid assembly inhibitor (CAI) peptide inhibited capsid assembly (36, 37), addition of increasing concentrations of ebselen resulted in a dose-dependent increase in the rate of CA multimerization. This same phenomenon was previously observed with the NTD capsid inhibitor PF74 (38). In contrast, the addition of raltegravir (INI) had no impact on HIV-1 capsid assembly (Fig. 6D). Altogether, these results suggest that ebselen acts at an early, postentry stage of the HIV-1 replication cycle by increasing the stability of the viral capsid during infection and impairing the uncoating process by a mechanism of action different from that of PF74. We cannot exclude the possibility that this increase in stability may result from aggregation of CTD protein as observed in NMR in the presence of a 2.4-fold excess of ebselen.
FIG 6.
Mechanism of action of Ebselen. (A to C) Impact of ebselen on early replication events. (A) Activity of ebselen was determined by HIV-1 single-round infection in HeLa-CD4-LacZ cells. β-Galactosidase activity was determined by quantitative CPRG assay. (B) The effect of ebselen on reverse-transcription products and HIV-1 integration was determined. HI, heat inactivated. Early and late RT products were determined by qPCR, and provirus integration was quantified by Alu-PCR followed by a qPCR. (C) The impact of ebselen on HIV-1 capsid stability was determined by a fate of capsid assay. (D) Impact of ebselen on in vitro capsid assembly. (E to G) Impact of ebselen on late viral replication events. (E) The effect of ebselen on Tat-mediated transactivation was determined in HeLa-CD4-LTR-Luc cells. Luciferase activity was measured at 48 h posttransfection of Tat-Flag. WT, wild type. (F) Activity of the indicated drugs on Gag expression and maturation was determined in 293T cells transfected with pNL4.3 (top). Gag expression was quantified by Western blotting with corresponding densitometry analysis (bottom). The impact of ebselen on infectivity of the viral particles produced in the experiment shown in panel F was determined in infection of reporter HeLa-CD4-LTR-Luc cells. Luciferase activity was measured at 48 h postinfection.
We further investigated the impact of ebselen on late replication events (after integration of the viral DNA into the host cell genome). First we analyzed the impact of ebselen on HIV transcription promoted by the viral transactivator Tat using HeLa-CD4-LTR-Luc cells that respond to Tat transactivation (Fig. 6E). We demonstrated that ebselen had no impact on the ability of Tat to activate HIV-1 transcription, while didehydro-cortistatin A (dCA), a potent Tat inhibitor (39), displayed inhibitory activity of 70%. We then analyzed the impact of ebselen on Gag expression and maturation after transfection of HEK 293T cells with pNL4.3 as well as on the infectivity of the viral particles produced. The Gag protein expression and maturation were similar in the presence of ebselen, raltegravir (INI), or the DMSO control, while, as expected, two protease inhibitors (ritonavir and lopinavir) affected the maturation process (Fig. 6F). The viral particles produced in the presence of ebselen and raltegravir showed levels of infectivity similar to those of the virions produced in the DMSO control, while ritonavir and lopinavir abolished the infectivity of the virions by interfering with the maturation process (Fig. 6G). Taken together, these results demonstrate that ebselen has no impact on late events in the HIV-1 life cycle.
Evaluation of analogs of ebselen.
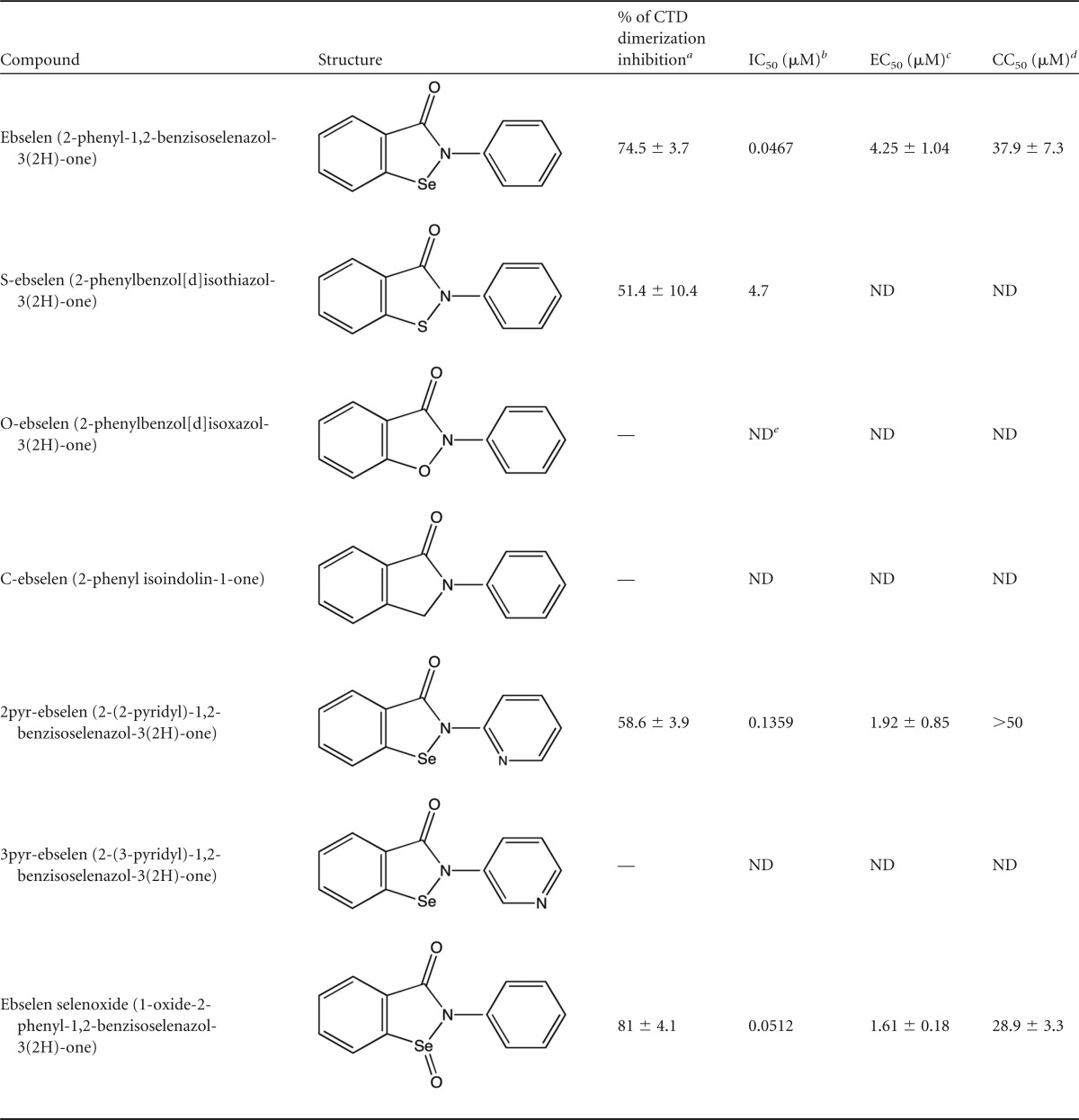
Based on our NMR and LC-ESI-MS results and as reported in the literature (27–29), ebselen forms a selenylsulfide (-Se-S-) linkage with cysteine residues. To confirm the importance of the selenium atom in ebselen, we analyzed analogs with selenium replaced by sulfur (S-ebselen), oxygen (O-ebselen), carbon (C-ebselen) (40), or a selenoxide group (ebselen oxide) or with the phenyl ring converted to pyridine (2pyr-ebselen and 3pyr-ebselen) (41) (Table 1). The inhibitory activity of these analogs on CTD dimerization was evaluated by TR-FRET assay. Only the S-ebselen, 2pyr-ebselen, and ebselen oxide analogs retained the inhibitory activity of ebselen, while O-ebselen, C-ebselen, and 3pyr-ebselen had no impact on dimerization. Since ebselen oxide and 2pyr-ebselen displayed IC50s in the same range as the ebselen IC50 in TR-FRET assay, we further studied these compounds in single-round infections and determined their cytotoxicity. Ebselen oxide and 2pyr-ebselen presented slightly lower EC50s than ebselen (1.61 ± 0.18 μM and 1.92 ± 0.85 μM versus 4.07 ± 0.93 μM, respectively); however ebselen oxide was slightly more toxic than ebselen in HeLa-CD4 cells (CC50 of 28.9 ± 3.3 μM versus 37.9 ± 7.3 μM, respectively). Taking these results together, ebselen modification to ebselen oxide or 2pyr-ebselen slightly improved ebselen's potency. Collectively, these results reveal the importance of ebselen's selenium atom in its antiviral activity.
TABLE 1.
Structure and activity of ebselen analogs
a Maximal percent inhibitory activities of ebselen and its analogs were measured in TR-FRET assays at final concentrations of 10 μM. —, inhibition by less than 25%.
b IC50s of analogs were determined by a dose-response inhibition of CTD dimerization in TR-FRET assays.
c EC50s of ebselen, 2pyr-ebselen, and ebselen oxide were determined in HIV-1 single-round infections in HeLa-CD4-LTR-LacZ cells.
d CC50 values were determined by MTT assays on HeLa-CD4-LTR-LacZ cells for a period of 48 h.
e ND, not determined.
Impact of ebselen on other retroviruses and nonretroviruses.
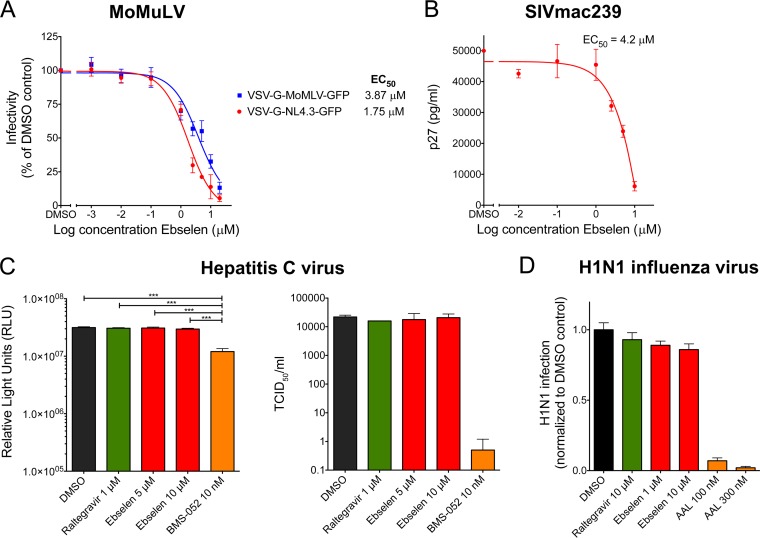
We assessed the specificity of ebselen to inhibit infection by Moloney murine leukemia virus (MoMuLV) and simian immunodeficiency (SIVmac239) virus. Ebselen inhibited MoMuLV and HIV-1, with EC50s of 3.87 μM and 1.75 μM, respectively (Fig. 7A), and it inhibited SIV, with an EC50 of 4.2 μM (Fig. 7B). The inhibition of SIVmac239 by ebselen was expected since the SIV CTD sequence is conserved relative to that of HIV-1 and also contains the two cysteine residues, Cys198 and Cys218 (42). However, little homology exists between MoMuLV and HIV-1 capsids. MoMuLV capsid contains a single cysteine residue at position 55 and a unique carboxy terminus containing charge-rich residues (arginine and glutamic acid), critical for proper viral assembly (43, 44). These results suggest that the binding of ebselen to capsid may also have a conformational component.
FIG 7.
Impact of ebselen on MoMuLV, SIV, HCV, and influenza viruses. (A) Impact of ebselen on the retrovirus MoMuLV in TE671 cells infected with VSV-G-MoMuLV-GFP and VSV-G-NL4.3-GFP viruses. The percentage of GFP-positive cells was determined by flow cytometry. (B) Impact of ebselen on SIV infection in CEM174 cells infected with SIVmac239 virus. Viral supernatants were assayed for their p27 antigen content using a sandwich ELISA kit. (C) Impact of ebselen on HCV by replication and infectivity assays in Huh-7.5 cells containing an HCV reporter virus (pSG-Rluc-2a-neo-NS3-5B JFH1) by measuring luciferase activity (left) or infected with a cell culture-adapted strain of JC1 (JC1.1) by determining the 50% tissue culture infective dose/ml using an anti-NS5A antibody. (D) Impact of ebselen on influenza virus (H1N1) in HeLa-CD4 cells determined by flow cytometry using a monoclonal antibody to H1N1.
We further evaluated the effect of ebselen on nonretroviruses such as hepatitis C (HCV) and influenza viruses, and in both cases, ebselen showed no activity (Fig. 7C and D). These results suggest ebselen to be rather specific for retroviruses. In a recent report, ebselen was shown to inhibit HCV NS3 helicase binding to nucleic acid by interacting with cysteine residues, with an EC50 of 30 ± 17 μM and a CC50 of 34 ± 4 μM. Thus, the inhibitory activity of ebselen could partly be due to cellular toxicity (45).
Conclusions.
Protein-protein interactions play key roles in a wide range of biological processes and are attractive targets for the design of novel therapeutics (46). However, the development of inhibitors that target these interactions is far more challenging than targeting enzyme active sites. Notably, none of the drugs currently used in antiretroviral therapy target the HIV-1 capsid protein. Given the capsid's key importance in productive infection, it seems reasonable to promote drug development against the capsid. HIV is well known for its high mutation rate and, consequently, rapid site-directed drug resistance; thus, developing inhibitors insensitive to such viral mutational evolution is important. Targeting the viral capsid seems appropriate since this protein has been shown to mutate less readily and presents 70% sequence conservation between isolates (47).
To this end, we conducted a TR-FRET-based HTS assay to identify inhibitors of HIV-1 capsid dimerization. This screening successfully identified ebselen as an inhibitor of capsid dimerization and HIV-1 replication. NMR confirmed perturbation of residues in the dimerization domain of CTD when ebselen is present at more than 2-fold the CTD molar concentration. Using cellular assays, we observed that this small molecule acts on the early postentry step of the viral cycle prior to reverse transcription by stabilizing the incoming viral capsid. However, we cannot exclude the possibility that this increase in stability may result from aggregation of CTD protein, as observed in NMR in the presence of a 2.4-fold excess of ebselen. Moreover, ebselen was initially found as an inhibitor of dimerization in TR-FRET assays using only the CTD of capsid. In this context it is also a possibility that aggregation might have led to a loss in fluorescence intensity. Interestingly, ebselen inhibits diverse retroviruses but has no impact on nonrelated viruses such as HCV and influenza viruses. In addition, the selenium atom in ebselen was shown to be important for its activity. However, ebselen has been identified in several biological assays, suggesting lack of selectivity (reviewed in reference 26); therefore, chemical innovation around the ebselen pharmacophore, while retaining the key selenium atom, will be necessary for increasing specificity for retroviral inhibition. A phase 2 clinical trial testing ebselen (SPI-1005) to prevent and treat noise-induced hearing loss has been conducted. Ebselen demonstrated clinically relevant reduction in temporary threshold shift (TTS) induced by loud-sound exposure, and the doses of ebselen administered (200, 400, and 600 mg twice daily for 4 days) were well tolerated. Therefore, despite its lack of specificity, ebselen is safely tolerated (48).
This study is a proof-of-concept employing a TR-FRET-based HTS platform to identify a small molecule that targets the viral capsid. The identification of ebselen as an inhibitor of HIV-1 replication further highlights the validity of the HIV-1 capsid as a promising drug target. Moreover, as the use of covalent inhibitors is growing in drug discovery (reviewed in reference 49), this study lays the groundwork for future drug design involving covalently linked capsid inhibitors that target HIV-1 replication.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the NIH AIDS reagent program, Division of AIDS, NIAID, NIH, for the HIV-1 p24 monoclonal antibody (183-H12-5C) from Bruce Chesebro and Kathy Wehrly and for providing all the HIV-1 primary isolates and drug-resistant viruses. Analogs of Ebselen, S-, O-, and C-Ebselen, were a gift from Barbara Slusher, Johns Hopkins University School of Medicine, Baltimore, MD. HeLa-CD4-LTR-LacZ and HeLa-CD4-LTR-Luc cells were provided by Uriel Hazan, Université de Cachan, France. Human hepatocarcinoma cell line Huh7.5 cells were provided by Charles Rice of Rockefeller University. We thank Massimo Caputi for providing the GST-CTD and CTD-Flag constructs. CAI peptide was a gift from Asim K. Debnath.
We also honor the memory of our friend and colleague Donny Strosberg, who initiated this project.
We declare that we have no conflicts of interest.
Funding Statement
National Institute of Allergy and Infectious Diseases (NIAID) provided funding for this work under grant number R01AI100685.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02574-15.
REFERENCES
- 1.Ambrose Z, Aiken C. 2014. HIV-1 uncoating: connection to nuclear entry and regulation by host proteins. Virology 454–455:371–379. doi: 10.1016/j.virol.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulme AE, Perez O, Hope TJ. 2011. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci U S A 108:9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forshey BM, von Schwedler U, Sundquist WI, Aiken C. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hulme AE, Kelley Z, Okocha EA, Hope TJ. 2015. Identification of capsid mutations that alter the rate of HIV-1 uncoating in infected cells. J Virol 89:643–651. doi: 10.1128/JVI.03043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 7.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan A, Campbell EM, Brandariz-Nunez A, Diaz-Griffero F. 2014. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. doi: 10.1186/s12977-014-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. 2013. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 10.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. 2004. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol 78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez CS, Eccles JD, Still A, Sloan RE, Barklis RL, Tsagli SM, Barklis E. 2011. Determinants of the HIV-1 core assembly pathway. Virology 417:137–146. doi: 10.1016/j.virol.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, Gronenborn AM. 2009. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 14.Pornillos O, Ganser-Pornillos BK, Yeager M. 2011. Atomic-level modelling of the HIV capsid. Nature 469:424–427. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin R, Tzou YM, Krishna NR. 2011. Structure of a monomeric mutant of the HIV-1 capsid protein. Biochemistry 50:9457–9467. doi: 10.1021/bi2011493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. 2013. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thenin-Houssier S, Valente ST. HIV-1 capsid inhibitors as antiretroviral agents. Curr HIV Res, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith PF, Ogundele A, Forrest A, Wilton J, Salzwedel K, Doto J, Allaway GP, Martin DE. 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-O-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob Agents Chemother 51:3574–3581. doi: 10.1128/AAC.00152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Baelen K, Salzwedel K, Rondelez E, Van Eygen V, De Vos S, Verheyen A, Steegen K, Verlinden Y, Allaway GP, Stuyver LJ. 2009. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob Agents Chemother 53:2185–2188. doi: 10.1128/AAC.01650-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garzon MT, Lidon-Moya MC, Barrera FN, Prieto A, Gomez J, Mateu MG, Neira JL. 2004. The dimerization domain of the HIV-1 capsid protein binds a capsid protein-derived peptide: a biophysical characterization. Protein Sci 13:1512–1523. doi: 10.1110/ps.03555304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Curreli F, Zhang X, Bhattacharya S, Waheed AA, Cooper A, Cowburn D, Freed EO, Debnath AK. 2011. Antiviral activity of alpha-helical stapled peptides designed from the HIV-1 capsid dimerization domain. Retrovirology 8:28. doi: 10.1186/1742-4690-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kota S, Scampavia L, Spicer T, Beeler AB, Takahashi V, Snyder JK, Porco JA, Hodder P, Strosberg AD. 2010. A time-resolved fluorescence-resonance energy transfer assay for identifying inhibitors of hepatitis C virus core dimerization. Assay Drug Dev Technol 8:96–105. doi: 10.1089/adt.2009.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung J, Byeon IJ, Ahn J, Concel J, Gronenborn AM. 2010. 1H, 15N and 13C assignments of the dimeric C-terminal domain of HIV-1 capsid protein. Biomol NMR Assign 4:21–23. doi: 10.1007/s12104-009-9198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong HC, Shin R, Krishna NR. 2008. Solution structure of a double mutant of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Biochemistry 47:2289–2297. doi: 10.1021/bi7022128. [DOI] [PubMed] [Google Scholar]
- 26.Azad GK, Tomar RS. 2014. Ebselen, a promising antioxidant drug: mechanisms of action and targets of biological pathways. Mol Biol Rep 41:4865–4879. doi: 10.1007/s11033-014-3417-x. [DOI] [PubMed] [Google Scholar]
- 27.Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. 2012. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem Biol 7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakurai T, Kanayama M, Shibata T, Itoh K, Kobayashi A, Yamamoto M, Uchida K. 2006. Ebselen, a seleno-organic antioxidant, as an electrophile. Chem Res Toxicol 19:1196–1204. doi: 10.1021/tx0601105. [DOI] [PubMed] [Google Scholar]
- 29.Favrot L, Lajiness DH, Ronning DR. 2014. Inactivation of the Mycobacterium tuberculosis antigen 85 complex by covalent, allosteric inhibitors. J Biol Chem 289:25031–25040. doi: 10.1074/jbc.M114.582445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDermott J, Farrell L, Ross R, Barklis E. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J Virol 70:5106–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambrecht N, Corbett Z, Bayle D, Karlish SJ, Sachs G. 1998. Identification of the site of inhibition by omeprazole of a α-β fusion protein of the H,K-ATPase using site-directed mutagenesis. J Biol Chem 273:13719–13728. doi: 10.1074/jbc.273.22.13719. [DOI] [PubMed] [Google Scholar]
- 32.Didenko T, Liu JJ, Horst R, Stevens RC, Wuthrich K. 2013. Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr Opin Struct Biol 23:740–747. doi: 10.1016/j.sbi.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. 2005. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol 12:678–682. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Luban J, Diaz-Griffero F. 2014. The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol Biol 1087:29–36. doi: 10.1007/978-1-62703-670-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M. 2014. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc Natl Acad Sci U S A 111:18625–18630. doi: 10.1073/pnas.1419945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barklis E, Alfadhli A, McQuaw C, Yalamuri S, Still A, Barklis RL, Kukull B, Lopez CS. 2009. Characterization of the in vitro HIV-1 capsid assembly pathway. J Mol Biol 387:376–389. doi: 10.1016/j.jmb.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanman J, Sexton J, Sakalian M, Prevelige PE Jr. 2002. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J Virol 76:6900–6908. doi: 10.1128/JVI.76.14.6900-6908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. 2010. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog 6:e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mousseau G, Clementz MA, Bakeman WN, Nagarsheth N, Cameron M, Shi J, Baran P, Fromentin R, Chomont N, Valente ST. 2012. An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host Microbe 12:97–108. doi: 10.1016/j.chom.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas AG, Rojas C, Tanega C, Shen M, Simeonov A, Boxer MB, Auld DS, Ferraris DV, Tsukamoto T, Slusher BS. 2013. Kinetic characterization of ebselen, chelerythrine and apomorphine as glutaminase inhibitors. Biochem Biophys Res Commun 438:243–248. doi: 10.1016/j.bbrc.2013.06.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pino MA, Pietka-Ottlik M, Billack B. 2013. Ebselen analogues reduce 2-chloroethyl ethyl sulphide toxicity in A-431 cells. Arch Ind Hyg Toxicol 64:77–86. [DOI] [PubMed] [Google Scholar]
- 42.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J 17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheslock SR, Poon DT, Fu W, Rhodes TD, Henderson LE, Nagashima K, McGrath CF, Hu WS. 2003. Charged assembly helix motif in murine leukemia virus capsid: an important region for virus assembly and particle size determination. J Virol 77:7058–7066. doi: 10.1128/JVI.77.12.7058-7066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang MQ, Goff SP. 2003. Defects in virion production caused by mutations affecting the C-terminal portion of the Moloney murine leukemia virus capsid protein. J Virol 77:3339–3344. doi: 10.1128/JVI.77.5.3339-3344.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukherjee S, Weiner WS, Schroeder CE, Simpson DS, Hanson AM, Sweeney NL, Marvin RK, Ndjomou J, Kolli R, Isailovic D, Schoenen FJ, Frick DN. 2014. Ebselen inhibits hepatitis C virus NS3 helicase binding to nucleic acid and prevents viral replication. ACS Chem Biol 9:2393–2403. doi: 10.1021/cb500512z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jubb H, Higueruelo AP, Winter A, Blundell TL. 2012. Structural biology and drug discovery for protein-protein interactions. Trends Pharmacol Sci 33:241–248. doi: 10.1016/j.tips.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 47.Li G, Verheyen J, Rhee SY, Voet A, Vandamme AM, Theys K. 2013. Functional conservation of HIV-1 Gag: implications for rational drug design. Retrovirology 10:126. doi: 10.1186/1742-4690-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kil JLE, Griffiths S, Lobarinas E, Spankovich C, Antonelli PJ, Le Prell C. 2014. Efficacy of SPI-1005 for prevention of noise-induced hearing loss: phase 2 clinical trial results. Otolaryngol Head Neck Surg 151(Suppl):P83–P84. doi: 10.1177/0194599814541627a172. [DOI] [Google Scholar]
- 49.Bauer RA. 2015. Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discov Today 20:1061–1073. doi: 10.1016/j.drudis.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat Med 7:631–634. [DOI] [PubMed] [Google Scholar]
- 51.Kortagere S, Madani N, Mankowski MK, Schon A, Zentner I, Swaminathan G, Princiotto A, Anthony K, Oza A, Sierra LJ, Passic SR, Wang X, Jones DM, Stavale E, Krebs FC, Martin-Garcia J, Freire E, Ptak RG, Sodroski J, Cocklin S, Smith AB III. 2012. Inhibiting early-stage events in HIV-1 replication by small-molecule targeting of the HIV-1 capsid. J Virol 86:8472–8481. doi: 10.1128/JVI.05006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.