Abstract
Two laboratory mutants of NDM-1 were generated by replacing the isoleucine at position 35 with threonine and serine residues: the NDM-1I35T and NDM-1I35S enzymes. These mutants were well characterized, and their kinetic parameters were compared with those of the NDM-1 wild type. The kcat, Km, and kcat/Km values calculated for the two mutants were slightly different from those of the wild-type enzyme. Interestingly, the kcat/Km of NDM-1I35S for loracarbef was about 14-fold higher than that of NDM-1. Far-UV circular dichroism (CD) spectra of NDM-1 and NDM-1I35T and NDM-1I35S enzymes suggest local structural rearrangements in the secondary structure with a marked reduction of α-helix content in the mutants.
INTRODUCTION
The NDM-1 metallo-β-lactamase (MBL) was first described in a urinary Klebsiella pneumoniae isolate recovered from a Swedish patient who traveled to New Delhi and who had received medical care in India (1). This is the most recent MBL to have widely spread around the world among enterobacterial strains, Pseudomonas aeruginosa (2), Acinetobacter baumannii (3), Morganella morganii (4), Alcaligenes faecalis, Vibrio cholerae, and Stenotrophomonas maltophilia (5, 6). NDM-1-producing bacteria have been recovered from many infection sites as hospital-acquired and community-acquired infections but also in environmental samples (7). Since its finding, 12 NDM variants have been identified (http://www.lahey.org/Studies/). The factor that has influenced the wide geographic spread of blaNDM-1 gene is its localization on complex plasmids that mediate the transfer of this resistant determinant under the selective pressure of antibiotic therapy (8, 9). Metallo-β-lactamases show a broad-spectrum substrate profile; they are resistant to classical β-lactamase inhibitors and hydrolyze carbapenems very efficiently. MBLs require one or two zinc ions to catalyze the hydrolysis of β-lactams. It is commonly suggested that the zinc ion acts as a Lewis acid to stabilize the transient tetrahedral intermediate formed by the nucleophilic attachment of a hydroxide ion to the carboxyl group of the β-lactam ring (10). NDM-1 enzyme shows a great ability to hydrolyze all β-lactam antibiotics (11). Several crystal structures of NDM-1 have been solved, and the enzyme displays the typical αβ/βα fold of MBLs (12, 13, 14). NDM-1 belongs to subclass B1, and it contains a binuclear Zn active site surrounded by several loops responsible for substrate binding and specificity (13). Several studies pointed out the attention focused on the L3 loop, which could be involved in the hydrophobic contacts with substrates through the presence of an aromatic residue. Moreover, its great flexibility makes the loop able to drive substrate into the active site. The role of the L3 loop has been studied in such subclass B1 MBLs as IMP-1 and VIM-2 (15), but no experimental data are available for the NDM-1 enzyme. In comparison to the IMP and VIM variants, NDM-1 has a longer N terminus which forms two extra strands that pack on the L3 loop through an isoleucine residue at position 35 (BBL numbering). Thus, the role of this loop in the NDM-1 enzyme could be related to the influence of isoleucine 35 (I35). In fact, as previously reported, the I35 in NDM-1 is positioned at the tip of the β1 strand and seems to play an important role both in the extension of the hydrophobicity of the L3 loop and in the substrate binding and specificity (13). In order to elucidate the importance of residue 35, a nonpolar residue of isoleucine in this position was replaced by two polar residues, serine (S) and threonine (T). In particular, our aim was to understand how I35 could influence the permanence of the hydrophobicity of the L3 loop and substrate binding and if it perturbs the enzyme structure.
(Part of this study was presented at the 12th β-Lactamase Meeting, June 28 to July 1 2014, Gran Canaria, Canary Islands, Spain.)
MATERIALS AND METHODS
Mutant design.
NDM-1I35T and NDM-1I35S mutants were generated by site-directed mutagenesis using the overlap extension method (16). Each mutation was introduced into a PCR amplicon using mutagenic primers in combination with external primers NDM_for and NDM_rev (Table 1) and pFM-NDM-1 plasmid as the template. Two partially overlapping DNA fragments generated by PCR were subsequently used in an overlap extension reaction coupled to amplification of the entire coding sequence with external primers. Each fragment was sequenced using an ABI Prism 310 single-capillary automated sequencer (Life Technologies, Monza, Italy).
TABLE 1.
Oligonucleotides used for site-directed mutagenesis and sequencing
| Primer and category | Sequencea |
|---|---|
| Mutagenic | |
| I35T_for | 5′-GACGACTGGCCAGCAAATG-3′ |
| I35T_rev | 5′-CATTTGCTGGCCAGTCGTC-3′ |
| I35S_for | 5′-GACGAGTGGCCAGCAAATG-3′ |
| I35S_rev | 5′-CATTTGCTGGCCACTCGTC-3′ |
| Amplification | |
| NDM_for | 5′-GGGGGCATATGGGTGAAATCCGCCCGA-3′ |
| NDM_rev | 5′-GGGGGCTCGAGTCAGCGCAGCTTGTCGGC-3′ |
The mutated positions are underlined. The restriction site sequences are in bold.
Cloning of blaNDM-1, blaNDM-1I35T, and blaNDM-1I35S determinants.
Genes blaNDM-1 (wild type), blaNDM-1I35T, and blaNDM-1I35S were cloned with and without signal peptide in pET-24(a) vector using the NdeI and XhoI restriction sites to obtain plasmids pSP-NDM-1, pSP-NDM-1I35T, and pSP-NDM-1I35S (with signal peptide) and pFM-NDM-1, pFM-NDM-1I35T, and pFM-NDM-1I35S (without signal peptide). Escherichia coli NovaBlue strain was used as a nonexpression host for the initial cloning. The recombinant plasmids were then transferred into E. coli JM109(DE3) for enzyme expression. The authenticity of cloned mutant genes was verified by sequencing both strands of the three recombinant plasmids using a single-capillary automated sequencer (ABI Prism 310; Life Technologies, Italy).
Antimicrobial susceptibility tests.
The phenotypic profile has been characterized by a microdilution method using a bacterial inoculum of 5 × 105 CFU/ml according to Clinical and Laboratory Standards Institute (CLSI) performance standards (17). Nitrocefin was kindly provided by Shariar Mobashery (Notre Dame University, South Bend, IN, USA). Meropenem was from AstraZeneca (Milan, Italy). Imipenem, ertapenem, and cefoxitin were from Merck Sharp & Dohme (Rome, Italy). Biapenem was from Cyanamid (Catania, Italy). Loracarbef and cefaclor were from Eli Lilly and Co. (Indianapolis, IN, USA). Ceftazidime was from GlaxoSmithKline (Verona, Italy). Biapenem was from Wyeth-Lederle (Catania, Italy). The other antimicrobial agents used in this study were purchased from Sigma-Aldrich (Milan, Italy).
Expression and purification of NDM-1, NDM-1I35T, and NDM-1I35S enzymes.
E. coli JM109(DE3) cells containing the recombinant plasmids pFM-NDM-1, pFM-NDM-1I35T, and pFM-NDM-1I35S were grown in 1 liter of tryptic soy broth (TSB) medium with kanamycin (50 μg/ml) at 37°C in an orbital shaker (180 rpm). Each culture was grown to achieve an A600 of approximately 0.5, and 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added. After addition of IPTG, the cultures were incubated for 16 h at 22°C, under aerobic conditions. Cells were harvested by centrifugation at 8,000 rpm for 10 min at 4°C and washed twice with 20 mM Tris-HCl buffer (pH 7.0) (buffer A). Crude enzymes were obtained by treatment with lysozyme at a concentration of 2 mg/ml for 30 min at 30°C following by sonication on ice (5 cycles at 60 W for 1 min and 2 min of break). The lysate was centrifuged at 30,000 rpm for 30 min, and the cleared supernatant was recovered and loaded onto a Sepharose-Q fast-flow column equilibrated with buffer A. The column was extensively washed to remove unbound proteins, and the β-lactamase was eluted with a linear gradient of 20 mM Tris-HCl (pH 7.0)–NaCl (0.5 M) (buffer B). The fractions containing β-lactamase activity were pooled, concentrated 20-fold using an Amicon concentrator (YM 10 membrane; Millipore, Bedford, MA, USA), and loaded onto Sephacryl S-100 equilibrated with buffer A. The pure fractions were pooled and dialyzed in 20 mM HEPES buffer (pH 7.0) containing 0.01 mg/ml bovine serum albumin (BSA) (buffer C) for further experiments.
Electrophoretic analysis and protein determination.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by following the method of Laemmli (18) with a fixed 12.5% (wt/vol) polyacrylamide gel in the presence of 0.1% SDS, using a Mini-Protean III apparatus (Bio-Rad Laboratories, Richmond, CA). After electrophoresis, the proteins bands were stained with Coomassie brilliant blue R-250. The Mr value was determined by comparison of its relative mobility with that of a standard protein mixture. Protein content was determined by the Bradford method with bovine serum albumin as the standard (19).
Determination of kinetic parameters.
β-Lactamase activity was routinely measured by spectrophotometric assay following the hydrolysis of 100 μM imipenem. One unit of β-lactamase activity was defined as the amount of enzyme which hydrolyzes 1 μmol of substrate per min at 25°C in buffer C, containing 0.01 mg/ml of BSA (bovine serum albumin) for enzyme stability.
Steady-state kinetic experiments were performed following the hydrolysis of 16 β-lactams (data not shown) at 25°C in buffer C plus 20 μM ZnCl2. The data were collected with a Perkin-Elmer Lambda 25 spectrophotometer (Perkin-Elmer Italia, Monza, Italy). Kinetic parameters were determined under initial-rate conditions using GraphPad Prism6 software to generate Michaelis-Menten curves or by analyzing the complete hydrolysis time courses (20, 21). Each kinetic value represents the mean of the results of five different measurements; the error rate was below 5%.
The effect of zinc content on NDM-1, NDM-1I35T, and NDM-1I35S activity was evaluated by following the hydrolysis of 100 μM imipenem using concentrations of ZnCl2 ranging from 0 μM to 100 μM. The initial hydrolysis rate (v0) was expressed in millimoles per minute per milligram. The volume of the reaction mixture was 1 ml. The effect of temperature on enzyme stability was evaluated by measuring the residual activity of the enzymes versus that seen with 100 μM imipenem at 30°C, 40°C, and 50°C. The enzymes were incubated for up to 120 min in buffer C.
Fluorescence spectroscopy.
Fluorescence spectra of the purified enzymes (200 nM in buffer C) at 20°C were recorded in the 300-to-460-nm range following excitation at 280 nm, using a Perkin-Elmer LS50B fluorescence spectrometer (Perkin-Elmer, Monza, Italy) and a scan rate of 200 nm min−1. Excitation and emission slit widths were both 5 nm. Spectra were recorded five times, the results were averaged, and the fluorescence background of the buffer was subtracted.
CD.
Far-UV circular dichroism (CD) spectra (185 to 260 nm) were recorded with a Jasco J-810 spectropolarimeter at 20°C using 20 mM sodium phosphate (pH 7) and a 1-mm-path-length quartz Suprasil cell (Hellma), with protein concentrations of ca. 0.06 to 0.1 mg/ml. Data from four scans (10 nm/min, 1-nm bandwidth, 0.2-nm data pitch, and 1-s data integration time [DIT]) were averaged, baselines were subtracted, and no smoothing was applied. Data are presented as the residue ellipticity ([ϴ]MRW) (MRW, mean residue weight) calculated using the molar concentration of protein and number of residues.
Secondary-structure analyses using the CDSSTR (22, 23), CONTINLL (24, 25), and SELCON3 (26, 27) algorithms were performed on the CD data with the Dichroweb analysis server (28, 29), using both reference data sets 3 and 6. The results from the three algorithms were averaged, and the standard deviations of the results of comparisons of the calculated secondary structures are given in Results (Table 2). The accuracy of the analysis was checked using the goodness-of-fit parameter (normalized root mean square deviation [NRMSD]) values. With both CDSSTR and CONTINLL, NRMSD values below 0.1 indicated a good correspondence between the calculated secondary-structure data and the experimental CD data. In contrast, SELCON3 yielded higher NRMSD values (ranging between 0.09 and 0.16); nevertheless, the data obtained with this algorithm and with the other two were identical within the error limit.
TABLE 2.
Calculated secondary structure derived from the far-UV CD spectra
| Enzyme | Secondary-structure values |
|||
|---|---|---|---|---|
| Helices (%) | Strands (%) | Turns (%) | Unordered (%) | |
| NDM-1 wta | 33 ± 2 | 17 ± 1 | 20 ± 2 | 30 ± 2 |
| NDM-1 I35S | 19 ± 1 | 32 ± 7 | 21 ± 0.4 | 28 ± 6 |
| NDM-1 I35T | 11 ± 2 | 34 ± 3 | 21 ± 2 | 34 ± 3 |
wt, wild type.
Molecular modeling.
The X-ray structure of NDM-1 in complex with cephalexin (PDB code 4RL2) (30) was used for the initial positioning of loracarbef in the active site of NDM-1 before proceeding with a standard energy minimization protocol with the program YASARA (31). This protocol consists of steepest-descent minimization followed by simulated annealing of the ligand and protein side chains. Simulation parameters consist of the use of a Yasara2 force field (32), a cutoff distance of 7.86 Å, particle mesh Ewald (PME) long-range electrostatics (33), periodic boundary conditions, and a water-filled simulation cell.
RESULTS
NDM-1I35T and NDM-1I35S mutants were obtained by site-directed mutagenesis by using NDM-1 as the template. The NDM-1 enzyme is unstable when it is produced with its native signal peptide (34). For this reason, blaNDM-1, blaNDM-1I35T, and blaNDM-1I35S genes were cloned into pET-24(a) vector without signal peptide for protein expression and purification. The wild-type and mutant enzymes were purified from E. coli JM109(DE3)/pFM-NDM-1, E. coli JM109(DE3)/pFM-NDM-1I35T, and E. coli JM109(DE3)/pFM-NDM-1I35S by two chromatographic steps which yielded the enzyme as more than 95% pure, as evaluated by SDS-PAGE analysis.
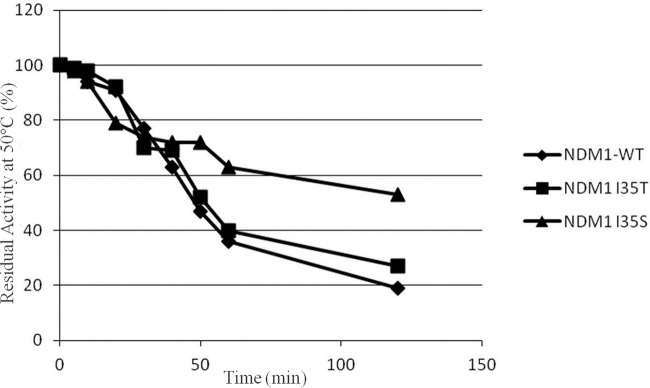
Before starting with analysis of the kinetic parameters, analysis of the stability of enzymes NDM-1, NDM-1I35T, and NDM-1I35S was performed by incubating the enzymes at 30°C, 40°C, and 50°C for up to 120 min in buffer C (without zinc ions). At 30°C and 40°C, all three enzymes followed the same trend, and their residual activity after 120 min of incubation was about 60%. Indeed at 50°C, NDM-1 and NDM-1I35T showed, after 120 min of incubation, 20% and 30% residual activity, respectively. In contrast, NDM-1I35S showed a residual activity level of approximately 50% under the same conditions (Fig. 1). The experiments were replicated but using a buffer C with the addition of 20 μM ZnCl2. At 30°C, 40°C, and 50°C, after 120 min of incubation, the residual activity seen with all three enzymes ranged between 90% and 100% (data not shown). Therefore, the three enzymes were in all cases stable in the presence of zinc.
FIG 1.
Thermal stability of NDM-1 mutants in comparison with that of the NDM-1 enzyme at 50°C. Data are mean values of the results of three measurements. The standard deviation was always lower than 10%. The experiments were performed as described in the text. WT, wild type.
The conformations of the NDM-1, NDM-1I35T, and NDM-1I35S enzymes were studied by analysis of fluorescence spectra using a 200 nM concentration of each enzyme. The most intense fluorescence peaks were at about 345 nm, but the three enzymes showed different levels of fluorescence intensity (data not shown). The arbitrary fluorescence unit values determined for NDM-1, NDM-1I35T, and NDM-1I35S were about 240, 140, and 296, respectively. The secondary-structure content was studied by far-UV CD spectra (Fig. 2) and analyzed by using three different algorithms. The levels of helical content calculated from these spectra were 33% ± 2% for NDM-1, 19% ± 1% for NDM-1I35S, and 11% ± 2% for NDM-1I35T (Table 2). Comparing with NDM-1, the NDM-1I35S and NDM-1I35T mutants showed 10% and 20% reductions in α-helix content, respectively.
FIG 2.
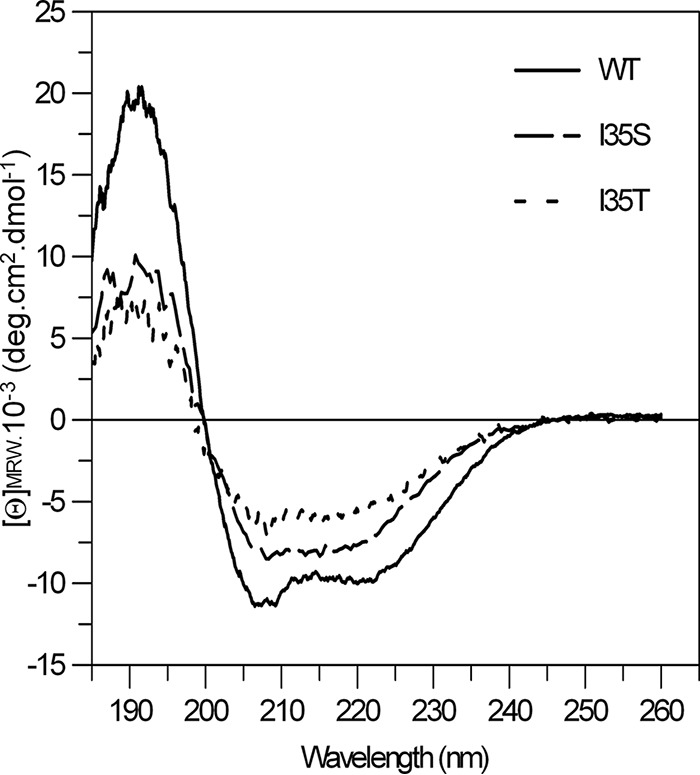
Far-UV CD spectra of the NDM-1 wild-type strain and the NDM-1I35S and NDM-1I35T mutants. Data were obtained at 20°C using 20 mM phosphate buffer, with protein concentrations of ca. 2 to 4 μM.
The effect of the zinc ion was assessed by spectrophotometric assay using 100 μM imipenem as the substrate and ZnCl2 concentrations ranging from 0 to 100 μM. As shown in Table 3, an increase in specific activity was observed in NDM-1 and NDM-1I35T at 20 μM ZnCl2, but addition of Zn+2 at higher concentrations did not affect the activity versus that seen with imipenem. In contrast, the addition of ZnCl2 did not alter the imipenem hydrolysis in the NDM-1I35S mutant. On the basis of these results, a concentration of 20 μM ZnCl2 was used for kinetic assays.
TABLE 3.
Initial rates of hydrolysis for NDM-1, NDM-1I35T, and NDM-1I35S at different concentrations of ZnCl2
| Enzyme |
v0 (mmol min−1 mg−1)a |
|||
|---|---|---|---|---|
| 0 μM Zn2+ | 20 μM Zn2+ | 50 μM Zn2+ | 100 μM Zn2+ | |
| NDM-1 | 59 | 130 | 134 | 130 |
| NDM-1I35T | 27 | 80 | 70 | 70 |
| NDM-1I35S | 99 | 99 | 107 | 103 |
Rates of hydrolysis (v0) are expressed in millimoles per milligram of imipenem (100 μM) hydrolyzed per minute per milligram of enzyme.
The NDM-1, NDM-1I35T, and NDM-1I35S enzymes were tested versus 16 β-lactam substrates that were all hydrolyzed by the three enzymes with the exception of aztreonam (Table 4). Benzylpenicillin and carbenicillin are good substrates for the three enzymes, with similar Km and kcat values, with the exception of kcat for NDM-1I35T, where the values are reduced by 50%. Even when the kcat values calculated for imipenem and biapenem for NDM-1I35S were higher than those calculated for NDM-1 and NDM-1I35T, their catalytic efficiencies were similar. The Km of NDM-1I35S for biapenem was 1,000 μM, about 10-fold higher than that of NDM-1. Regarding meropenem, the catalytic efficiencies of NDM-1I35T and NDM-1I35S mutants were lower than that of NDM-1. This is due to the low kcat values of the mutants in comparison to the wild-type enzyme.
TABLE 4.
Kinetic parameters of the NDM-1, NDM-1I35T, and NDM-1I35S enzymes for some β-lactams
| Substrate | NDM-1 |
NDM-1I35T |
NDM-1I35S |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM s−1) | |
| Benzylpenicillin | 250 ± 10 | 105 | 0.42 | 250 ± 10 | 58 | 0.23 | 380 ± 23 | 130 | 0.34 |
| Carbenicillin | 285 ± 5 | 108 | 0.38 | 650 ± 36 | 62 | 0.09 | 680 ± 30 | 163 | 0.24 |
| Imipenem | 35 ± 1 | 64 | 1.83 | 42 ± 3 | 24.5 | 0.58 | 90 ± 6 | 107 | 1.19 |
| Meropenem | 80 ± 2 | 75 | 0.94 | 90 ± 5 | 27 | 0.30 | 60 ± 4 | 35 | 0.58 |
| Biapenem | 120 ± 4 | 30 | 0.25 | 300 ± 8 | 19 | 0.06 | 1,000 ± 40 | 141 | 0.14 |
| Cefazolin | 20 ± 1 | 42 | 2.10 | 14 ± 2 | 12 | 0.86 | 21 ± 3 | 28 | 0.90 |
| Cefoxitin | 26 ± 1 | 23 | 0.88 | 18 ± 1 | 11 | 0.61 | 65 ± 4 | 44 | 0.68 |
| Cefaclor | 30 ± 2 | 31 | 1.03 | 35 ± 3 | 12 | 0.34 | 14 ± 1 | 6 | 0.43 |
| Loracarbef | 40 ± 2 | 42 | 1.05 | 9 ± 1 | 30 | 3.33 | 4 ± 0.5 | 56 | 14.0 |
| Moxalactam | 22 ± 1 | 15 | 0.68 | 50 ± 7 | 7 | 0.14 | 60 ± 5 | 17 | 0.28 |
| Cefotaxime | 14 ± 1 | 20 | 1.43 | 8 ± 3 | 5 | 0.63 | 3 ± 0.3 | 11 | 3.67 |
| Ceftazidime | 50 ± 4 | 18.5 | 0.37 | 44 ± 5 | 6 | 0.14 | 40 ± 2 | 11 | 0.28 |
| Cefepime | 35 ± 5 | 13 | 0.37 | 40 ± 3 | 6 | 0.15 | 32 ± 1 | 6 | 0.19 |
| Aztreonam | NHa | NH | NH | NH | NH | NH | NH | NH | NH |
| Sulbactam | 1,400 ± 50 | 50 | 0.036 | 1,100 ± 45 | 19 | 0.017 | 1,300 ± 32 | 66 | 0.05 |
| Nitrocefin | 15 ± 3 | 10 | 0.67 | 21 ± 4 | 6 | 0.29 | 20 ± 2 | 7 | 0.35 |
NH, not hydrolyzed.
The NDM-1, NDM-1I35T, and NDM-1I35S enzymes were probed against first-, second-, third-, and fourth-generation cephalosporins. Compared with NDM-1, the mutant enzymes showed a decrease of kcat and kcat/Km values for cefazolin. In the case of second-generation cephalosporins, different results were observed for cephamycin (cefoxitin), carbacephems (cefaclor and loracarbef), and oxacephem (moxalactam). The kcat values for NDM-1, NDM-1I35T, and NDM-1I35S for cefoxitin were 23 s−1, 11 s−1, and 44 s−1, respectively, but the catalytic efficiency values are similar. Toward cefaclor and moxalactam, the three enzymes showed the same behavior: a reduction of kcat and kcat/Km values was observed. Interestingly, the NDM-1I35S mutant showed a kcat/Km value versus loracarbef about 14-fold higher than that of NDM-1. A significant reduction of the Km value in NDM-1I35S with respect to the wild type was also observed. The fourth-generation cephalosporin tested, cefepime, was less hydrolyzed by the mutants than by NDM-1. Aztreonam was not hydrolyzed by the three enzymes. Sulbactam, a serine-β-lactamase inhibitor, was well hydrolyzed by the three enzymes but with high (>1,000 μM) Km values.
In vitro susceptibility to various β-lactams was investigated for E. coli JM109(DE3)/pSP-NDM-1, E. coli JM109(DE3)/pSP-NDM-1I35S, and E. coli JM109(DE3)/pSP-NDM-1I35T in comparison with the E. coli JM109(DE3)/pET-24(a) recipient. Results of these experiments showed that production of the NDM-1, NDM-1I35T, and NDM-1I35S enzymes was readily able to confer resistance to all of the β-lactams used with the exception of aztreonam (Table 5).
TABLE 5.
Patterns of β-lactam resistance mediated by NDM-1, NDM-1I35S, and NDM-1I35T in E. coli JM109(DE3) in comparison with the pattern in E. coli JM109(DE3)/pET-24(a)
| β-Lactam | MIC (μg/ml) |
|||
|---|---|---|---|---|
| E. coli JM109(DE3)/pSP-NDM-1 | E. coli JM109(DE3)/pSP-NDM-1I35S | E. coli JM109(DE3)/pSP-NDM-1I35T | E. coli JM109(DE3)/pET-24(a) | |
| Imipenem | >64 | >64 | >64 | 0.5 |
| Meropenem | >64 | >64 | >64 | 0.5 |
| Biapenem | 16 | 16 | 64 | 0.125 |
| Cefazolin | 256 | 128 | 256 | <0.0625 |
| Cefotaxime | 128 | 32 | 64 | <0.0625 |
| Ceftazidime | 256 | 128 | 256 | <0.0625 |
| Cefoxitin | 8 | 16 | 32 | <0.125 |
| Cefepime | 32 | 16 | 32 | <0.0625 |
| Loracarbef | 128 | 128 | 128 | <0.0625 |
| Cefaclor | 64 | 64 | 128 | 0.5 |
| Moxalactam | 64 | 32 | 64 | <0.125 |
| Benzylpenicillin | 512 | 256 | >512 | 0.5 |
| Carbenicillin | 512 | 256 | >512 | 0.5 |
DISCUSSION
NDM-1 metallo-β-lactamase is an extended-spectrum enzyme with an ability to hydrolyze most β-lactams, with the exception of monobactams. Unlike such other subclass B1 examples as IMP and VIM enzymes, NDM-1 has a long N terminus forming two extra strands designated β1′ and β1″ (13). In addition, NDM-1 has a wider L3 loop. This flexible loop is present in most subclass B1 MBLs, and it is involved in substrate binding and specificity (13). In contrast to IMP and VIM variants, in NDM-1 enzyme an isoleucine residue at position 35 connects the two extra β-strands positioned at the N terminus of the enzyme. Isoleucine 35 is at the tip of the β1 strand, and it is oriented toward the active site and seems to be correlated with substrate binding and the hydrophobicity of the L3 loop (13). In the present study, two NDM-1 laboratory mutants, obtained by replacing the isoleucine at position 35 (I35) with a serine (NDM-1I35S mutant) or a threonine (NDM-1I35T mutant), were generated to elucidate the role of isoleucine in the structure, substrate binding, and hydrophobicity of the enzyme. To date, experimental studies regarding this residue in NDM-1 have not been available. The wild type and the mutants were overexpressed and purified without signal peptide because of their instability, as previously reported by Kim and coworkers (34). The temperature stabilities of the three enzymes at 30°C and 40°C were similar. However, when the enzymes were incubated at 50°C the NDM-1I35S mutant was more stable than the wild-type and NDM-1I35T enzymes. Fluorescence assays showed changes in the intensities of the spectra among the three enzymes (data not shown) even if any shift in the maximum emission wavelength (lambda max) was observed in the mutants. The absence of change in the lambda max indicates that the exposure of tryptophan residues to solvent does not change in the mutants. This means that the overall three-dimensional (3D) structure was unchanged. Combined with far-UV CD data, this suggests that conformational changes are restricted to the local conformation of the backbone (i.e., the secondary structure) and do not perturb significantly the 3D structure.
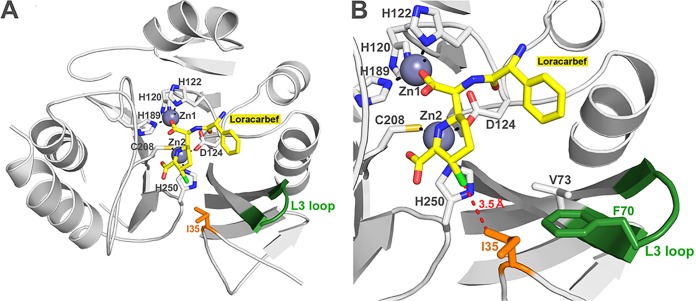
Kinetic data on NDM-1I35T and NDM-1I35S show that, even though a hydrophobic residue such as isoleucine was replaced by two polar residues (serine and threonine), the enzymes maintained high catalytic efficiencies. The I35 is packed against the L3 loop and is located in the proximity of the substrate binding site (13). It is only 5.2 Å away from the two methyl groups bound on the C6 carbon of ampicillin in the NDM-1–ampicillin complex structure (PDB code 3Q6X) (data not shown). Despite the introduction of polar residues, differences between the two mutants in the activities directed toward the substrates tested were observed. In particular, in comparison to NDM-1, NDM-1I35T exhibited lower kcat and catalytic efficiencies toward all β-lactams tested with the exception of loracarbef. This decrease of enzyme activity could be due to a different orientation of the threonine versus the active site because of its less bulky side chain compared to isoleucine. Nevertheless, the substitution of the I35 into a threonine results in retention of most of the shape. Compared to NDM-1I35T, the mutation of I35 into a serine adds some flexibility to the L3 loop. The introduction of a serine residue at this position maintains the catalytic efficiencies for all β-lactams tested, except loracarbef. For loracarbef, the kcat/Km value is 14-fold higher than that of NDM-1. The increase of catalytic efficiency is due to the lower Km value (4 μM) compared to NDM-1 (Km = 40 μM). The serine residue seems to react better with loracarbef. This is likely due to the proximity of the chlorine atom of this substrate and the hydroxyl group of the serine (Fig. 3), leading to the potential formation of a hydrogen bond during substrate hydrolysis responsible for the higher affinity observed. Interestingly, NDM-1 showed similar levels of behavior in terms of Km, kcat, and kcat/Km toward loracarbef and cefaclor, two second-generation cephalosporins with the same R1 and R2 groups (data not shown). In contrast, the introduction of polar residues changes the kcat/Km values of mutants versus loracarbef only and not versus cefaclor. The slightly differences of conformation among the six-member rings of loracarbef and cefaclor therefore seem sufficient to prevent an efficient interaction during cefaclor hydrolysis. The catalytic efficiency of NDM-1I35S toward biapenem is only slightly lower than that of the wild-type enzyme. It is, however, the result of an 8-fold Km increase compensated by a 4-fold kcat increase. This behavior could be due to the reduced compatibility between the bulky and rigid charged side chain of biapenem and the more flexible and polar side chain of a serine at position 35. In conclusion, mutation at position 35 induces some structural changes, which can be assigned to a 10% to 20% loss of α-helical structure. The I35 is important for the enzyme structure but not for substrate binding and catalysis. However, the mutants remain folded and well organized but the secondary-structure modification that occurs locally at the site of the mutation seems to influence the activity against only some substrates (biapenem and loracarbef).
FIG 3.
Molecular modeling of the NDM-1 in complex with hydrolyzed loracarbef. (A) NDM-1 in complex with loracarbef. (B) Overview of loop L3 focused on residue Ile-35. The distance between Ile-35 and loracarbef is shown as a red dashed line.
ACKNOWLEDGMENTS
We thank Anna Toso (Toronto Catholic District School Board, Toronto, Canada) for the language revision of the manuscript.
F.K. is a research associate of the FRS-FNRS (Brussels, Belgium). Research in Liège is supported in part by the Belgian program of Interuniversity Attraction Poles initiated by the Federal Office for Scientific Technical and Cultural Affaires (PAI no. P7/44).
We declare that we have no conflicts of interest.
REFERENCES
- 1.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carattoli A, Fortini D, Galetti R, Garcia-Fernandez A, Nardi G, Orazi D, Capone A, Majolino I, Proia A, Mariani B, Petrosillo N. 2013. Isolation of NDM-1-producing Pseudomonas aeruginosa sequence type ST235 from a stem cell transplant patient in Italy, May 2013. Euro Surveill 18:pii=20633. doi: 10.2807/1560-7917.ES2013.18.46.20633. [DOI] [PubMed] [Google Scholar]
- 3.Sartor AL, Raza MW, Abbasi SA, Day KM, Perry JD, Paterson DL, Sidjabat HE. 2014. Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob Agents Chemother 58:5589–5593. doi: 10.1128/AAC.02425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olaitan AO, Diene SM, Gupta SK, Adler A, Assous MV, Rolain JM. 2014. Genome analysis of NDM-1 producing Morganella morganii clinical isolate. Expert Rev Anti Infect Ther 12:1297–1305. doi: 10.1586/14787210.2014.944504. [DOI] [PubMed] [Google Scholar]
- 5.Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, Rolain J. 2014. New Delhi metallo-β-lactamase around the world: an eReview using Google maps. Euro Surveill 22:pii=20809. doi: 10.2807/1560-7917.ES2014.19.20.20809. [DOI] [PubMed] [Google Scholar]
- 6.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 7.Kutumbaka KK, Hans S, Mategko J, Nadala C, Buser GL, Cassidy MP, Beldavs ZG, Weissman SJ, Morey KE, Vega R, Samadpour M. 2014. Draft genome sequence of blaNDM-1-positive Escherichia coli O25b-ST131 clone isolated from an environmental sample. Genome Announc 2:pii:e00462–14. doi: 10.1128/genomeA.00462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnin RA, Poirel L, Carattoli A, Nordmann P. 2012. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PloS One 7:e34752. doi: 10.1371/journal.pone.0034752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dolejska M, Villa L, Poirel L, Nordmann P, Carattoli A. 2013. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J Antimicrob Chemother 68:34–39. doi: 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- 10.Page MI, Badarau A. 2008. The mechanisms of catalysis by metallo beta-lactamases. Bioinorg Chem Appl 2008:576297. doi: 10.1155/2008/576297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas PW, Zheng M, Wu S, Guo H, Liu D, Xu D, Fast W. 2011. Characterization of purified New Delhi metallo-β-lactamase-1. Biochemistry 50:10102–10113. doi: 10.1021/bi201449r. [DOI] [PubMed] [Google Scholar]
- 12.King D, Strynadka N. 2011. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci 20:1484–1491. doi: 10.1002/pro.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang HO, Hau Q. 2011. Crystal structure of NDM-1 reveals a common β-lactam hydrolysis mechanism. FASEB J 25:2574–2582. doi: 10.1096/fj.11-184036. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Cunningham MA, Mire J, Tesar C, Sacchettini J, Joachimiak A. 2013. NDM-1, the ultimate promiscuous enzyme: substrate recognition and catalytic mechanism. FASEB J 27:1917–1927. doi: 10.1096/fj.12-224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meini MR, Llarrull LI, Vila AJ. 2014. Evolution of metallo-beta-lactamases: trends revealed by natural diversity and in vitro evolution. Antibiotics 3:285–316. doi: 10.3390/antibiotics3030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using a polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. Seventh edition. Document M7-A7, 26 (2). CLSI, Wayne, PA, USA. [Google Scholar]
- 18.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Segel IH. 1976. Biochemical calculations, 2nd ed, p 236–241. John Wiley & Sons, New York, NY. [Google Scholar]
- 21.De Meester F, Joris B, Reckinger G. 1987. Automated analysis of enzyme inactivation phenomena. Application to β-lactamases and DD-peptidases. Biochem Pharmacol 36:2393–2403. [DOI] [PubMed] [Google Scholar]
- 22.Manavalan P, Johnson WC Jr. 1987. Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal Biochem 167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 23.Sreerama N, Woody RW. 2000. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem 287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 24.Provencher SW, Glöckner J. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 25.van Stokkum IH, Spoelder HJ, Bloemendal M, van Grondelle R, Groen FC. 1990. Estimation of protein secondary structure and error analysis from circular dichroism spectra. Anal Biochem 191:110–118. doi: 10.1016/0003-2697(90)90396-Q. [DOI] [PubMed] [Google Scholar]
- 26.Sreerama N, Woody RW. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem 209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 27.Sreerama N, Venyaminov SY, Woody RW. 1999. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci 8:370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitmore L, Wallace BA. 2004. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res 32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitmore L, Wallace BA. 2008. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers 89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 30.Feng H, Ding J, Zhu D, Liu X, Xu X, Zhang Y, Wang DC, Liu W. 2014. Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins. J Am Chem Soc 136:14694–14697. doi: 10.1021/ja508388e. [DOI] [PubMed] [Google Scholar]
- 31.Krieger E, Darden T, Nabuurs SB, Finkelstein A, Vriend G. 2004. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- 32.Krieger E, Joo K, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K. 2009. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins 77:114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. 1995. A smooth particle mesh Ewald method. J Chem Phys 103:8577. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 34.Kim Y, Tesar C, Mire J, Jedrzejczak R, Binkowski A, Babnigg G, Sacchettini J, Joachimiak A. 2011. Structure of apo- and monometalated forms of NDM-1-A highly potent carbapenem-hydrolyzing metallo-β-lactamase. PloS One 6:e24621. doi: 10.1371/journal.pone.0024621. [DOI] [PMC free article] [PubMed] [Google Scholar]