Abstract
We report for the first time the isolation of CTX-M-15-producing Escherichia coli strains belonging to sequence type (ST) 410, ST224, and ST1284 in commercial swine in Brazil. The blaCTX-M-15 gene was located on F-::A9::B1 and C1::A9::B1 IncF-type plasmids, surrounded by a new genetic context comprising the IS26 insertion sequence truncated with the ISEcp1 element upstream of blaCTX-M-15. These results reveal that commercial swine have become a new reservoir of CTX-M-15-producing bacteria in South America.
TEXT
The rapid spread of the blaCTX-M-15 gene, which encodes the currently most widely distributed extended-spectrum β-lactamase (ESBL) enzyme in Gram-negative bacteria, is a global challenge to human and veterinary medicine. In this context, food-producing animals have acquired an important role as reservoirs for CTX-M-15-producing bacteria, which can be transmitted to the community (1, 2). In this report, we describe for the first time the isolation of CTX-M-15-producing Escherichia coli in commercial swine in Brazil, highlighting a new reservoir of CTX-M-15-producing isolates with zoonotic potential in South America.
In 2012, during a study conducted to assess the occurrence of ESBL-producing bacteria in swine production, eight (3%) ceftiofur-resistant CTX-M-15-producing E. coli isolates were recovered from 267 fecal swabs collected from male and female nursery (40 days) and finishing (90 days) pigs from 28 farms in seven Brazilian states. In this regard, ceftiofur-resistant strains were isolated by using MacConkey agar supplemented with 2 μg/ml of ceftiofur. E. coli isolates were identified by the MALDI Biotyper (Bruker Daltonics, Germany), and the antibiotic resistance profiles were determined by the Kirby-Bauer method, with MICs determined by the microdilution technique by using Sensititre ESBL-confirmatory MIC plates (Trek Diagnostic Systems, Thermo Fisher) and/or the agar dilution method (3, 4). ESBL production was screened by the double-disk synergy test, and the presence of blaCTX-M-type genes was examined by PCR amplification and sequencing. E. coli isolates were further characterized by phylogenetic grouping (5) and multilocus sequence typing (MLST) (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). Next, plasmids were extracted and used to transform electrocompetent E. coli TOP10 recipient cells (Invitrogen) by electroporation. The transformant E. coli TOP10 strains were selected on Mueller-Hinton agar supplemented with 2 μg/ml of cefotaxime and further analyzed by replicon typing (6), plasmid MLST experiments (http://pubmlst.org/plasmid/), and partial plasmid sequencing to elucidate the blaCTX-M-15 genetic environment.
The eight ceftiofur-resistant E. coli isolates produced CTX-M-15 and exhibited high MICs of human and veterinary cephalosporins (Table 1). Additionally, E. coli strains were resistant to ciprofloxacin, enrofloxacin, norfloxacin, tetracycline, sulfonamide-trimethoprim, and gentamicin (7). Otherwise, all isolates remained susceptible to amikacin, cephamycins, and carbapenems. The isolates belonged to three clonal lineages of sequence type (ST) 224 (5 strains from farm X in Minas Gerais State, southeastern Brazil), ST410 (CC23; 1 strain from farm Y in Minas Gerais State), and ST1284 (2 strains from farm Z in Paraná State, southern Brazil). While none of the eight isolates belonged to phylogroup B2, phylogroups A and B1 were identified among CTX-M-15-producing E. coli strains. For E. coli ST224 and ST410 strains, the blaCTX-M-15 gene was successfully transferred to TOP10 E. coli isolates by transformation, being associated with the presence of IncF incompatibility plasmid groups, with sizes ranging from 40 to 90 kb (Table 1). Additional resistance to gentamicin, sulfonamides, and tetracycline was cotransferred, and the presence of aacA4, aac(3)-IIa, aadA1, sul1, and tetA genes was confirmed in both donor and receptor E. coli strains, whereas PCR analysis of 16S rRNA methylase genes was negative. On the other hand, fluoroquinolone resistance was not cotransferred, and, indeed, no plasmid-mediated quinolone resistance (PMQR) genes were found.
TABLE 1.
Epidemiological and microbiological characteristics of CTX-M-15-producing E. coli strains isolated from commercial swine, E. coli TOP10 (transformed) and E. coli TOP10
| E. coli strain | Farm/stateb/yr | ST/phylogroup | MIC (μg/ml)c for: |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CEF | CTX | CRO | EFT | CPD | CAZ | FEP | FOX | IPM | MER | CIP | ENO | NOR | GEN | AMI | TET | SUL | SXT | |||
| 180A | X/MG/2012 | 224/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 16 | 8 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 16 | ≥128 | ≥512 | ≥32 |
| T-180A | ≥32 | ≥32 | 32 | 8 | 16 | ≥64 | 4 | 2 | ≤4 | ≤0.5 | ≤1 | 0.0035 | 0.007 | 0.06 | ≤4 | 2 | 4 | ≤12 | ≤2 | ||
| 180B | X/MG/2012 | 224/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 16 | 16 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 16 | ≥128 | ≥512 | ≥32 |
| 181 | X/MG/2012 | 224/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 8 | 8 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 16 | ≥128 | ≥512 | ≥32 |
| T-181 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 8 | 8 | ≤0.5 | ≤1 | 0.0035 | 0.007 | 0.12 | ≥32 | 2 | ≥128 | ≥512 | ≤2 | ||
| 187a | X/MG/2012 | 224/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 16 | 16 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 16 | ≥128 | ≥512 | ≥32 |
| T-187a | ≥32 | ≥32 | ≥128 | 128 | ≥128 | ≥64 | 16 | 8 | ≤4 | ≤0.5 | ≤1 | 0.0035 | 0.0035 | 0.06 | ≥32 | 2 | 4 | ≥512 | ≤2 | ||
| 188 | X/MG/2012 | 224/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 16 | 16 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 8 | ≥128 | ≥512 | ≥32 |
| T-188 | ≥32 | ≥32 | ≥128 | ≥256 | 128 | ≥64 | 16 | 16 | 8 | ≤0.5 | ≤1 | 0.0035 | 0.007 | 0.12 | ≥32 | 2 | 4 | ≥512 | ≤2 | ||
| 223Ba | Y/MG/2012 | 410/A | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 32 | 16 | 16 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≤4 | 8 | ≥128 | ≥512 | ≥32 |
| T-223Ba | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 32 | 16 | ≤4 | ≤0.5 | ≤1 | 0.0035 | 0.0035 | 0.12 | ≤4 | 8 | ≥128 | ≥512 | ≤2 | ||
| 468 | Z/PR/2012 | 1284/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | 16 | 16 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 16 | ≥128 | ≥512 | ≥32 |
| 470 | Z/PR/2012 | 1284/B1 | ≥32 | ≥32 | ≥128 | ≥256 | ≥128 | ≥64 | 16 | ≥32 | 8 | ≤0.5 | ≤1 | ≥64 | ≥64 | ≥128 | ≥32 | 8 | ≥128 | ≥512 | ≥32 |
| TOP10 | ≤8 | ≤8 | ≤0.2 | ≤1 | 0.5 | 1 | 0.5 | ≤1 | ≤4 | ≤0.5 | ≤1 | 0.0035 | 0.0035 | 0.06 | ≤4 | 2 | 4 | ≤12 | ≤2 | ||
Representative IncF-positive plasmids from E. coli ST224 and ST410 were selected for replicon sequence typing, obtaining the FAB allele formulas F-::A9::B1 and C1::A9::B1, respectively.
MG, Minas Gerais State; PR, Paraná State.
MICs were determined by microdilution technique using Sensititre ESBL-confirmatory MIC plates (TREK Diagnostic Systems, Thermo Fisher) or agar dilution methods; resistance is indicated in bold by using CLSI criteria (3, 4). AMP, ampicillin; CEF, cephalothin; CTX, cefotaxime; CRO, ceftriaxone; EFT, ceftiofur; CPD, cefpodoxime; CAZ, ceftazidima; FEP, cefepime; FOX, cefoxitin; IPM, imipenem; MEM, meropenem; CIP, ciprofloxacin; ENO, enrofloxacin; NOR, norfloxacin; GEN, gentamicin; AMI, amikacin; TET, tetracycline; SUL, sulfonamide; SXT, sulfamethoxazole-trimethoprim.
Moreover, representative IncF-positive plasmids from E. coli ST224 and ST410 were selected for partial plasmid sequencing and replicon sequence typing, yielding the FAB allele formulas F-:;A9:;B1 and C1:;A9:;B1, respectively (BioProject accession numbers PRJNA279532 and PRJNA291430). In this regard, reports on the FAB formula of ST410 (C1:;A9:;B1) have been rare among CTX-M-15 producers, since most studies have been limited to blaCTX-M-type identification, where plasmid characterization is restricted to Inc type and size determination (8). Therefore, this FAB formula has most likely been underestimated.
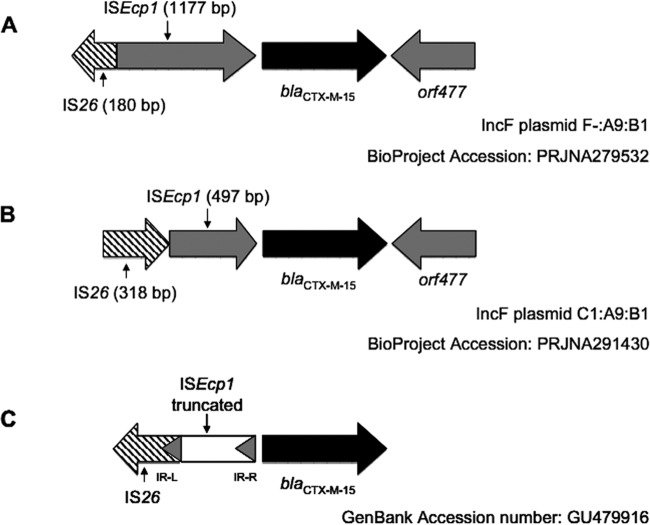
Finally, these results revealed that both plasmids carried an ISEcp1 truncated by an IS26 transposase upstream of the blaCTX-M-15 gene and an intact orf477 gene downstream from blaCTX-M-15 (Fig. 1).
FIG 1.
Schematic representations of the genetic environment surrounding the blaCTX-M-15 gene in swine E. coli isolates. (A) The plasmid blaCTX-M-15 gene from E. coli 181 ST224 (BioProject accession number PRJNA279532). (B) The plasmid blaCTX-M-15 gene from E. coli 223B ST410 (BioProject accession number PRJNA291430). (C) Genetic environment surrounding blaCTX-M-15 found in an E. coli strain belonging to ST410, isolated from clinical and food samples from southern Spain (8) (GenBank accession number GU479916). Sections, open reading frames and genes surrounding the blaCTX-M-15 gene; arrows, orientation of each coding sequence. Gene names are shown under the corresponding section.
In this study, we report for the first time the emergence of blaCTX-M-15-carrying E. coli in commercial healthy pigs in Brazil. In this regard, the identification of CTX-M-15-producing E. coli from commercial swine has until now been reported only from Asian and European countries (9, 10). Of particular interest is the description of CTX-M-15-producing E. coli belonging to ST410 (CC23), which was previously isolated from broiler and clinical samples, providing evidence for the transmission of CTX-M-15-producing E. coli between animals and humans (11). Indeed, CTX-M-15-producing E. coli ST410 (CC23), phylogroup A, has been predominant among ESBL-producing E. coli isolates from hospitals in Brazil (12). In contrast, E. coli strains belonging to ST224 have been identified in inpatients and outpatients and associated with the production of different clinically important β-lactamases, such as NDM-1, KPC-2, and even CTX-M-15 (13–15), and more recently, the production of CTX-M-8 in E. coli ST224 isolated from dairy buffalo was documented in Brazil (16). Representative E. coli strains belonging to ST410 and ST224 present distinct genetic arrangements upstream of the blaCTX-M-15 gene (Fig. 1). In this context, the ST223B strain presents the ISEcp1 flanked by IS26 upstream the blaCTX-M-15 gene in a similar structure described previously (17) but with a deletion of 727 bp in the ISEcp1 sequence. The blaCTX-M-15 gene was previously found together with ISEcp1 truncated by an IS26 element in E. coli strains belonging to ST410 from both clinical and food samples from southern Spain (8); however, IS26 was identified in the opposite orientation (Fig. 1). On the other hand, a new genetic context of blaCTX-M-15 was observed in E. coli strains belonging to ST224, which presents a 1,177-bp ISEcp1 truncated by an incomplete IS26 transposase upstream of the blaCTX-M-15 gene, similar to genetic structures found in CTX-M-15-producing E. coli isolated from bovine mastitis in the United Kingdom (18).
Finally, the detection of phylogenetic groups A and B1 in this study suggests that selection of silent blaCTX-M-15 carriers among commensal E. coli in healthy commercial swine is ongoing, which is a worrisome prospect, since ESBL-producing E. coli of commensal origin can play a key role as opportunistic pathogens in humans and other animals that can serve as hosts. Most likely, as previously hypothesized, therapeutic and prophylactic use of ceftiofur in the swine industry may be contributing to the selection and recovery of enteric E. coli with resistance to cephalosporin drugs (19), where blaCTX-M-type ESBL genes can rapidly disseminate among healthy pigs. However, given that all isolates were also resistant to fluoroquinolones, the use of enrofloxacin may have exerted a selection pressure. In this regard, in Brazil, both ceftiofur and enrofloxacin are used for the treatment of enteric, urinary, or systemic infections, and in some herds, ceftiofur is used for systematic prophylaxis in 1-day-old piglets.
In summary, surveillance of antimicrobial resistance in bacteria from food-producing animals and derived food products needs to be a priority. Moreover, strategies for the rational use of antimicrobial agents in food animals need to be undertaken urgently, in order to inhibit the release of bacteria harboring clinically important resistance genes. In this regard, the dissemination of cephalosporin-resistant bacteria carrying blaCTX-M-15 has the potential to impact both veterinary and human therapeutic treatment options.
ACKNOWLEDGMENT
FAPESP and CNPq research grants are gratefully acknowledged. A.M.M. and N.L. are research grant fellows of CNPq. K.C.S. received a postgraduate fellowship from FAPESP (process 2012/08332-0).
REFERENCES
- 1.Trott D. 2013. β-Lactam resistance in Gram-negative pathogens isolated from animals. Curr Pharm Des 19:239–249. doi: 10.2174/138161213804070339. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, Stephan R, Power K, Yan Q, Hächler H, Fanning S. 2014. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother 69:2658–2668. doi: 10.1093/jac/dku206. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; 2nd informational supplement. CLSI document VETO1-S2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 5.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 7.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.López-Cerero L, Egea P, Rodríguez-Baño J, Pascual A. 2011. Similarities between the genetic environments of blaCTX-M-15 in Escherichia coli from clinical and food samples from Spain and overseas travellers. J Antimicrob Chemother 66:2177. doi: 10.1093/jac/dkr274. [DOI] [PubMed] [Google Scholar]
- 9.Tian GB, Wang HN, Zou LK, Tang JN, Zhao YW, Ye MY, Tang JY, Zhang Y, Zhang AY, Yang X, Xu CW, Fu YJ. 2009. Detection of CTX-M-15, CTX-M-22, and SHV-2 extended-spectrum beta-lactamases (ESBLs) in Escherichia coli fecal-sample isolates from pig farms in China. Foodborne Pathog Dis 6:297–304. doi: 10.1089/fpd.2008.0164. [DOI] [PubMed] [Google Scholar]
- 10.Hammerum AM, Jakobsen L, Olsen SS, Agersø Y. 2012. Characterization of CTX-M-14- and CTX-M-15-producing Escherichia coli of porcine origin. J Antimicrob Chemother 67:2047–2049. doi: 10.1093/jac/dks148. [DOI] [PubMed] [Google Scholar]
- 11.López-Cerero L, Egea P, Serrano L, Navarro D, Mora A, Blanco J, Doi Y, Paterson DL, Rodríguez-Baño J, Pascual A. 2011. Characterisation of clinical and food animal Escherichia coli isolates producing CTX-M-15 extended-spectrum β-lactamase belonging to ST410 phylogroup A. Int J Antimicrob Agents 37:365–367. doi: 10.1016/j.ijantimicag.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Peirano G, Asensi MD, Pitondo-Silva A, Pitout JD. 2011. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin Microbiol Infect 17:1039–1043. doi: 10.1111/j.1469-0691.2010.03440.x. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Li W, Wang J, Pan J, Sun S, Yu Y, Zhao B, Ma Y, Zhang T, Qi J, Liu G, Lu F. 2013. Identification and characterization of the first Escherichia coli strain carrying NDM-1 gene in China. PLoS One 8:e66666. doi: 10.1371/journal.pone.0066666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baraniak A, Grabowska A, Izdebski R, Fiett J, Herda M, Bojarska K, Żabicka D, Kania-Pudło M, Młynarczyk G, Żak-Puławska Z, Hryniewicz W, Gniadkowski M, KPC-PL Study Group. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob Agents Chemother 55:5493–5499. doi: 10.1128/AAC.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. 2011. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect 17:1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 16.Aizawa J, Neuwirt N, Barbato L, Neves PR, Leigue L, Padilha J, Pestana de Castro AF, Gregory L, Lincopan N. 2014. Identification of fluoroquinolone-resistant extended-spectrum β-lactamase (CTX-M-8)-producing Escherichia coli ST224, ST2179 and ST2308 in buffalo (Bubalus bubalis). J Antimicrob Chemother 69:2866–2869. doi: 10.1093/jac/dku218. [DOI] [PubMed] [Google Scholar]
- 17.Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N. 2011. Variation in the genetic environments of blaCTX-M-15 in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012. doi: 10.1093/jac/dkr041. [DOI] [PubMed] [Google Scholar]
- 18.Timofte D, Maciuca IE, Evans NJ, Williams H, Wattret A, Fick JC, Williams NJ. 2014. Detection and molecular characterization of Escherichia coli CTX-M-15 and Klebsiella pneumoniae SHV-12 β-lactamases from bovine mastitis isolates in the United Kingdom. Antimicrob Agents Chemother 58:789–794. doi: 10.1128/AAC.00752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutz EA, McCarty MJ, Mollenkopf DF, Funk JA, Gebreyes WA, Wittum TE. 2011. Ceftiofur use in finishing swine barns and the recovery of fecal Escherichia coli or Salmonella spp. resistant to ceftriaxone. Foodborne Pathog Dis 8:1229–1234. doi: 10.1089/fpd.2011.0925. [DOI] [PubMed] [Google Scholar]