Abstract
Dietary consumption of long-chain omega-3 polyunsaturated fatty acids (n-3 PUFA) may protect against cardiometabolic disease through modulation of systemic and adipose inflammation. However it is often difficult to detect the subtle effects of n-3 PUFA on inflammatory biomarkers in traditional intervention studies. We aimed to identify novel n-3 PUFA modulated gene expression using unbiased adipose transcriptomics during evoked endotoxemia in a clinical trial of n-3 PUFA supplementation. We analyzed adipose gene expression using RNASeq in the fenofibrate and omega-3 fatty acid modulation of endotoxemia (FFAME) trial of healthy individuals at three timepoints: before and after n-3 PUFA supplementation (n=8; 3600mg/day EPA/DHA) for 6 weeks compared with placebo (n=6), as well as during a subsequent evoked inflammatory challenge (LPS 0.6 ng/kg I.V.). As expected, supplementation with n-3 PUFA vs. placebo alone had only modest effects on adipose tissue gene expression e.g., increased expression of immediate early response 2 (IER2). In contrast, the transcriptomic response to evoked endotoxemia was significantly modified by n-3 PUFA supplementation, with several genes demonstrating significant n-3 PUFA gene-nutrient interactions e.g., enhanced transcriptional responses in specific immune genes IER5L, HES1, IL1RN, CCL18, IL1RN, IL7R, IL8, CCL3 and others. These data highlight potential mechanisms whereby n-3 PUFA consumption may enhance the immune response to an inflammatory challenge. In conclusion, unbiased transcriptomics during evoked inflammation reveals novel immune modulating functions of n-3 PUFA nutritional intervention in a dynamic pathophysiological setting.
Keywords: n-3 PUFA, inflammation, LPS, RNASeq, evoked endotoxemia, adipose tissue
INTRODUCTION
Inflammation plays a central role in the human homeostatic response to environmental stimuli in the setting of pathogenic infection and injury as well as during chronic dietary and lifestyle stresses.[1] The ability to regulate an appropriate inflammatory response for a specific stimulus is crucial to maintain health and prevent the development and progression of disease. Obesity and diet-related adipose tissue inflammation is an important factor in chronic inflammatory cardiometabolic diseases.[2, 3]
Dietary components which modulate inflammation are of great interest in disease prevention and treatment. Long-chain omega-3 polyunsaturated fatty acids (n-3 PUFA) have been extensively studied for their anti-inflammatory and cardio-protective effects.[4–6] Supplementation with n-3 PUFA is thought to confer benefit against inflammatory cardiometabolic disease. However studies remain inconclusive and conflicting, likely due to the challenges inherent in human nutritional intervention studies,[7] population and context specific effects, and genetic differences between individuals in their responsiveness to dietary components.[8] In vitro studies have shown n-3 PUFA can reduce inflammation in adipocytes and macrophages.[6] It is thought that n-3 PUFA might exert disease protection through tissue-specific anti-inflammatory effects in adipose tissue,[9] yet anti-inflammatory effects in clinical and population studies are weak or inconsistent.[10]
Due to the subtle effects of nutritional interventions in the resting state, application of evoked phenotype modeling to nutrigenomic studies may have increased power to detect the effects of nutritional interventions that are most relevant to dynamic human pathophysiologies.[11, 12] We and others apply low-dose systemic lipopolysaccharide (LPS) administration (endotoxemia) as a model to study the inflammatory and metabolic impact of activation of innate immunity in humans and have described the response to LPS in healthy volunteers; the evoked transient systemic inflammation, adipose inflammation and insulin resistance resembles the chronic abnormalities seen in human cardiometabolic disease.[13–15] Furthermore, we have shown that LPS induces tissue-specific transcriptomic changes, of relevance to both inflammatory responses and to cardiometabolic disease risk.[16, 17]
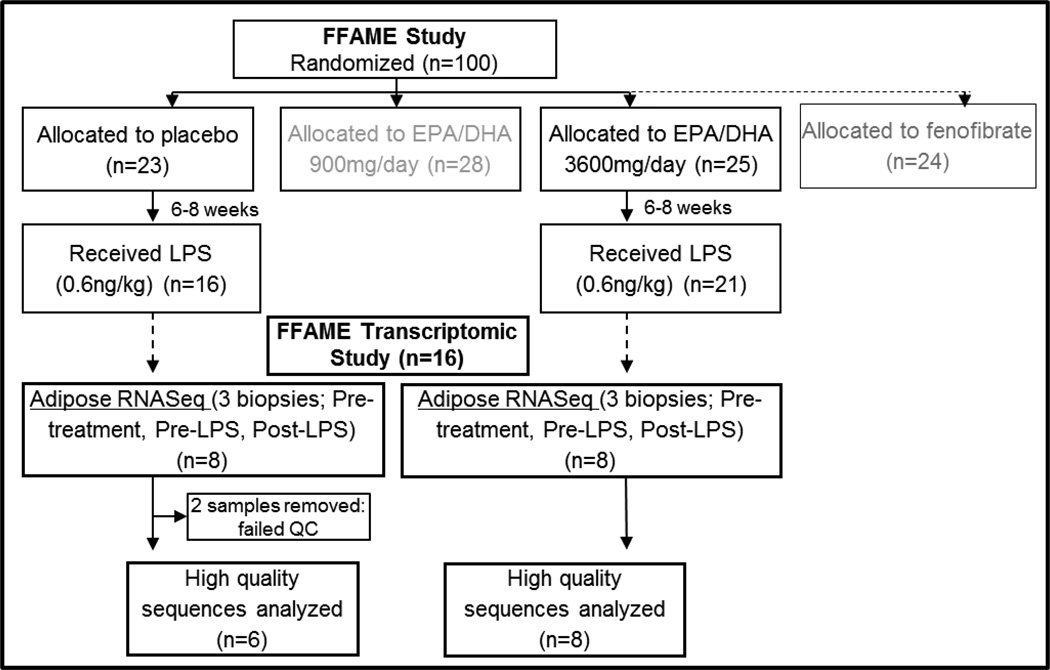
The fenofibrate and omega-3 fatty acid modulation of endotoxemia (FFAME) Study recruited healthy volunteers (N=80) to a University of Pennsylvania Clinical and Translational Research Center (UPenn CTRC) protocol.[13] Subjects were randomized to supplementation with n-3 PUFA or placebo, and completed an endotoxin challenge (LPS 0.6ng/kg) after 6–8 weeks treatment. Treatment with “high”-dose n-3 PUFA (3600mg/day EPA/DHA) led to a significant reduction in the febrile response to LPS, and a trend towards decreased cytokine response.[13] We hypothesized that treatment with n-3 PUFA would also alter the transcriptomic response to LPS in adipose tissue, and selected a subset of individuals (n=16) for adipose tissue transcriptomic analysis using RNA sequencing (RNASeq). We sequenced RNA from adipose tissue biopsies at 3 timepoints in all individuals: before intervention, following ~6 weeks of supplementation with n-3 PUFA (3600mg EPA/DHA, n=8) or placebo (n=6), and then following evoked endotoxemia (LPS 0.6 ng/kg), to identify novel immune-responsive genes altered by n-3 PUFA supplementation.
SUBJECTS AND METHODS
The FFAME Study: Human very low-dose endotoxemia
The FFAME Study recruited healthy volunteers (N=80) to an inpatient endotoxemia protocol at the University of Pennsylvania (UPenn), clinicaltrials.gov NCT01048502 as described.[13] Subjects were randomized to double-blind placebo-controlled treatment arms: placebo (corn oil), fish-oil derived omega-3-acid ethyl ester eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) (Lovaza, GlaxoSmithKline; 465mg EPA + 375mg DHA) supplemented at either “low-dose” (1/day, 900mg) or “high-dose” (4/day, 3600mg), or fenofibrate (Tricor, Abbott Laboratories) 145 mg/day. Each capsule contained α-tocopherol (4mg) as an antioxidant. The fenofibrate and n-3 PUFA arms of the trial were analyzed and reported separately.[13, 18] The treatment period ranged from 6 to 8 weeks, to allow for scheduling of the baseline and inpatient visits. The median treatment time was 48 days. All subjects completed an inpatient lipopolysaccharide (LPS) challenge at the UPenn Clinical and Translational Research Center (CTRC). Following overnight acclimatization, 0.6ng/kg U.S. standard reference endotoxin (lot No. CCRE-LOT-1+2; Clinical Center, Pharmacy Department at the National Institutes of Health, Bethesda, MD) was administered. Subjects remained confined to their bed during the post-LPS phase. Samples of gluteal subcutaneous adipose tissue were obtained at randomization, post-treatment pre-LPS, and four hours following LPS, as previously described.[16] The four hour post-LPS adipose biopsy was chosen based on our previous work highlighting significant tissue expression changes, concurrent with the systemic inflammatory response.[16] Adipose biopsies were snap-frozen for subsequent RNA extraction. The trial was conducted with the approval of the UPenn Institutional Review Board, and all participants provided written informed consent.
FFAME Transcriptomic sub-study
We previously reported that high, but not low-dose EPA/DHA significantly reduced the inflammatory response to LPS.[13] To interrogate this difference further, a subset of FFAME study participants were selected from the “high-dose” n-3 PUFA (3600mg/day EPA/DHA; n=8) and placebo (n=8) groups for in-depth adipose transcriptomic analyses using high-throughput RNA sequencing (RNASeq). Based on our previous work, [17] this number is well-powered to detect differences in gene and lincRNA expression. To minimize variation attributable to race or sex differences, we only included individuals of European Ancestry, 50% female, with high quality RNA. As described below, 2 subjects were removed from the placebo group during RNASeq quality control (QC) analysis. This included one male and one female subject, maintaining sex balance between the groups. There were no differences between the groups in baseline characteristics (Table 1). An overview of the FFAME transcriptomic study is shown in Figure 1.
Table 1.
Baseline characteristics of the FFAME Transcriptomic Sub-study sample
| Placebo (n=8) | n-3 PUFA (n=8) | ||||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | P Value | |
| Gender (n male/female) | 4/4 | 4/4 | |||
| Age (years) | 30.5 | 8.0 | 24.3 | 5.0 | 0.08 |
| BMI (kg/m2) | 23.95 | 3.7 | 23.35 | 2.9 | 0.73 |
| Blood Pressure (mmHg) | 114/68 | 15/9 | 114/69 | 4/5 | 0.99 |
| IL-6 (pg/ml) | 1.14 | 0.6 | 1.16 | 0.6 | 0.96 |
| TNFα (pg/ml) | 1.26 | 0.9 | 1.50 | 0.6 | 0.54 |
| IL-1RA (pg/ml) | 144.2 | 28.3 | 176.3 | 67.6 | 0.24 |
| IL-10 (pg/ml) | 0.81 | 0.1 | 0.89 | 0.3 | 0.48 |
| MCP-1 (pg/ml) | 142.1 | 41.0 | 139.5 | 38.5 | 0.90 |
BMI, Body mass index; IL-6, Interleukin 6; TNFα, Tumor necrosis factor alpha; IL-1RA, Interleukin 1 receptor agonist; IL-10, Interleukin 10; MCP-1, monocyte chemoattractant protein 1. P value obtained from independent samples T test.
Figure 1. Overview of the FFAME Transcriptomic Study.
European ancestry subjects receiving “high-dose” n-3 PUFA (3600mg/day EPA/DHA; n=8) vs. placebo (n=8) were selected for adipose tissue RNASeq at baseline (pre-treatment), after 6–8 weeks supplementation (pre-LPS), and 4-hours following LPS (post-LPS).
RNA extraction and expression analysis
RNA from FFAME Study adipose tissue biopsies (3 samples per subject, 48 samples total) was extracted using the RNeasy total RNA kit (Qiagen Inc., Valencia, CA).[17] RNA concentration and quality was assessed using an Agilent BioAnalyzer (Agilent, Santa Clara, CA). Strand-specific poly-A RNA libraries (TruSeq RNA Sample Preparation Kit, Illumina, San Diego, CA) were prepared as described.[17] Libraries were sequenced on an Illumina HiSeq 2000 sequencer, with 6 samples per lane (~30 million 2×101 bp paired-end reads per sample after filtering).
Bioinformatics and statistical analysis
RNASeq data were aligned to the hg19 reference genome using STAR 2.3.0e[19] with default options. Reads were required to be uniquely mapped and read pairs were required to be mapped to the same chromosome with a mapping distance < 500 kb between two reads. Expression levels (Fragments Per Kilobase of transcript per Million mapped reads; FPKM) were estimated for all known protein-coding genes, as well as all described long intergenic non-coding RNA (lincRNA) using a composite set of 54,944 distinct autosomal lincRNAs derived from four published lincRNA datasets.[20–23] LincRNAs which overlapped between the four datasets were merged if one exon of a lincRNA overlapped at least 50% of an exon from another lincRNA. RNA-SeQC[24] was used to assess quality of each sample. All samples within the n-3 PUFA group passed QC, with expected number of reads. Within the placebo subjects, two subjects were removed for poor sequencing quality leaving 6 subjects in the placebo group. For each protein-coding gene or lincRNA, the estimated expression level from pre-to post-treatment, or from pre-to post-LPS was compared within groups using the cuffdiff option in Cufflinks version 2.1.1. Gene-nutrient interaction analyses were carried out using linear regression models including interaction terms for treatment*time and treatment*LPS to detect the effects of supplementation alone, and supplementation effects on LPS response respectively. P values were adjusted for transcriptome-wide false discovery rate (FDR), with adjusted P<0.05 considered as significant. P values are reported after adjustment unless otherwise stated. P values for baseline group differences in clinical variables were obtained from independent samples T test (IBM SPSS Statistics 22). Functional enrichment and pathway analysis was carried out using DAVID.[25]
RESULTS
Supplementation for 6–8 weeks with n-3 PUFA or placebo in healthy individuals has limited effects on adipose tissue gene expression
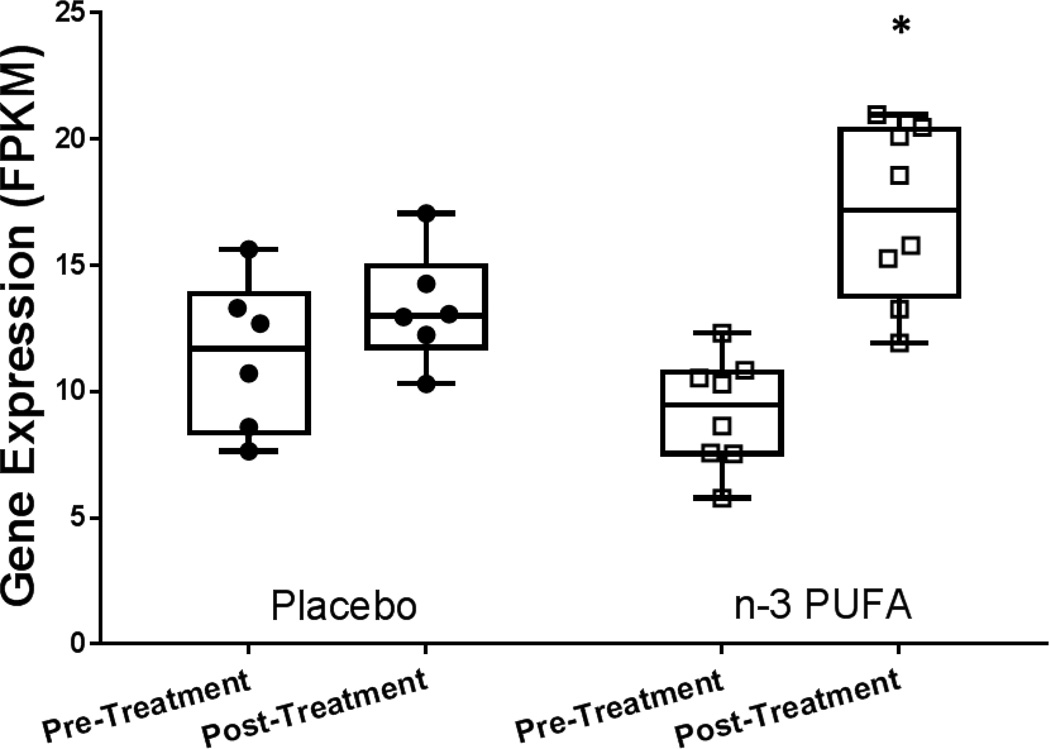
Gene expression was analyzed by RNASeq in adipose tissue before and after supplementation with n-3 PUFA or placebo. Out of 79,763 genes and lincRNAs expressed at either time-point, less than 0.1% were significantly altered within groups over the treatment period. The majority were genes with very modest expression changes consistent with stochastic changes in expression over the 6–8 weeks (Supplement Table 1). Between-group interaction analysis of adipose tissue gene expression from the pre-to post-treatment period confirmed limited effect of supplementation on gene expression in healthy individuals. There were no gene-nutrient interactions that reached significance after FDR correction. Within genes that had borderline significance for gene-nutrient expression (unadjusted P<0.05), IER2 (immediate early response 2) displayed evidence of gene-nutrient interaction (nominal P=0.01), and was significantly up-regulated in the n-3 PUFA group (P=0.02) and unchanged in the placebo group (P=0.2) (Figure 2).
Figure 2. Expression of immediate early response 2 (IER2) was significantly up-regulated in adipose tissue after 6–8 weeks supplementation with n-3 PUFA (adj. P=5×10−5), with evidence for gene-nutrient interaction compared with placebo treatment (nominal P=0.01).
Expression measured as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). * indicates significant difference from pre-post treatment P<0.05.
LPS elicits significant changes in gene expression in both n-3 PUFA and placebo-treated individuals
As expected, there were substantially more DE genes following LPS. Of a total number of 79,763 genes and lincRNAs expressed, there were 419 DE genes in the n-3 PUFA group, while in the placebo group there were 643 DE genes. Details of all genes significantly altered in either group are presented in Supplement Table 2. Of the DE genes, many overlapped, as would be expected given the known physiological effects of LPS on the inflammatory response.[13] Common to both groups, 184 genes were up-regulated, and 81 genes were down-regulated. These broadly comprised genes that we have previously shown to be increased in adipose in response to LPS [16], with little difference between the groups for many known inflammatory response genes (e.g. CX3CL1, up 9.6-fold in placebo P=0.002, up 9.8-fold in n-3 PUFA, P=0.003; ICAM1, up 14.5-fold in placebo P=0.002, up 13.4 fold in n-3 PUFA, P=0.003). Of 526 genes that were significantly altered in one group but not the other, 90% were changed in the same direction in both groups, and approximately half reached nominal significance (i.e. unadjusted P<0.05) in the other group.
Evidence for differential LPS response in n-3 PUFA compared with Placebo
There was evidence for attenuation of LPS-induced gene expression with n-3 PUFA supplementation. PPARG, a known n-3 PUFA responsive gene in adipose[26] was up-regulated significantly post-LPS in the placebo group (2.2-fold P=0.002) and non-significantly up-regulated in the n-3 PUFA group (1.3-fold, P=0.2). Similarly the fatty acid desaturase, FADS1, was significantly up-regulated in placebo (2-fold, P=0.02) but unchanged in n-3 PUFA (1.0-fold, P=0.99). Notably, the prostaglandin synthase gene PTGDS was significantly up-regulated post-LPS in the n-3 PUFA group (1.8-fold, P=0.01) but not in the placebo arm (1.1-fold, P=0.9). Interestingly, three genes were significantly DE in both groups but in different directions; CCL18, SERPINA1, and RGS2 were significantly down-regulated in response to LPS in placebo (down ~2-fold, P<0.05) but were significantly up-regulated in n-3 PUFA (up 2-fold, P<0.05). These genes are known for their role in immune system function.[27–29] A further 56 genes were also altered in opposite directions (with significance in at least one group). A summary of differentially-regulated genes with evidence for functionality is presented in Table 2.
Table 2.
Genes with evidence for differential regulation post-LPS in placebo and n-3 PUFA
| Placebo | n-3 PUFA | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Gene Name | FPKM Pre-LPS |
FPKM Post-LPS |
Adjusted P Value |
FPKM Pre-LPS |
FPKM Post-LPS |
Adjusted P Value |
Functional Enrichment (pathway analysis) |
Additional known relevant functions |
| Significantly down-regulated post-LPS in placebo, up in n-3 PUFA | |||||||||
| CCL18 | chemokine (C-C motif) ligand 18 | 9.8 | 3.9 | 0.009 | 6.2 | 13.3 | 0.042 | Immune Response; Secreted Protein; Signaling Protein | immunosuppression |
| CCL3 | chemokine (C-C motif) ligand 3 | 6.7 | 1.7 | 0.002 | 2.3 | 3.4 | 0.866 | Immune Response; Secreted Protein; Signaling Protein | |
| RGS2 | regulator of G-protein signaling 2, 24kDa | 11.7 | 6.6 | 0.011 | 8.9 | 16.9 | 0.010 | adipocyte differentiation | |
| SERPINA1 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | 2.8 | 0.9 | 0.002 | 1.0 | 2.2 | 0.020 | Secreted; Signaling Protein | associated with lipid rafts |
| APLN | apelin | 1.0 | 0.4 | 0.026 | 0.8 | 1.3 | 0.638 | Immune Response; Secreted Protein; Signaling Protein | blood pressure, angiogenesis |
| FCGR3A | Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | 9.4 | 5.5 | 0.019 | 6.0 | 6.8 | 0.999 | Immune Response; Secreted Protein; Signaling Protein | |
| FCN1 | ficolin (collagen/fibrinogen domain containing) 1 | 5.7 | 2.1 | 0.002 | 2.2 | 4.6 | 0.134 | Immune Response; Secreted Protein; Signaling Protein | |
| HES1 | hes family bHLH transcription factor 1 | 17.3 | 7.0 | 0.002 | 12.9 | 12.4 | 0.999 | ||
| IL1RN | interleukin 1 receptor antagonist | 11.0 | 5.1 | 0.009 | 5.5 | 6.6 | 0.999 | Immune Response; Secreted Protein; Signaling Protein | |
| IL7R | interleukin 7 receptor | 1.1 | 0.5 | 0.042 | 0.6 | 0.7 | 0.999 | Immune Response; Secreted Protein; Signaling Protein | |
| IL8 | interleukin 8 | 1.0 | 0.2 | 0.032 | 0.4 | 0.8 | 0.399 | Immune Response; Secreted Protein; Signaling Protein | |
| LCP1 | lymphocyte cytosolic protein 1 (L-plastin) | 26.2 | 16.9 | 0.010 | 21.3 | 22.0 | 0.999 | Immune Response | |
| TREM1 | triggering receptor expressed on myeloid cells 1 | 1.2 | 0.4 | 0.037 | 0.3 | 0.7 | 0.417 | Immune Response; Secreted Protein; Signaling Protein | |
| Significantly up-regulated post-LPS in placebo down in n-3 PUFA | |||||||||
| FADS1 | fatty acid desaturase 1 | 20.9 | 40.5 | 0.023 | 26.9 | 27.8 | 0.999 | biosynthesis of unsaturated fatty acids | |
| PPARG | peroxisome proliferator-activated receptor gamma | 172.2 | 378.4 | 0.002 | 212.6 | 294.7 | 0.176 | Immune Response | adipose n-3 PUFA receptor |
| Significantly up-regulated post-LPS in n-3 PUFA, down in placebo | |||||||||
| FCGR3B | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | 1.2 | 0.9 | 0.975 | 0.4 | 5.4 | 0.003 | Immune Response; Secreted Protein; Signaling Protein | |
| IER5L | Immediate Early Response 5-Like | 3.4 | 2.9 | 0.986 | 2.6 | 5.0 | 0.003 | early cellular response | |
| PTGDS | prostaglandin D2 synthase | 13.9 | 16.1 | 1.000 | 18.2 | 32.2 | 0.012 | Secreted; Signaling Protein | prostaglandin synthesis |
Evidence for lincRNA modulation by n-3 PUFA
Expression of several lincRNAs was significantly altered by LPS, with modest evidence for differential regulation by n-3 PUFA. LincRNAs that were significantly up-regulated in the placebo group but not n-3 PUFA are shown in Table 3. Expression of these lincRNAs was generally lower than protein-coding genes (FPKM 0.2 – 3.1). These lincRNAs were identified by Hanguaer et al.[21] as novel non-coding transcripts, and their functions are not yet known.
Table 3.
Long intergenic non-coding RNAs altered post-LPS in placebo, but not in n-3 PUFA
| LincRNA ID | # exons | Genomic Location | Placebo | n-3 PUFA | ||
|---|---|---|---|---|---|---|
| Fold Change | P value | Fold Change | P Value | |||
| FPKM1_group_31576_transcript_1 | 1 | chr8:12614212-12614832 | 7.3 | 0.02 | 1.7 | 0.7 |
| FPKM1_group_20742_transcript_3 | 1 | chr22:26912031-26912972 | 6.6 | 0.02 | 2.6 | 0.2 |
| FPKM1_group_4015_transcript_1 | 2 | chr10:113893707-113894533 | 6.4 | 0.2 | 3.4 | 0.6 |
| FPKM1_group_7876_transcript_1 | 1 | chr13:41241815-41243199 | 4.7 | 0.002 | 2.3 | 0.3 |
n-3 PUFA treatment alters the adipose transcriptional response to evoked endotoxemia
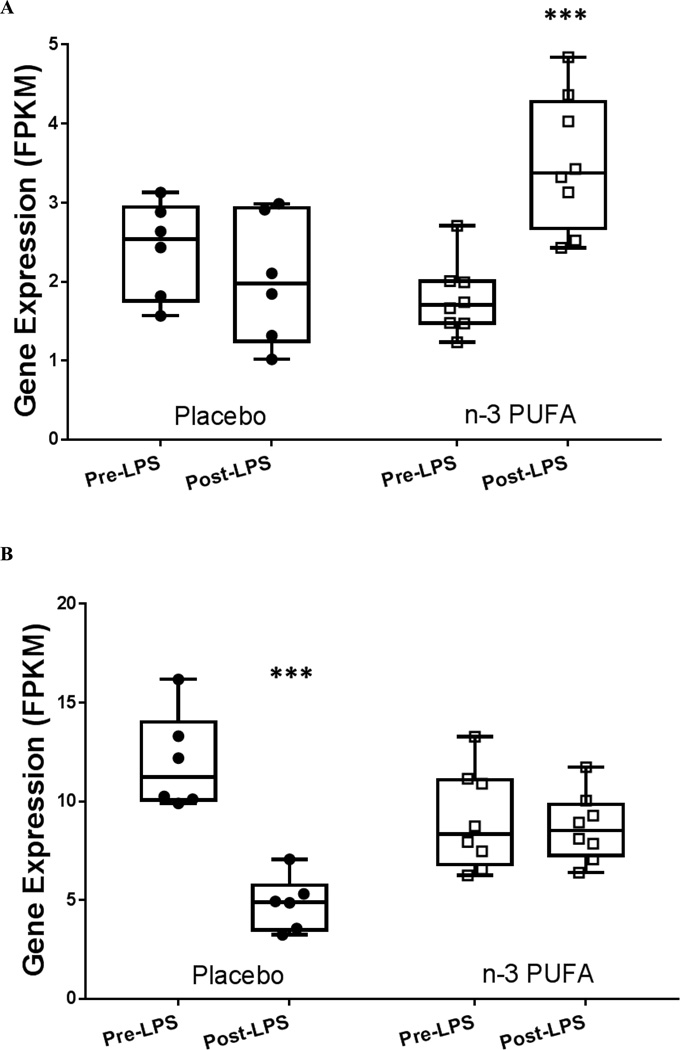
Interaction analyses revealed significant LPS-dependent gene-nutrient effects in 4 genes at P<0.001 (FDR significance threshold), with 416 genes reaching nominal significance at P<0.05. IER5L (immediate early response 5-like) displayed a significant gene-nutrient interaction (nominal P=0.0006) (Figure 3A). The related family member IER2 was identified as n-3 PUFA-modulated in the basal pre-LPS analysis. Expression of HES1 (hes family basic helix-loop-helix transcription factor 1) previously shown to be altered following EPA treatment in neural stem cells,[30] was also significantly altered (nominal P=4.8×10−5)(Figure 3B).
Figure 3. There were significant gene-nutrient interactions for expression of several genes following LPS administration.
(A) Immediate early response 5-like (IER5L) was significantly up-regulated in n-3 PUFA (adj. P=0.003) but not placebo (interaction nominal P=0.0006). (B) Hes family basic helix-loop-helix transcription factor 1 (HES1) was significantly down-regulated in placebo (adj. P=0.003), but not in n-3 PUFA (interaction nominal P=4.8×10−5). Expression measured as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). *** indicates significant difference from pre-post LPS P<0.005.
Pathway analysis highlights n-3 PUFA effect on genes involved in immune response
We carried out pathway and functional analysis using DAVID, including all genes that were differentially regulated between n-3 PUFA and placebo, defined as genes that were significantly altered in one or both groups, and were altered in opposite directions (N=59 genes). Of these genes, 86% were down-regulated in placebo, and unchanged or up-regulated in n-3 PUFA. The differentially-regulated genes were predominately associated with immune response (Enrichment score 4.1; Benjamini-adjusted P=2.6×10−3), including APLN, IL1RN, IL7R, IL8, CCL3, FCGR3A, FCGR3B, FCN1, LCP1, TREM1. Further, there was significant enrichment for secreted proteins (Enrichment score 3.45, Benjamini-adjusted P=4×10−4), and proteins involved in signaling (Enrichment score 3.4; Benjamini-adjusted P=4.2×10−3). These genes are summarized in Table 2. Overall, these data support a pattern of reduced down-regulation of immune-related genes in adipose after n-3 PUFA supplementation during evoked inflammation, potentially altering the systemic inflammatory response through altered secretion of immune-related signaling proteins.
DISCUSSION
We present the first transcriptomic analysis of adipose tissue in a trial of n-3 PUFA supplementation during evoked adipose inflammation induced by experimental endotoxemia. Detection of tissue-specific gene expression changes resulting from n-3 PUFA supplementation is challenging. This study utilized an evoked phenotype model to enhance discovery of anti-inflammatory effects of n-3 PUFA on gene expression. We found that n-3 PUFA treatment alone had limited effect on adipose tissue gene expression, and contrast this with significant n-3 PUFA-mediated effects on gene expression during evoked inflammation. In this manuscript we highlight adipose genes and pathways induced or repressed in inflammation that are modulated by n-3 PUFA supplementation and may represent candidates for nutrient targeting of inflammatory cardiometabolic stress in humans.
As expected, we detected few changes in adipose tissue gene expression following n-3 PUFA supplementation. The dearth of clear differences in global gene expression between n-3 PUFA and placebo in the resting state, i.e. without the presence of disease or other stimulation, highlights the difficulty in detecting changes due to nutritional interventions, particularly in studies of biomarkers in the general population. IER2, immediate early response 2, was one gene with evidence of n-3 PUFA regulation in adipose tissue before administration of LPS. While little is known about this gene, it is reported to promote tumor cell motility and metastasis,[31] and is up-regulated in adipose stromal cells following vitamin C treatment.[32] Remarkably, another IER family member, IER5L, has a significant n-3 PUFA-gene interaction in response to LPS. Immediate early genes are primary response genes that are rapidly up-regulated in response to external stimuli, without requiring protein synthesis, and have been particularly studied with respect to cancer.[33] As understudied immediate early response genes, IER2 and IER5L may be involved in a novel nutrient-responsive gene pathway in adipose that warrants further mechanistic and translational study.
In the FFAME trial, both the n-3 PUFA and the placebo groups exhibited a robust inflammatory response to LPS, although we found that the physiological response to LPS was attenuated by n-3 PUFA treatment, with a reduced febrile inflammatory response.[13] In the present study, many common group only, many were altered in the same direction and were of nominal significance in the other genes were modulated by LPS in both groups, including inflammatory and cell adhesion genes CX3CL1, ICAM1 and many others. Furthermore, of genes reaching statistical significance for change in one group, indicating that endotoxemia has a similar broad effect on adipose in both placebo and n-3 PUFA. This is not surprising given that LPS induces a robust inflammatory response, and that the effects of n-3 PUFA may be more gene and pathway specific rather than driving global modification of innate immune function.
We observed specific effects of n-3 PUFA on LPS-induced adipose transcriptomic responses that were not apparent in the basal state, including inflammatory and key adipose metabolic genes. Relative to placebo, n-3 PUFA reduced the LPS-induction of PPARG but had no effect on its pre-LPS expression. PPARγ has been well-studied as a lipid-responsive nuclear transcription factor,[34] of particular importance in adipocyte function and cardiometabolic disease.[35–37] The fatty acid desaturase FADS1, a key enzyme in PUFA generation, was induced by LPS in the placebo group but not the n-3 PUFA group. This might reflect sufficient n-3 PUFA levels in adipose of the n-3 PUFA group thus abrogating the need for FADS1 induction and increased PUFA synthesis in states of inflammatory stress. In contrast, the prostaglandin synthase gene PTGDS, rate-limiting in the production of the arachidonic acid derived prostaglandin D2, which is required for integrated inflammatory and febrile responses to stress, was induced by endotoxemia only in the n-3 PUFA group but not in those receiving placebo. It is possible the lower proportional amount of arachidonic acid present in adipose following EPA and DHA supplementation resulted in lower bioavailability of the AA-derived prostaglandins, and that PTGDS was up-regulated in n-3 PUFA to compensate for lower PGD2 precursor availability.
Interestingly, of genes modulated in different directions between the n-3 PUFA and placebo groups following endotoxemia, most were down-regulated in placebo and up-regulated or unchanged in the placebo group. These were significantly enriched for genes and pathways known to be involved in innate and adaptive immunity and specifically encoded for proteins known or predicted to be secreted. This suggests that n-3 PUFA supplementation prior to inflammatory stress may lead to dis-inhibition of genes that are normally down-regulated during acute inflammation. Alternatively, n-3 PUFA may enhance the resolution phase of inflammation. Expression of several lincRNAs, many of which modulate nuclear transcriptional networks in cis or trans, also differed between the two groups. While intriguing, the biological significance of these novel n-3 PUFA mediated inflammatory lincRNAs remains unknown.
This study has a number of strengths. We and others have shown that human experimental endotoxemia is a powerful model of innate immune activation of specific relevance to complex cardiometabolic and inflammatory disease.[38] Application of this model in a nutritional setting has considerable utility for novel discovery. Applying an immune stimulus in healthy individuals allows for the detection of genes involved in early response to inflammation, rather than in an established inflammatory disease setting, and is thus of particular relevance for disease prevention. We studied a clinically relevant n-3 PUFA supplementation dose (3600mg/day EPA/DHA) that is commonly used in practice for management of hypertriglyceridemia. Adipose tissue is of crucial relevance in obesity and cardiometabolic disease. We applied transcriptomics through high-throughput RNASeq to obtain high quality unbiased estimates of protein coding gene and lincRNA expression. This study also has limitations. The number of subjects studied was relatively small, however this study is the largest known study applying RNASeq to adipose tissue within the context of a nutritional intervention in humans. We also did not examine n-3 PUFA effects in other tissues. Findings of this small hypothesis-generating experimental trial in healthy individuals require replication and generalization in relevant clinical settings as well as functional studies to promote mechanistic understanding and clinical translation.
In conclusion, supplementation with a high-dose n-3 PUFA in healthy individuals has limited effects on gene expression in adipose tissue at rest, but modulates the adipose transcriptomic response during inflammatory stress. Our findings suggest that supplementation with n-3 PUFA before an inflammatory challenge modifies specific inflammatory and metabolic responses and reveals specific adipose genes and pathways as candidates for nutrient targeting of inflammatory and metabolic stress in human health and cardiometabolic stress.
Supplementary Material
Acknowledgments
Funding: The project described was supported by the National Institutes of Health (NIH) through Grant UL1RR024134 and P20-DK 019525 (both to the University of Pennsylvania) as well as a NIH-NHLBI SCCOR Project grant (P50-HL-083799) and a pilot grant from Penn Genomes Frontiers Institute to MPR. JFF was supported by a postdoctoral fellowship grant from the American Heart Association (12POST11840017). JFF is also supported by 15SDG24890015. MPR is also supported by R01-HL-111694, R01-HL-113147, R01-DK-090505, U01-HL-108636 and K24-HL-107643. ML is also supported by R01-GM-108600. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. GlaxoSmithKline provided Lovaza and matching placebo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions to manuscript
Designed research (JFF, MPR); Conducted research (JFF); Analyzed data (JFF, CX, YH, ML); Wrote paper (JFF, MPR); Have primary responsibility for final content (JFF, MPR).
No authors declare a conflict of interest
REFERENCES
- 1.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 2.Calder PC, Ahluwalia N, Brouns F, Buetler T, Clement K, Cunningham K, et al. Dietary factors and low-grade inflammation in relation to overweight and obesity. The British journal of nutrition. 2011;106(Suppl 3):S5–S78. doi: 10.1017/S0007114511005460. [DOI] [PubMed] [Google Scholar]
- 3.de Luca C, Olefsky JM. Stressed out about obesity and insulin resistance. Nature medicine. 2006;12:41–42. doi: 10.1038/nm0106-41. discussion 2. [DOI] [PubMed] [Google Scholar]
- 4.Carpentier YA, Portois L, Malaisse WJ. n-3 fatty acids and the metabolic syndrome. The American journal of clinical nutrition. 2006;83:1499S–1504S. doi: 10.1093/ajcn/83.6.1499S. [DOI] [PubMed] [Google Scholar]
- 5.Psota TL, Gebauer SK, Kris-Etherton P. Dietary omega-3 fatty acid intake and cardiovascular risk. The American journal of cardiology. 2006;98:3i–18i. doi: 10.1016/j.amjcard.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Calder PC. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. The American journal of clinical nutrition. 2006;83:1505S–1519S. doi: 10.1093/ajcn/83.6.1505S. [DOI] [PubMed] [Google Scholar]
- 7.Temple NJ. Nutrition. Vol. 18. Burbank: Los Angeles County, Calif; 2002. Nutrition and disease: challenges of research design; pp. 343–347. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson JF, Phillips CM, McMonagle J, Perez-Martinez P, Shaw DI, Lovegrove JA, et al. NOS3 gene polymorphisms are associated with risk markers of cardiovascular disease, and interact with omega-3 polyunsaturated fatty acids. Atherosclerosis. 2010;211:539–544. doi: 10.1016/j.atherosclerosis.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Morine MJ, Tierney AC, van Ommen B, Daniel H, Toomey S, Gjelstad IM, et al. Transcriptomic coordination in the human metabolic network reveals links between n-3 fat intake, adipose tissue gene expression and metabolic health. PLoS computational biology. 2011;7:e1002223. doi: 10.1371/journal.pcbi.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skulas-Ray AC. Omega-3 fatty acids and inflammation: a perspective on the challenges of evaluating efficacy in clinical research. Prostaglandins Other Lipid Mediat. 2015;116–117:104–111. doi: 10.1016/j.prostaglandins.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Franco LM, Bucasas KL, Wells JM, Nino D, Wang X, Zapata GE, et al. Integrative genomic analysis of the human immune response to influenza vaccination. eLife. 2013;2:e00299. doi: 10.7554/eLife.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson JF, Ryan MF, Gibney ER, Brennan L, Roche HM, Reilly MP. Dietary isoflavone intake is associated with evoked responses to inflammatory cardiometabolic stimuli and improved glucose homeostasis in healthy volunteers. Nutr Metab Cardiovasc Dis. 2014;24:996–1003. doi: 10.1016/j.numecd.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson JF, Mulvey CK, Patel PN, Shah RY, Doveikis J, Zhang W, et al. Omega-3 PUFA supplementation and the response to evoked endotoxemia in healthy volunteers. Mol Nutr Food Res. 2014;58:601–613. doi: 10.1002/mnfr.201300368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson JF, Patel PN, Shah RY, Mulvey CK, Gadi R, Nijjar PS, et al. Race and gender variation in response to evoked inflammation. J Transl Med. 2013;11:63. doi: 10.1186/1479-5876-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suffredini AF, Fromm RE, Parker MM, Brenner M, Kovacs JA, Wesley RA, et al. The cardiovascular response of normal humans to the administration of endotoxin. N Engl J Med. 1989;321:280–287. doi: 10.1056/NEJM198908033210503. [DOI] [PubMed] [Google Scholar]
- 16.Shah R, Lu Y, Hinkle CC, McGillicuddy FC, Kim R, Hannenhalli S, et al. Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes. 2009;58:2211–2219. doi: 10.2337/db09-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Ferguson JF, Xue C, Ballantyne RL, Silverman IM, Gosai SJ, et al. Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:902–912. doi: 10.1161/ATVBAHA.113.303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulvey CK, Ferguson JF, Tabita-Martinez J, Kong S, Shah RY, Patel PN, et al. Peroxisome Proliferator-Activated Receptor-alpha Agonism With Fenofibrate Does Not Suppress Inflammatory Responses to Evoked Endotoxemia. J Am Heart Assoc. 2012;1:e002923. doi: 10.1161/JAHA.112.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS genetics. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics (Oxford, England) 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 26.Mejia-Barradas CM, Del-Rio-Navarro BE, Dominguez-Lopez A, Campos-Rodriguez R, Martinez-Godinez M, Rojas-Hernandez S, et al. The consumption of n-3 polyunsaturated fatty acids differentially modulates gene expression of peroxisome proliferator-activated receptor alpha and gamma and hypoxia-inducible factor 1 alpha in subcutaneous adipose tissue of obese adolescents. Endocrine. 2014;45:98–105. doi: 10.1007/s12020-013-9941-y. [DOI] [PubMed] [Google Scholar]
- 27.Schutyser E, Richmond A, Van Damme J. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. Journal of leukocyte biology. 2005;78:14–26. doi: 10.1189/jlb.1204712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guttman O, Baranovski BM, Schuster R, Kaner Z, Freixo-Lima GS, Bahar N, et al. Acute-phase protein alpha1-anti-trypsin: diverting injurious innate and adaptive immune responses from non-authentic threats. Clinical and experimental immunology. 2015;179:161–172. doi: 10.1111/cei.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moratz C, Harrison K, Kehrl JH. Regulation of chemokine-induced lymphocyte migration by RGS proteins. Methods Enzymol. 2004;389:15–32. doi: 10.1016/S0076-6879(04)89002-5. [DOI] [PubMed] [Google Scholar]
- 30.Katakura M, Hashimoto M, Okui T, Shahdat HM, Matsuzaki K, Shido O. Omega-3 polyunsaturated Fatty acids enhance neuronal differentiation in cultured rat neural stem cells. Stem cells international. 2013;2013:490476. doi: 10.1155/2013/490476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neeb A, Wallbaum S, Novac N, Dukovic-Schulze S, Scholl I, Schreiber C, et al. The immediate early gene Ier2 promotes tumor cell motility and metastasis, and predicts poor survival of colorectal cancer patients. Oncogene. 2012;31:3796–3806. doi: 10.1038/onc.2011.535. [DOI] [PubMed] [Google Scholar]
- 32.Kim JH, Kim WK, Sung YK, Kwack MH, Song SY, Choi JS, et al. The molecular mechanism underlying the proliferating and preconditioning effect of vitamin C on adipose-derived stem cells. Stem cells and development. 2014;23:1364–1376. doi: 10.1089/scd.2013.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Healy S, Khan P, Davie JR. Immediate early response genes and cell transformation. Pharmacology & therapeutics. 2013;137:64–77. doi: 10.1016/j.pharmthera.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Sampath H, Ntambi JM. Polyunsaturated fatty acid regulation of genes of lipid metabolism. Annual review of nutrition. 2005;25:317–340. doi: 10.1146/annurev.nutr.25.051804.101917. [DOI] [PubMed] [Google Scholar]
- 35.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annual review of biochemistry. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 37.Madsen L, Petersen RK, Kristiansen K. Regulation of adipocyte differentiation and function by polyunsaturated fatty acids. Biochimica et biophysica acta. 2005;1740:266–286. doi: 10.1016/j.bbadis.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Patel PN, Shah RY, Ferguson JF, Reilly MP. Human Experimental Endotoxemia in Modeling the Pathophysiology, Genomics, and Therapeutics of Innate Immunity in Complex Cardiometabolic Diseases. Arteriosclerosis, thrombosis, and vascular biology. 2014 doi: 10.1161/ATVBAHA.114.304455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.