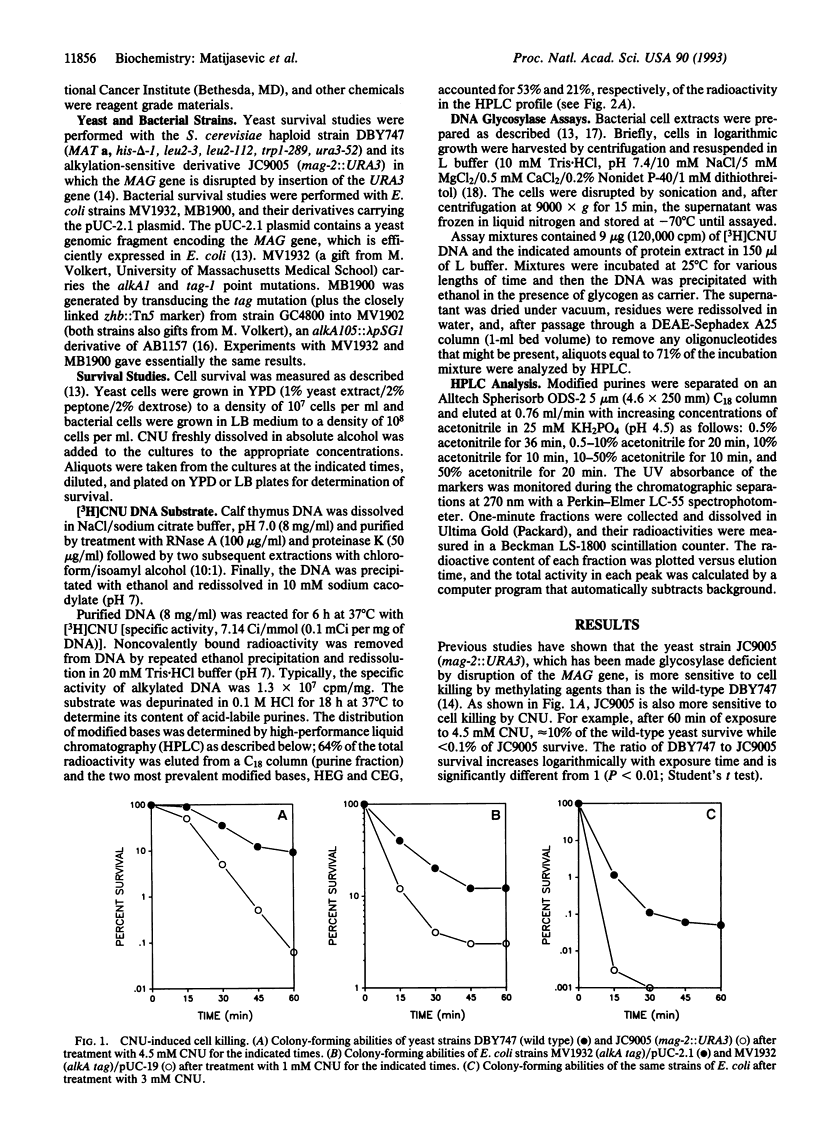
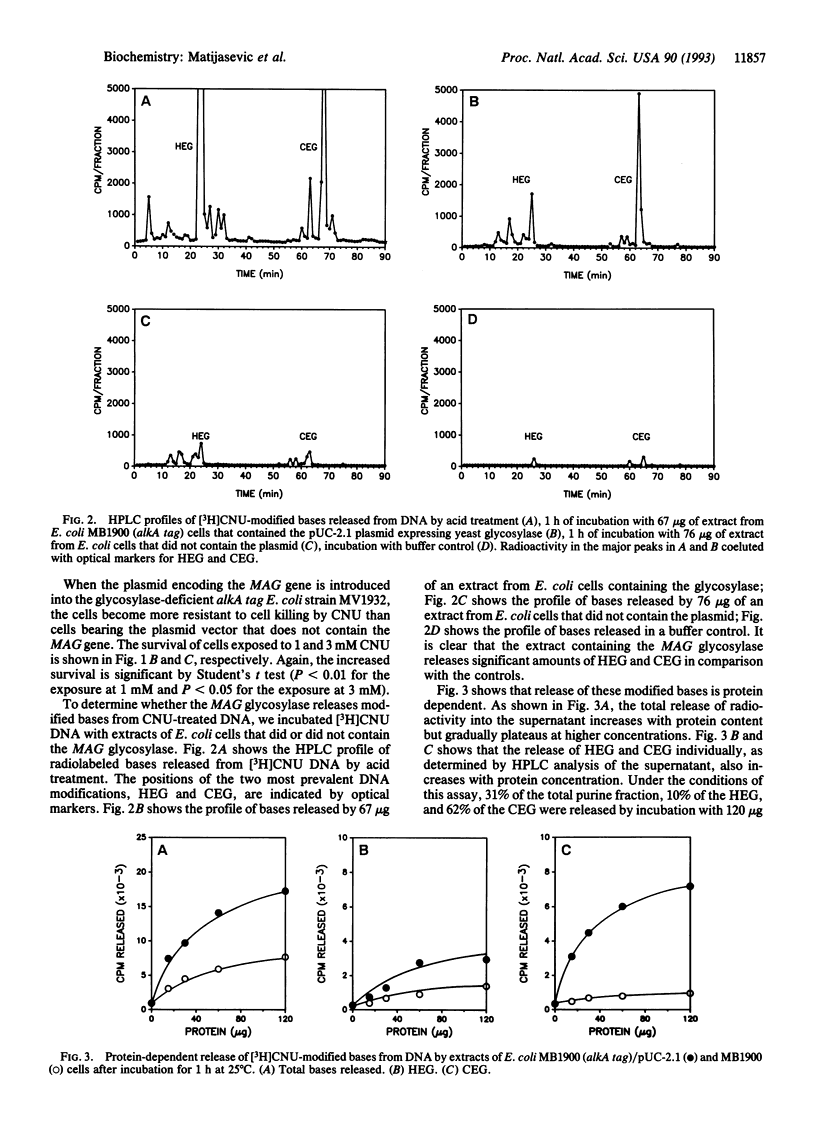
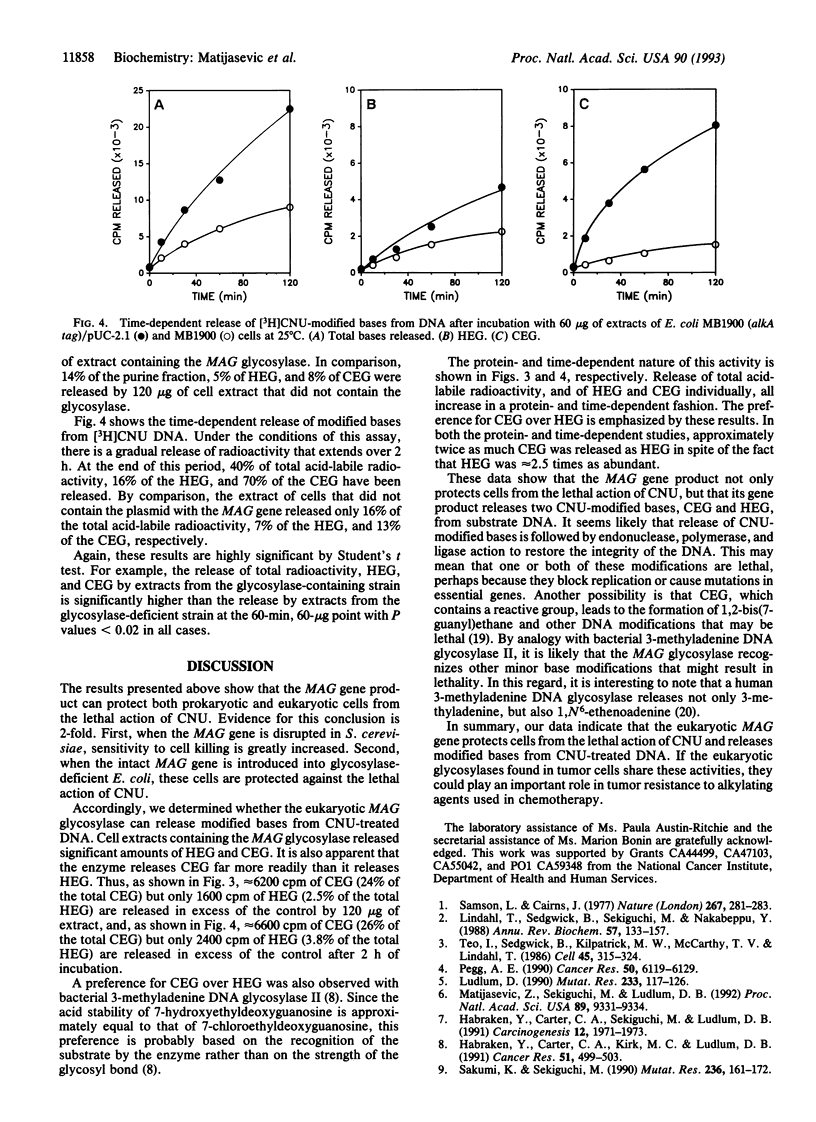
Abstract
A eukaryotic 3-methyladenine DNA glycosylase gene, the Saccharomyces cerevisiae MAG gene, was shown to prevent N-(2-chloroethyl)-N-nitrosourea toxicity. Disruption of the MAG gene by insertion of the URA3 gene increased the sensitivity of S. cerevisiae cells to N-(2-chloroethyl)-N-nitrosourea, and the expression of MAG in glycosylase-deficient Escherichia coli cells protected against the cytotoxic effects of N-(2-chloroethyl)-N-nitrosourea. Extracts of E. coli cells that contain and express the MAG gene released 7-hydroxyethylguanine and 7-chloroethylguanine from N-(2-chloroethyl)-N-nitrosourea-modified DNA in a protein- and time-dependent manner. The ability of a eukaryotic glycosylase to protect cells from the cytotoxic effects of a haloethylnitrosourea and to release N-(2-chloroethyl)-N-nitrosourea-induced DNA modifications suggests that mammalian glycosylases may play a role in the resistance of tumor cells to the antitumor effects of the haloethylnitrosoureas.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen J., Derfler B., Maskati A., Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990 Dec;9(13):4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Samson L. Induction of S.cerevisiae MAG 3-methyladenine DNA glycosylase transcript levels in response to DNA damage. Nucleic Acids Res. 1991 Dec 11;19(23):6427–6432. doi: 10.1093/nar/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson L. C., Laurent G., Sharkey N. A., Kohn K. W. DNA cross-linking and monoadduct repair in nitrosourea-treated human tumour cells. Nature. 1980 Dec 25;288(5792):727–729. doi: 10.1038/288727a0. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Carter C. A., Kirk M. C., Ludlum D. B. Release of 7-alkylguanines from N-(2-chloroethyl)-N'-cyclohexyl-N-nitrosourea-modified DNA by 3-methyladenine DNA glycosylase II. Cancer Res. 1991 Jan 15;51(2):499–503. [PubMed] [Google Scholar]
- Habraken Y., Carter C. A., Sekiguchi M., Ludlum D. B. Release of N2,3-ethanoguanine from haloethylnitrosourea-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Carcinogenesis. 1991 Oct;12(10):1971–1973. doi: 10.1093/carcin/12.10.1971. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Ludlum D. B. Release of chloroethyl ethyl sulfide-modified DNA bases by bacterial 3-methyladenine-DNA glycosylases I and II. Carcinogenesis. 1989 Mar;10(3):489–492. doi: 10.1093/carcin/10.3.489. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Sedgwick B., Sekiguchi M., Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu Rev Biochem. 1988;57:133–157. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- Ludlum D. B. DNA alkylation by the haloethylnitrosoureas: nature of modifications produced and their enzymatic repair or removal. Mutat Res. 1990 Nov-Dec;233(1-2):117–126. doi: 10.1016/0027-5107(90)90156-x. [DOI] [PubMed] [Google Scholar]
- Matijasevic Z., Bodell W. J., Ludlum D. B. 3-Methyladenine DNA glycosylase activity in a glial cell line sensitive to the haloethylnitrosoureas in comparison with a resistant cell line. Cancer Res. 1991 Mar 1;51(5):1568–1570. [PubMed] [Google Scholar]
- Matijasevic Z., Sekiguchi M., Ludlum D. B. Release of N2,3-ethenoguanine from chloroacetaldehyde-treated DNA by Escherichia coli 3-methyladenine DNA glycosylase II. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9331–9334. doi: 10.1073/pnas.89.19.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisson P. E., Lawrence C. W. The isolation and characterization of an alkylating-agent-sensitive yeast mutant, ngs1. Mutat Res. 1986 May;165(3):129–137. doi: 10.1016/0167-8817(86)90047-7. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990 Oct 1;50(19):6119–6129. [PubMed] [Google Scholar]
- Rebeck G. W., Smith C. M., Goad D. L., Samson L. Characterization of the major DNA repair methyltransferase activity in unadapted Escherichia coli and identification of a similar activity in Salmonella typhimurium. J Bacteriol. 1989 Sep;171(9):4563–4568. doi: 10.1128/jb.171.9.4563-4568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Singer B., Antoccia A., Basu A. K., Dosanjh M. K., Fraenkel-Conrat H., Gallagher P. E., Kuśmierek J. T., Qiu Z. H., Rydberg B. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Kilpatrick M. W., McCarthy T. V., Lindahl T. The intracellular signal for induction of resistance to alkylating agents in E. coli. Cell. 1986 Apr 25;45(2):315–324. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- Tong W. P., Ludlum D. B. Formation of the cross-linked base, diguanylethane, in DNA treated with N,N'-bis(2-chloroethyl)-N-nitrosourea. Cancer Res. 1981 Feb;41(2):380–382. [PubMed] [Google Scholar]
- Volkert M. R., Nguyen D. C., Beard K. C. Escherichia coli gene induction by alkylation treatment. Genetics. 1986 Jan;112(1):11–26. doi: 10.1093/genetics/112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]



