Abstract
The ability to visualize RNA in situ is essential to dissect mechanisms for the temporal and spatial regulation of gene expression that drives development. Although considerable attention has been focused on transcriptional control, studies in model organisms like Drosophila have highlighted the importance of post-transcriptional mechanisms - most notably intracellular mRNA localization - in the formation and patterning of the body axes, specification of cell fates, and polarized cell functions. Our understanding of both types of regulation has been greatly advanced by technological innovations that enable a combination of highly quantitative and dynamic analysis of RNA. This review presents two methods, single molecule fluorescence in situ hybridization for high resolution quantitative RNA detection in fixed Drosophila oocytes and embryos and genetically encoded fluorescent RNA labeling for detection in live cells.
1. Introduction
The visualization of mRNA expression patterns is fundamental to deciphering the regulation and function of genes that control animal development. With its wealth of genetic and molecular tools, as well as tissue accessibility, Drosophila has long served as a model system for elucidating the temporal and spatial patterns of gene transcription that give rise to the segmental body plan. Studies in Drosophila have also led the way toward understanding the importance of intracellular mRNA localization in generating cellular and developmental asymmetry. Much work has focused on the analysis of maternal transcripts, whose localization in the oocyte and/or early embryo are essential for the establishment and patterning of the body axes and the specification and development of the germline [1, 2]. In addition, a variety of localization patterns and functions for RNA localization in differentiated cells are coming to light, highlighting the versatility of this post-transcriptional regulatory mechanism [3]. Notably, large scale fluorescence in situ hybridization screens found that 71% of 3000 transcripts analyzed in the Drosophila embryo, and 22% of nearly 6000 analyzed in the ovary, are subcellularly localized [4, 5].
Following their synthesis, localized mRNAs must be directed to the appropriate machinery for delivery to the correct region of the cell. As a general paradigm, RNAs are transported as ribonucleoprotein particles (RNPs); in most known examples, the RNPs attach to molecular motors for directed, cytoskeletal-based transport but in some cases they move by diffusion and become locally entrapped [1, 6]. RNPs are built through the interactions of both transcript-specific and more general factors with sequence elements or structural motifs in the transcript, and the particular set of RNP components is thought to determine RNP behavior. They may also be remodeled or augmented for different stages of the localization process, for example through the recruitment of adaptors to motors for transport or proteins involved in anchoring at the target destination [3, 7]. The mechanisms governing the formation of these RNPs, their specific RNA and protein content, and their dynamic behavior over the life of a transcript are areas of active investigation.
Our mechanistic understanding of mRNA localization has advanced as methods to detect RNA in situ have improved and expanded. The earliest in situ hybridization experiments to analyze RNA distributions in Drosophila oocytes and embryos were performed using radiolabeled probes applied to tissue sections [8, 9]. Indirect detection of probes containing digoxigenin or biotin-conjugated bases by enzyme-based immunohistochemistry, which greatly increased efficacy and sensitivity and could be applied to whole mount embryo preparations, soon became the method of choice [10]. These methods provided basic information about the location of a particular transcript within a cell as well as the first insights into RNA localization mechanisms, revealing the effects of genetic or pharmacological perturbations on the RNA distribution [11–15]. The advent of fluorescence in situ hybridization (FISH) improved spatial resolution and facilitated multiplex RNA detection (for a comprehensive review of FISH, see Levsky and Singer [16]). Further adaptations allowed FISH to be combined with immunofluorescence, permitting co-detection of RNA and protein. However, neither enzyme-based immunohistochemical detection nor detection by typical FISH probes synthesized with stochastically incorporated fluorophores allow for absolute RNA molecule quantification. The development of highly sensitive FISH methods capable of detecting single RNA molecules – single molecule FISH (smFISH) – now make it possible to quantify gene expression in situ. For the field of mRNA localization, the ability to detect transcripts quantitatively and to map their positions with high resolution by smFISH has opened the door to determining the precise molecular contents and assembly mechanisms of RNPs that mediate various stages in the life of an mRNA and its travels within a cell.
In situ hybridization is limited to a static view of the cell at a particular time, leaving the events that produce the final observed RNA distribution to conjecture. A full understanding of dynamic processes like mRNA localization requires the ability to visualize RNA molecules in live cells, in real time. Numerous methods have been developed to this end, including injection of in vitro synthesized fluorescently labeled transcripts and the application of conditionally fluorescent RNA-binding probes like molecular beacons, RNA aptamers, and RNA intercalating dyes (see Gaspar and Ephrussi for detailed review [17]). While these reagents can be readily delivered to cultured cells, introducing them into Drosophila oocytes and embryos is problematic, requiring microinjection or inefficient and potentially harmful permeabilization schemes. In contrast, genetically encoded fluorescent tagging methods based on the high affinity interaction of bacteriophage proteins with cognate RNA stem-loops [17] are particularly well suited for Drosophila given the ease of transgenesis.
Here we describe the application of smFISH and genetically encoded RNA tagging to the analysis of intracellular mRNA localization in Drosophila oocytes and embryo. Both of these methods can be combined with protein detection methods to determine the spatial relationships of RNA and protein. We also briefly discuss their use for measuring transcription and transcriptional dynamics. While we focus here on transcript visualization oocytes and early embryos, both smFISH and in vivo RNA tagging are amenable to use in differentiated tissues at later stages of development.
2. Methods for detection of RNA in fixed oocytes and embryos by smFISH
The smFISH technique developed by Raj et. al. [18] allows highly sensitive, quantitative RNA detection and can be easily multiplexed to monitor several RNA species simultaneously. In contrast to traditional FISH methods, which use one or several probes complementary to the target RNA that are generally hundreds of bases in length, smFISH uses many short oligonucleotide probes arrayed along the target RNA (Figure 1A). Each probe is coupled to a fluorophore and as a result, binding of the set of probes to the RNA produces a high-intensity point source that is detected as a diffraction limited spot; the sensitivity achieved by the high density of labeled probes affords single molecule detection. Moreover, because signal detection requires binding of a substantial number of probes, background due to non-specific probe binding is minimized; i.e., there is a high signal to noise ratio. These attributes allow accurate counting of RNA molecules within a cell. Importantly, the method is readily adaptable to many tissue and cell types [18]. The small size of the oligonucleotide probes has a particular advantage for the Drosophila ovary tissue by allowing efficient penetration of late-stage oocytes, which are largely inaccessible to traditional probes [19, 20] (Figure 2). The protocol described here is adapted from Raj and Tyagi [21], and has been optimized for detection of transcripts in Drosophila ovaries and embryos by S. Little [19, 22]. We refer the reader to the original protocol for the detailed rationale behind the procedures [21].
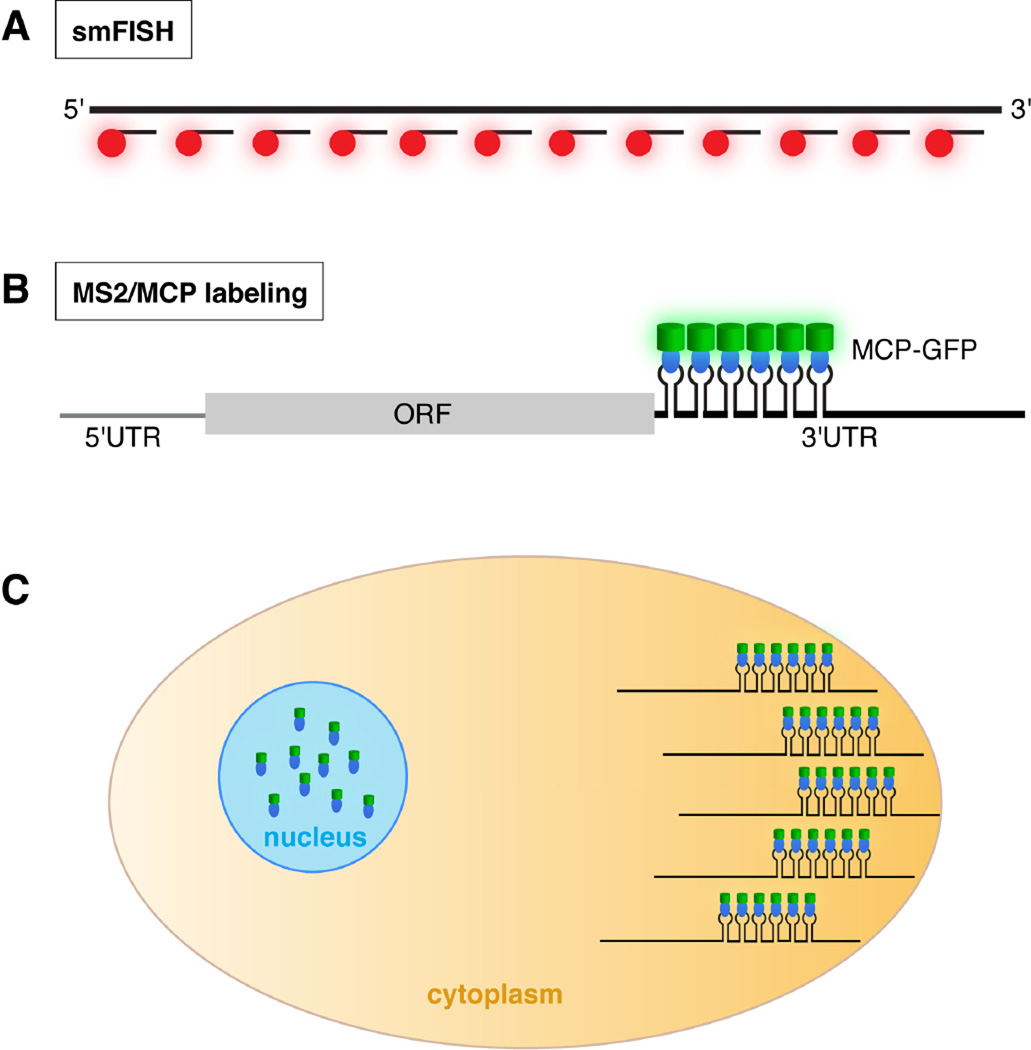
Fig. 1.
Detection of RNA by smFISH and the MS2/MCP labeling system. A. In smFISH, many short probes, each coupled to a fluorophore (red), hybridize along the length of the target RNA (at top). The large number of fluors decorating each RNA molecule results in high signal intensity with low background. B. When transcripts engineered with MS2 stem-loops are co-expressed with MCP-GFP (blue oval: MCP; green cylinder: GFP), binding of MCP-GFP to its cognate stem-loops results in fluorescently labeled mRNA. In this example, the MS2 stem-loops are located at the beginning of the 3’ UTR. C. Cartoon of a cell illustrating detection of a localized mRNA labeled using the MS2/MCP system. For mRNA localization studies, the MCP-FP contains a nuclear localization signal. As a result, MCP-FP molecules not bound to MS2-tagged transcripts are sequestered in the nucleus, reducing background fluorescence.
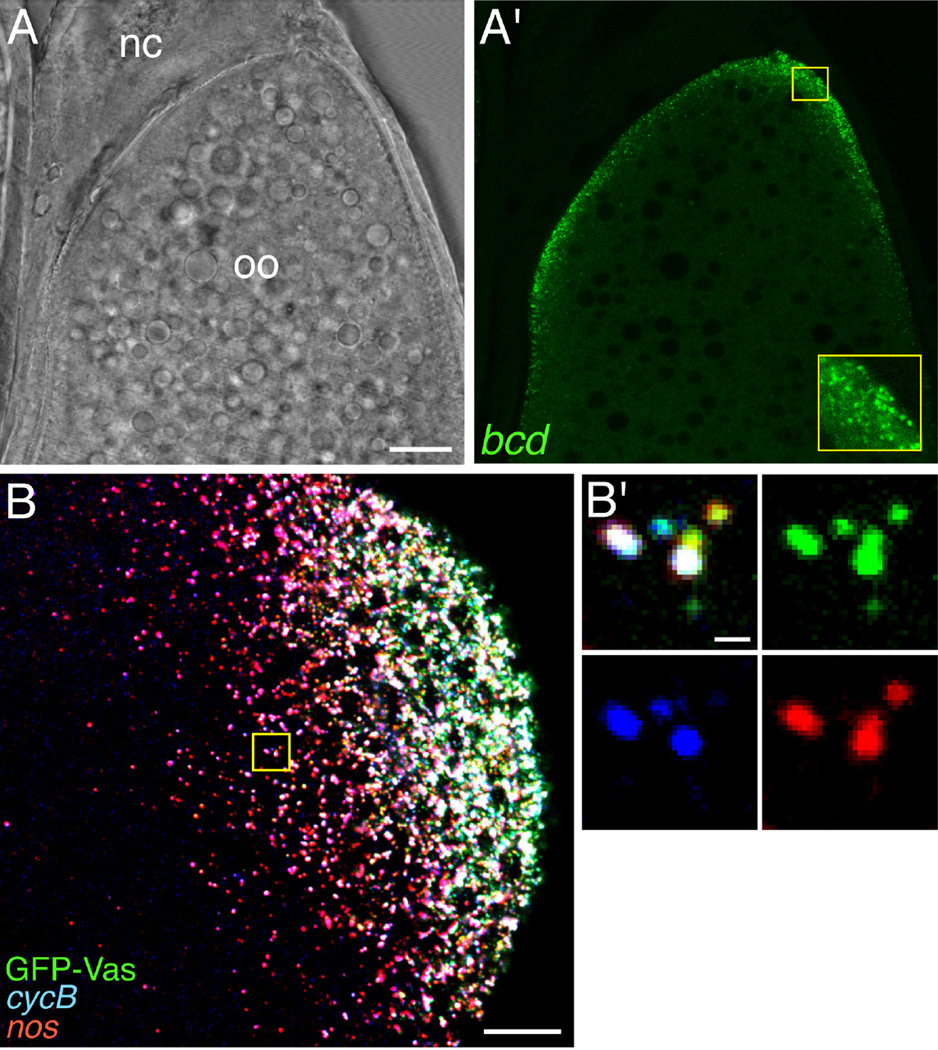
Fig. 2.
Detection of localized transcripts in Drosophila oocytes by smFISH. A, A’. A single confocal section of a stage 12 oocyte. By this stage, traditional in situ hybridization methods become unreliable due to the deposition of the vitelline membrane and chorion. The brightfield image (A) shows the position of the oocyte (oo) and the nurse cell remnants (nc) anterior to the oocyte; the fluorescence image (A’) shows smFISH detection of bcd mRNA localized along the anterior cortex of the oocyte. The inset in A’ is a 3-fold magnification of the small boxed region, showing individual bcd RNPs. B. Multiplex smFISH detection of cyclin B (cycB) and nos mRNAs together with the polar granule marker GFP-Vas at the posterior cortex of a stage 13 oocyte. A maximum confocal z-series projection spanning 8 µm is shown. B’. High magnification of the boxed region showing the heterogeneous RNA composition of the polar granules. Scale bars: 20 µm (A), 10 µm (B), 0.5 µm (B’). Panels B and B’ are reprinted by permission from Little et. al. [20].
2.1 Probe design and preparation
Sets of oligonucleotide probes targeting an RNA sequence can be designed using a free web-based program, Stellaris FISH Probe Designer, developed by Raj et al. [18] and available at http://www.biosearchtech.com/stellarisdesigner/. The program optimizes GC content while allowing for customization of probe length and spacing between individual probes. We routinely use 20mer DNA oligonucleotide probes, with a spacing of at least two nucleotides between adjacent probes. Each probe sets typically contains ≥48 probes; however, as few as 32 probes have yielded adequate signal in our experiments [20]. Although probes complementary to untranslated regions are more likely to be affected by sequence polymorphisms than probes complementary to the coding region, the inclusion of probes targeting the untranslated regions can help to maximize detection of short transcripts. Probes can be ordered from Biosearch Technologies pre-conjugated to a fluorophore, or with a 3’ amine modification, desalted, and delivered in water for in house coupling to fluorophores. In house coupling and purification is currently far more cost effective and affords the flexibility of generating probe sets with various fluorophores as needed. The conjugation and subsequent purification steps are described in detail by Raj and Tyagi [21].
While Alexa fluors (Molecular Probes, Invitrogen) and other dyes have been used for smFISH, we prefer Atto dye fluorophores (NHS ester for in house coupling; Sigma) for their greater photostability. Photobleaching can indeed be a problem when acquiring high quality image stacks necessary for quantification. Fluorophore wavelength is one of the major considerations for Drosophila tissues. The use of longer wavelength fluors (565 and 647) is advantageous by avoiding background from green autofluorescence of the yolk in oocytes and early embryos. In addition, these probes can be easily combined with DAPI staining or GFP-tagged proteins (Figure 2).
2.2 smFISH Procedure
The following protocol is optimized for ovaries and early (0–3 hr) embryos. The initial fixation procedure differs for ovaries and embryos; the subsequent hybridization steps of the protocol are identical.
2.2.1 Reagents/solutions
PBST: 1× PBS, 0.1% Tween-20
Fixative: 16% electron microscopy grade formaldehyde (EM Sciences), dilute to 4% with 1× PBS
Wash buffer (WB): 4× SSC, 35% deionized formamide, 0.1% Tween-20
Hybridization buffer (HB): 4× SSC, 35% deionized formamide, 0.1% Tween-20, 2 mM vanadyl ribonucleoside (New England Biolabs), 0.1 mg/ml salmon sperm DNA, 100 mg/ml dextran sulfate, 20 µg/ml RNase-free BSA (New England Biolabs)
Probes: Probe set stock concentration ranges from 0.2–2 µM of each oligonucleotide.
We typically dilute to a final concentration of 1 nM in 100 µl of HB. For shorter incubations or low abundance transcripts, it may be necessary to increase the concentration by two-fold or more.
Notes:
-
-
Care should be taken to keep reagents and solutions RNase free.
-
-
To deionize formamide, gently stir 100 ml formamide with 10 g mixed bed resin (Sigma #M8032) for 1 hr at RT. Remove resin by filtration, aliquot, and store at −20° C.
-
-
Except when otherwise specified, use ~0.5–1 ml of solution for washes and incubations.
-
-
Take care to remove all liquid during washes and incubations. Use of a drawn-out Pasteur pipette minimizes sample loss.
2.2.2 Ovary fixation
Dissect ovaries from well-fed females in PBS, and gently tease apart using forceps or fine dissecting needles to allow access of interior tissue to solutions.
Transfer ovaries to 1.5 ml microcentrifuge tube with a Pasteur pipette. Keep ovaries moving within the narrow barrel of the Pipette to prevent their sticking to the glass.
Remove PBS and add 400 µl of fixative. Incubate for 30 min. on a nutating mixer.
Remove fixative and wash 3 × 5min. in PBST on a nutating mixer.
Transfer ovaries into methanol stepwise, incubating 5 min. each in 7:3 PBST:MeOH, 1:1 PBST:MeOH, 3:7 PBST:MeOH on a nutating mixer.
Incubate ovaries in 100% methanol for 10 min. on a nutating mixer. At this point, ovaries can be stored in methanol at −20° C. However, we find sample and signal quality is best if the hybridization is performed immediately.
Notes: While a 20 minute fixation suffices, fixation for 30 minutes yields excellent results for late-stage oocytes and does not appear to be detrimental for earlier stages. Our standard protocol calls for stepping oocytes into methanol to assist with tissue permeabilization and maximize signal intensity. While we have not encountered difficulties with methanol when combining smFISH with visualization of GFP-tagged proteins or immunofluorescence, it may be desirable to eliminate the use of methanol to avoid quenching of GFP fluorescence for some GFP fusion proteins. Note that phalloidin cannot be used to counterstain methanol treated tissue because methanol destroys the conformation of F-actin required for phalloidin binding.
2.2.3 Embryo fixation
Rinse embryos with water and dechorionate in fresh 50% bleach for 1 min.
Wash thoroughly with water to remove bleach and blot dry.
Transfer embryos to a 20 ml glass scintillation vial containing 2 ml 4% formaldehyde + 8 ml heptane. Incubate for 20 min. with gentle rocking.
Allow liquid phases to separate, then completely remove the lower formaldehyde phase with a drawn-out Pasteur pipette.
Add 10 ml methanol and shake vigorously for 30–60 sec. to remove vitelline membrane. Devitellinized embryos will fall to the bottom of the vial. Note that devitellinization is often inefficient for pre-cellular embryos and for some mutant genotypes.
Transfer devitellinized embryos to a 1.5 ml microcentrifuge tube.
Wash 3× with methanol. At this point, embryos can be stored in methanol at −20° C. However, we find sample and signal quality is best if the hybridization is performed immediately.
2.3.4 Hybridization
Rehydrate tissue stepwise by incubation for 5 min. each in 3:7 PBST:MeOH, 1:1 PBST:MeOH, 7:3 PBST:MeOH on a nutating mixer.
Briefly rinse 4× with PBST.
Prehybridize for 10 min. in 500 µl of WB on a nutating mixer at RT.
Add 100 µl HB containing probes to each sample. Incubate at 37° C, 12–16 hr in the dark. A box placed in a 37° C incubator works well. Nutation or rocking of samples is not necessary during this step.
Remove probe and rinse briefly with 500 µl WB pre-warmed to 37° C.
Incubate 2 × 30 min. in 500 µl pre-warmed WB, in the dark at 37° C.
Rinse 4× with PBST.
Optional: Samples can be stained with DAPI (1:4000 from 10 µg/ml stock) for 2 min. at this point, then rinsed 4× with PBST.
Notes: To combine smFISH with immunofluorescence, re-fix ovaries or embryos in 4% formaldehyde for 20 min. following rehydration from methanol (Hybridization step 1). Standard immunofluorescence procedures can then be performed following the smFISH protocol. To combine smFISH with direct visualization of GFP fusion proteins, decrease the hybridization time to 2–4 hr. While this decreases the intensity of the smFISH signal, it preserves GFP fluorescence (Figure 2B). Increasing the amount of probe during hybridization can partially offset the reduction in hybridization time.
2.3.5 Mounting tissue for microscopy
Transfer ovaries or embryos in PBST to a glass slide using a Pasteur pipette. To facilitate imaging of ovaries, carefully separate ovarioles or individual egg chambers using fine tungsten needles. Carefully remove all PBST from the slide with a drawn-out Pasteur pipette or syringe with fine gauge needle. Immediately, add one drop of mounting medium since the tissues will begin to dry out. If necessary, gently spread ovaries and embryos – the fixed tissue is fragile – throughout the drop of medium, then cover with a #1.5 coverglass. Vectashield (Vector Laboratories), an aqueous anti-fade medium, works well and the coverglass can be sealed by painting a thin strip of clear nail polish along each edge. ProLong Gold (Cell Signaling Technology) can also be used, but requires time to cure prior to imaging. Signal quality is best if samples are imaged immediately; however, satisfactory images can often be obtained weeks after preparation.
2.3.6 Microscopy
Imaging of Drosophila ovary and embryo smFISH samples can be performed on a variety of microscopy platforms including scanning confocal microscopes, wide-field microscopes, and structured illumination microscopes [18, 22, 23]. We routinely use a scanning confocal microscope to acquire image stacks using high laser power and scan averaging at Nyquist sampling intervals without significant photobleaching. Detection efficiency can be determined by performing smFISH using probes that alternate in fluorophore color along the length of the transcript [20, 24].
3. In vivo fluorescent labeling of mRNA
The development of genetically encoded fluorescent tagging methods has revolutionized the visualization of transcripts in living cells. A popular approach, the MS2/MCP system, takes advantage of the high affinity binding of bacteriophage MS2 coat protein (MCP) to its cognate RNA stem-loop. In this method, multiple MS2 stem-loops are inserted into the RNA of interest, which is then co-expressed with an MCP-fluorescent protein fusion (MCP-FP). Binding of the MCP-FP to the stem-loops generates fluorescently labeled RNA, which can be visualized by fluorescence microscopy (Figure 1B). Moreover, the ability to detect MS2 tagged RNAs as distinct foci permits dynamic tracking of RNAs. The MS2/MCP system was first used to study ASH1 mRNA localization to the bud tip of yeast and provided evidence for ASH1 transport on motor proteins [25]. We subsequently adapted this system for transgenic use in Drosophila in order to visualize nanos (nos) mRNA during oogenesis, and showed that localization occurred by diffusion/entrapment [26]. It has since been used by us and others to investigate trafficking of a variety of mRNAs in oocytes, embryos, and neurons [19, 27–31] (Figures 3, 4) and a recent study has illustrated its utility for investigating localization of long non-coding RNAs [32]. The transgenic MS2/MCP system has also been used in Drosophila embryos to monitor native transcripts and transcriptional dynamics in individual nuclei cells and to track transcriptional activity within groups of cells [33, 34].
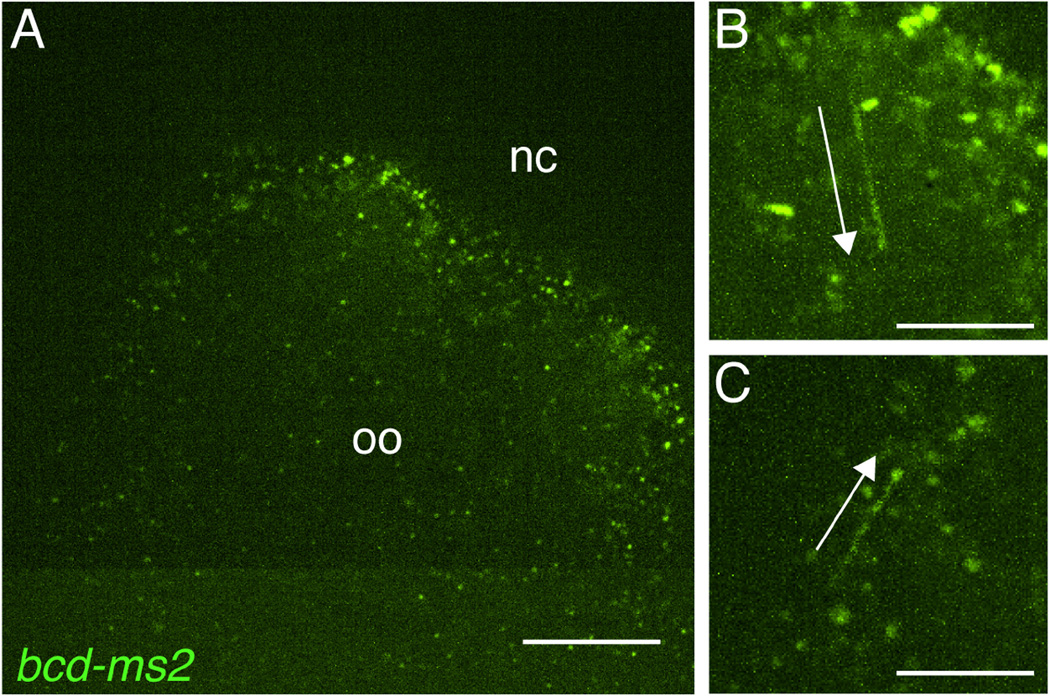
Fig. 3.
Tracking of dynamic bcd RNPs using the MS2/MCP system. A. Anterior region of a stage 12 oocyte co-expressing bcd-ms2 and MCP-GFP. A single frame from a 30 sec. movie acquired using spinning disc confocal microscopy is shown. Distinct RNPs containing bcd*GFP can be readily detected. The position of the nurse cell remnants (nc) that lie anterior to the oocyte (oo) is indicated. B, C. Time-series projections representing 3 sec. (15 frames of the movie), at 2× magnification. Arrows indicate RNPs undergoing directed transport. RNPs labeled by this method can be manually tracked or, if there is sufficient time resolution (typically >1 frame/sec.), automatically tracked using available computer software packages. Scale bars: 20 µm (A), 10 µm (B,C).
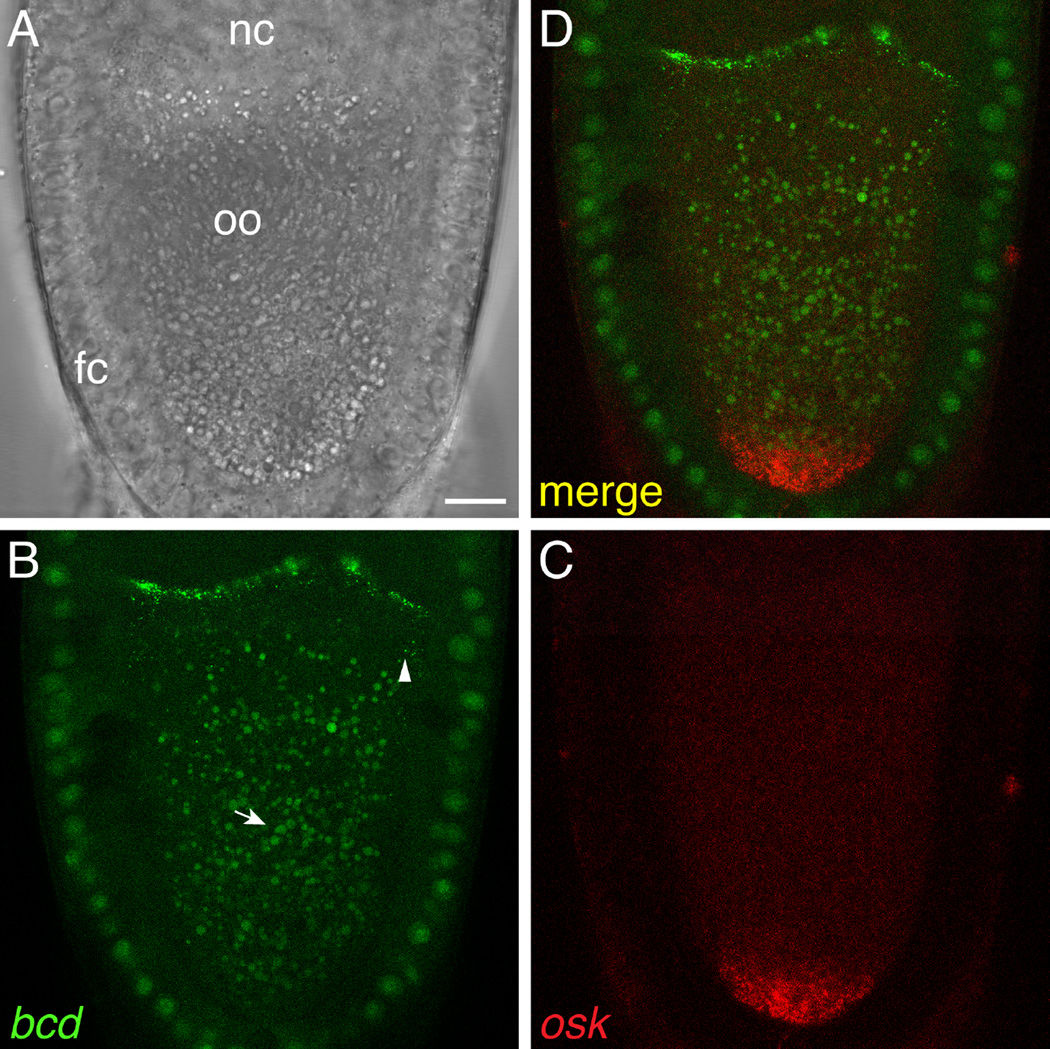
Fig. 4.
Simultaneous visualization of two different transcripts using the orthogonal MS2/MCP and PP7/PCP systems. Z-series projection (2 µm) of a live stage 10 oocyte expressing bcd-ms2 and osk-pp7 mRNAs together with MCP-GFP and PCP-mCherry, imaged by laser-scanning confocal microscopy. A. Brightfield image showing the location of the nurse cells (nc), oocyte (oo), and surrounding somatic follicle cell epithelium (fc). B. MCP-GFP specifically labels bcd-ms2, which is localized at the anterior of the oocyte, and not osk-pp7. Some bcd-ms2 particles can be seen at a distance from the anterior (arrowhead). In this experiment, MCP-GFP was expressed using the hsp83 promoter, which is active in both the germline and follicle cells. bcd-ms2 was expressed by the bcd promoter and is thus present only in the germline. Autofluorescent yolk granules are indicated by the arrow. C. PCP-mCherry specifically labels osk-pp7, which is localized at the posterior of the oocyte, and not bcd-ms2. PCP-mCherry was expressed using the nos promoter and is thus restricted to the germline. C. Merge of the two channels. Scale bar: 15 µm.
Other bacteriophage derived stem-loop/coat protein systems have been recently used in vivo, including boxB/AN and PP7/PP7 coat protein (PCP) [35–37]. PCP has a higher affinity than MCP for the cognate stem-loops so this system may increase sensitivity of RNA detection [38]. Because MCP and PCP use structurally distinct modes of RNA recognition, there is no cross-recognition of their RNA stem-loop targets [39]. Thus, MCP and PCP can be used in combination to detect two different mRNAs simultaneously (Figure 4). We have had good success using PP7/PCP to monitor mRNA transport and it has also been used for visualizing transcriptional and translational dynamics [36, 40]. While we focus on application of the MS2/MCP system, the following sections apply equivalently to use of the PP7/PCP system.
3.1 General approach to in vivo labeling
Use of the MS2/MCP system in Drosophila requires two components: the RNA of interest tagged with MS2 stem-loops and the MCP-FP (Figure 1B). To date, both the tagged RNA and MCP-FP are generated from transgenes introduced into flies by either P element-mediated transformation or FRT/phiC31 recombination, although the advent of CRISPR technology now raises the possibility of inserting MS2 stem-loops directly into the genomic locus. While we focus on the design of transgenic constructs, similar considerations will apply to genomic insertion of MS2 stem-loops. A common concern is whether tagging alters the behavior of the transcript. The insertion of the stem-loop array could affect mRNA stability, intracellular localization, and/or translation or its regulation. Confidence that tagging can be accomplished physiologically is best provided by the example of mice with MS2 stem-loops inserted at the endogenous β-actin locus. Homozygous knock-in mice are viable and fertile and visualization of MS2-tagged β-actin mRNA showed that it behaved as predicted from previous FISH analysis [41]. However, it has recently been reported that MS2 stem-loops inserted into 3’ untranslated regions (3’ UTRs) can inhibit 5’ to 3’ mRNA decay by the Xrn1 exoribonuclease in yeast [42], resulting in the accumulation of 3’ RNA decay fragments bound to MCP-FP.
In Drosophila, it is straightforward to determine whether tagging alters RNA function by testing the ability of an MS2-tagged transgene to rescue the phenotype of a mutation in corresponding gene. Because function does not necessarily reflect proper localization, demonstrating that the distribution of the tagged RNA and the distribution of the endogenous RNA observed by FISH are similar will increase confidence. More difficult to address is the question of whether motility parameters or RNP composition are altered by the binding of many MCP-FPs to the transcript. Given that mRNAs, and 3’ UTRs in particular, are decorated by large numbers of RNA-binding proteins, it seems probable that the MCP-FPs may make a relatively small contribution.
For pratical purposes, MS2-tagged RNA is generally expressed in addition to, rather than instead of, the endogenous wild-type RNA. Thus, it is important to be cognizant that competition between the tagged and wild-type RNA could cause untoward phenotypic effects or could limit the localized fluorescence signal in cases where the localization machinery is saturable.
3.1.1 Special materials and reagents
Plasmids containing different increments of MS2 and PP7 stem-loops are available at Addgene (www.addgene.org) [25, 36, 38, 40]. An updated version of the 24× MS2 stem-loops plasmid is engineered with repeats of two non-identical stem-loops to facilitate propagation in bacteria.
SURE 2 (Agilent Technologies) or Stbl2 (ThermoFisher Scientific) competent cells are recommended for manipulation and growth of stem-loop containing plasmids to minimize recombination and consequent deletion of stem-loops.
A wide variety of plasmids containing MCP and PCP sequences generated by numerous labs are available through Addgene. To our knowledge, MCP-FP transgenes currently in use in Drosophila derive from pG14-MS2-GFP [25] and PCP-FP transgenes from phage-ubc-nls-ha-pcp-gfp [40]. Both include a nuclear localization signal (NLS) attached to the coat protein. Small deletions engineered within the MCP and PCP sequences minimize multimerization needed for capsid assembly [39, 43]. Several transgenic strains expressing MCP-GFP or RFP under control of the maternally and zygotically active hsp83 promoter [26, 27] or zygotically active UASt sequences [44] are available at the Drosophila Bloomington Stock Center.
3.1.2 Expression of MS2-tagged mRNA
Key considerations for tagging the RNA are the position within the transcript where the stem-loop cassette is inserted and the number of stem-loops used, so as to maintain the functionality of the RNA. For mRNAs, the general trend has been to insert the stem-loops within the 3’ UTR to prevent interference with translation. However, mRNA localization and stability elements often reside within 3’ UTRs and avoiding these elements is essential for the analysis of localized RNAs. In the case of nos, where localization elements had been mapped, the MS2 stem-loops were placed in a region of the 3’UTR with no known regulatory function [26]. In cases where the positions of regulatory elements are unknown or widely distributed, a good strategy is to place the MS2 stem-loop cassette immediately downstream of the stop codon [29, 45] (Figure 1B). While this usually requires engineering a restriction site at the junction of the coding region and 3’ UTR, existing restriction sites in close proximity to the 5’ end of the 3’ UTR may also be suitable [27]. Note also that in studies using the MS2/MCP system to analyze transcription dynamics in early embryos, MS2 stem-loops have been placed 5’ to reporter coding sequences [33, 34]. Additionally, boxB stem-loops have been inserted at the 5’ end of reporter transgenes used to study the motility of oskar (osk) RNPs [37].
The first generation of MS2-tagged RNAs contained six MS2 stem-loops but subsequent tagging experiments in Drosophila have employed 12–24 stem-loops [29, 33, 34, 45, 46]. While each stem-loop is capable of binding two MCP-FP molecules, the degree of occupancy varies and it has been estimated that detection of single mRNA molecules requires at least 24 stem-loops [47]. However, the 24× MS2 cassette is large, and could potentially destabilize the transcript or interfere with 3’ UTR function. Another approach toward improving detection is to increase expression levels of the tagged mRNA. We generally prefer to express tagged transcripts under control of the endogenous promoter sequences to attain near physiological levels, but increasing the transgene copy number by 2–3 fold through P element hopping or recombination can dramatically improve detection, often without causing phenotypic consequences. Alternatively, tagged transcripts can be expressed at elevated levels under Gal4/UAS control using a strong Gal4 driver, but caution should be used in interpreting localization patterns.
3.1.3 Expression of MCP-FP fusion proteins
MCP can be fused at its C-terminus to a wide array of fluorescent, photoactivatable, and photoswitchable proteins. GFP and its variants continue to perform best for signal intensity and photostability, but mRFP and mCherry have been used successfully in the oocyte, early embryo, and peripheral neurons [27, 44, 48, 49]. Expression of MCP-FP is tailored by choosing an appropriate promoter. Because MCP-FP expression is not toxic in Drosophila, direct promoter fusions are feasible. For maternal expression, we favor either the nos or hsp83 promoter. The hsp83 promoter produces higher levels of MCP than the nos promoter but is also active in the somatic follicle cells surrounding the egg chamber, which can interfere with visualization of mRNA in the oocyte from early to mid-oogenesis. For zygotic expression, the Gal4/UAS system affords the most versatility [50, 51] but ubiquitous promoters, including hsp83, have also been used [51, 52].
A key consideration from the perspective of the MCP-FP fusion protein is minimization of background fluorescence due to MCP-FP that is not bound to the MS2-tagged mRNA. For studies of cytoplasmic mRNA localization, the contribution of unbound MCP-FP can be significantly reduced by attachment of an NLS, such as the SV40 NLS, to the MCP-FP. In this way, MCP-FP molecules that are not bound to tagged RNA are retained in the nucleus [25] (Figure 1C). For studies of transcriptional dynamics, however, it may be preferable to use an MCP lacking the NLS [33]. In such cases, the contribution of unbound MCP-FP can be minimized by optimizing the expression level of MCP-FP relative to that of the MS2-tagged RNA. This strategy is also relevant for visualization of cytoplasmic mRNA at late stages of oogenesis, when MCP-FP released from apoptosing nurse cell nuclei enters the oocyte along with the nurse cell cytoplasm. Recent modifications by Singer and colleagues may also improve the signal-to-noise ratio [38, 53]. One approach is use of a tandem dimer MCP. Because MCP binds to MS2 stem-loops as a dimer, the tandem dimer may help to reduce background and sensitivity of labeling [38]. Another approach involves tagging the RNA of interest with a cassette of alternating MS2 and PP7 stem-loops. In this strategy, MCP and PCP are fused the N- or C-terminus of Venus fluorescent protein, respectively; fluorescence is reconstituted only when CPs are bound to their cognate stem-loops in the tagged RNA [53].
3.1.4 Sample mounting and imaging
We have visualized MS2-tagged transcripts in living oocytes and embryos using laser scanning and spinning disk confocal microscopy, widefield microscopy with deconvolution, and 2-photon microscopy [19, 26, 27, 30, 46]. We have also had good results imaging mRNA transport in larval neurons by spinning disc confocal microscopy [44, 51]. The choice of microscope will depend on the specific specimen and the particular application. For example, visualizing microtubule-dependent transport of individual RNA particles is best performed on a system that allows image capture at rates greater than 1 frame/second without substantial photobleaching (Figure 3). Widefield and spinning disc confocal microscopy are good options, although we have also successfully used a confocal microscope with fast scanning capability. Widefield microscopy, which collects all of the emitted light, combined with deconvolution, can also be highly effective for detecting dim RNA particles [29, 30]. Two-photon microscopy can be advantageous for imaging movement of RNA particles deep within the oocyte or embryo [19].
Ovaries
For MS2-tagged mRNA visualization over short time periods (≤30 min.), we typically dissect and image isolated egg chambers in halocarbon 95 oil (Halocarbon Products Corporation) on a #1.5, 22 × 50 mm glass coverslip (Figure 3). Late-stage oocytes can also be dissected into Schneider’s media (Invitrogen), mounted in Schneider’s medium on glass slides and covered with a #1.5 glass coverslip [27]. We refer the readers to Parton et. al. [54] and Weil et. al. [55] for detailed guides to preparing samples, including isolation of individual egg chambers, for short term live imaging. For longer time-lapse experiments, late stage-oocytes (≥ stage 10b) are dissected in Schneider’s media (Invitrogen), transferred to uncoated #1.5 glass-bottom culture dishes (MatTek) in 200 µl of Schneider’s media, and covered with a 1 mm2 coverslip cut from a #1.5 glass coverslip with a diamond knife to hold them in place [26, 27]. This set up also allows for the introduction of media containing cytoskeletal or metabolic inhibitors. Stage 10b egg chambers remain viable and will continue development for >8 hrs. Prolonged imaging of stage 9 oocytes can be performed by using Schneider’s media containing insulin [56].
Embryos
Embryos should be first dechorionated and washed well with dH2O. After blotting dry, transfer to halocarbon 95 oil on a #1.5, 22 × 50 mm glass coverslip as for egg chambers. Whereas egg chambers will stick to the glass surface, it is often necessary to adhere embryos to the coverslip using sticky glue made by extracting glue from packing tape using heptane [57] or double-sided tape.
4. Concluding remarks
The smFISH technique described here has seen a wide range of applications in Drosophila, from analysis of polar granule formation and composition to quantification of transcription. With the added benefit of being accessible to late-stage oocytes, smFISH offers the ability to perform sensitive, high resolution, quantitative imaging. Moreover, it is ideal for multiplexing and compatible with direct and indirect protein detection methods. The simplicity and rapidity of the protocol, which can be performed in less than one day, could make it the method of choice even for routine ISH experiments. Currently, the major drawback is cost – both the probe sets and fluorophores are expensive. However, one probe set can be used for numerous rounds of fluorophore conjugations and once conjugated, a probe set will last years.
In vivo fluorescent RNA labeling provides both a powerful stand alone tool and a complement to smFISH. Whereas smFISH produces a static view, in vivo labeling allows for live imaging needed to analyze the spatiotemporal dynamics of RNA expression and RNA trafficking. In addition, by combining orthogonal labeling methods, like MS2/MCP and PP7/PCP, the behavior of two different transcripts can be monitored simultaneously in real time. Similarly, labeled RNAs and GFP fusion proteins can be co-visualized to follow RNA-protein complexes. With the continued development of new fluorescent proteins and modifications to existing labeling methods, the future is indeed bright for RNA imaging.
Highlights.
smFISH and genetically encoded fluorescent RNA tagging provide complementary tools to visualize RNA in situ
smFISH provides quantitative detection of transcripts at high resolution with particular advangages for Drosophila oocytes
Fluorescent RNA tagging in transgenic Drosophila allows dynamic analysis of RNA through live imaging
Both methods allow multiplexing and can be combined with protein visualization
Acknowledgments
We thank S. Little, W. Eagle, and M. Niepielko for their contributions to the protocols and for many helpful discussions, and G. Laevsky for microscopy assistance and advice. We thank W. Eagle, and M. Niepielko for comments on the manuscript. This work was funded by NIH grant R01GM067758 (E.R.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Becalska AN, Gavis ER. Development. 2009;136:2493–2503. doi: 10.1242/dev.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lasko P. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medioni C, Mowry K, Besse F. Development. 2012;139:3263–3276. doi: 10.1242/dev.078626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Elife. 2015;4 doi: 10.7554/eLife.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin KC, Ephrussi A. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchand V, Gaspar I, Ephrussi A. Curr Opin Cell Biol. 2012;24:202–210. doi: 10.1016/j.ceb.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Hafen E, Levine M, Garber RL, Gehring WJ. EMBO J. 1983;2:617–623. doi: 10.1002/j.1460-2075.1983.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nüsslein-Volhard C. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 11.St Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Development. 1989;107(Suppl):13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- 12.Ephrussi A, Dickinson LK, Lehmann R. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 13.Pokrywka NJ, Stephenson EC. Development. 1991;113:55–66. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Lehmann R. Cell. 1991;66:637–647. doi: 10.1016/0092-8674(91)90110-k. [DOI] [PubMed] [Google Scholar]
- 15.Neuman-Silberberg FS, Schüpbach T. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- 16.Levsky JM, Singer RH. J Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 17.Gaspar I, Ephrussi A. Wiley Interdiscip Rev Dev Biol. 2015;4:135–150. doi: 10.1002/wdev.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinsimer KS, Lee JJ, Thiberge SY, Gavis ER. Cell Rep. 2013;5:1169–1177. doi: 10.1016/j.celrep.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Little SC, Sinsimer KS, Lee JJ, Wieschaus EF, Gavis ER. Nat Cell Biol. 2015;17:558–568. doi: 10.1038/ncb3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj A, Tyagi S. Methods Enzymol. 2010;472:365–386. doi: 10.1016/S0076-6879(10)72004-8. [DOI] [PubMed] [Google Scholar]
- 22.Little SC, Tkačik G, Kneeland TB, Wieschaus EF, Gregor T. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. Nat Commun. 2015;6:7962. doi: 10.1038/ncomms8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little SC, Tikhonov M, Gregor T. Cell. 2013;154:789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 26.Forrest KM, Gavis ER. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 27.Weil TT, Forrest KM, Gavis ER. Dev Cell. 2006;11:251–262. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Estes PS, O’Shea M, Clasen S, Zarnescu DC. Mol Cell Neurosci. 2008;39:170–179. doi: 10.1016/j.mcn.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weil TT, Xanthakis D, Parton R, Dobbie I, Rabouille C, Gavis ER, Davis I. Development. 2010;137:169–176. doi: 10.1242/dev.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinsimer KS, Jain RA, Chatterjee S, Gavis ER. Development. 2011;138:3431–3440. doi: 10.1242/dev.065029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arib G, Cléard F, Maeda RK, Karch F. Mech Dev. 2015 doi: 10.1016/j.mod.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Garcia HG, Tikhonov M, Lin A, Gregor T. Curr Biol. 2013;23:2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. Proc Natl Acad Sci U S A. 2014;111:10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lange S, Katayama Y, Schmid M, Burkacky O, Bräuchle C, Lamb DC, Jansen RP. Traffic. 2008;9:1256–1267. doi: 10.1111/j.1600-0854.2008.00763.x. [DOI] [PubMed] [Google Scholar]
- 36.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh S, Marchand V, Gáspár I, Ephrussi A. Nat Struct Mol Biol. 2012;19:441–449. doi: 10.1038/nsmb.2257. [DOI] [PubMed] [Google Scholar]
- 38.Wu B, Chao JA, Singer RH. Biophys J. 2012;102:2936–2944. doi: 10.1016/j.bpj.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao JA, Patskovsky Y, Almo SC, Singer RH. Nat Struct Mol Biol. 2008;15:103–105. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halstead JM, Lionnet T, Wilbertz JH, Wippich F, Ephrussi A, Singer RH, Chao JA. Science. 2015;347:1367–1671. doi: 10.1126/science.aaa3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. Nat Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia JF, Parker R. RNA. 2015;21:1393–1395. doi: 10.1261/rna.051797.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peabody DS, Ely KR. Nucleic Acids Res. 1992;20:1649–1655. doi: 10.1093/nar/20.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brechbiel JL, Gavis ER. Curr Biol. 2008;18:745–750. doi: 10.1016/j.cub.2008.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaramillo AM, Weil TT, Goodhouse J, Gavis ER, Schupbach T. J Cell Sci. 2008;121:887–894. doi: 10.1242/jcs.019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lerit DA, Gavis ER. Curr Biol. 2011;21:439–448. doi: 10.1016/j.cub.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, Davis I. Nat Cell Biol. 2012;14:1305–1313. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi R, Wainwright SM, Liddell SJ, Pinchin SM, Horswell S, Ish-Horowicz D. G3 (Bethesda) 2014;4:749–760. doi: 10.1534/g3.114.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 51.Xu X, Brechbiel JL, Gavis ER. J Neurosci. 2013;33:14791–14800. doi: 10.1523/JNEUROSCI.5864-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardiol A, St Johnston D. Dev Biol. 2014;392:153–167. doi: 10.1016/j.ydbio.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu B, Chen J, Singer RH. Sci Rep. 2014;4:3615. doi: 10.1038/srep03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parton RM, Vallés AM, Dobbie IM, Davis I. Cold Spring Harb Protoc. 2010;2010:pdb.top75. doi: 10.1101/pdb.top75. [DOI] [PubMed] [Google Scholar]
- 55.Weil TT, Parton RM, Davis I. J Vis Exp. 2012 doi: 10.3791/3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad M, Jang AC, Starz-Gaiano M, Melani M, Montell DJ. Nat Protoc. 2007;2:2467–2473. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- 57.Bachmann A, Knust E. Methods Mol Biol. 2008;420:61–77. doi: 10.1007/978-1-59745-583-1_4. [DOI] [PubMed] [Google Scholar]