Abstract
The regulation of RNA transcription is central to cellular function. Changes in gene expression drive differentiation and cellular responses to events such as injury. RNA trafficking can also have a large impact on protein expression and its localization. Thus, the ability to image RNA transcription and trafficking in real time and in living cells is a worthwhile goal that has been difficult to achieve. The availability of “light-up” aptamers that cause an increase in fluorescence of their ligands when bound by the aptamer have shown promise for reporting on RNA production and localization in vivo. Here we have investigated two light-up aptamers (the malachite green aptamer and the Spinach aptamers) for their suitability as reporters of RNA expression in vivo using two eukaryotic cell types, yeast and mammalian. Our analysis focused on the aptamer ligands, their contributions to background noise, and the impact of tandem aptamer strings on signal strength and ligand affinity. Whereas the background fluorescence is very low in vitro, this is not always true for cell imaging. Our results suggest the need for caution in using light-up aptamers as reporters for imaging RNA. In particular, images should be collected and analyzed by operators blinded to the sample identities. The appropriate control condition of ligand with the cells in the absence of aptamer expression must be included in each experiment. This control condition establishes that the specific interaction of ligand with aptamer, rather than nonspecific interactions with unknown cell elements, is responsible for the observed fluorescent signals. High background signals due to nonspecific interactions of aptamer ligands with cell components can be minimized by using IMAGEtags (Intracellular Multiaptamer GEnetic tags), which signal by FRET and are promising RNA reporters for imaging transcription.
Keywords: RNA imaging, aptamer, DFHBI, malachite green, fluorescence, FRET
1. INTRODUCTION
Light-up aptamers, which create a fluorescent signal on binding their ligands, are promising sensors for in vitro homogeneous assays and for applications in vivo. Although many light-up strategies require that the aptamer be chemically modified with a fluorophore, which is not possible for in vivo reporting, a subcategory with in vivo application is the group of aptamers for which binding with the aptamer increases ligand fluorescence. These aptamers include those that bind triphenylmethanes [1], bisbenzimides [2], imidazoles [3], and the benzothiazolidene asymmetric cyanine dye, thiazole orange [4]. When linked to other aptamers that drive their ability to bind ligand, these light-up aptamers can report on the presence of analytes in vitro that range from metabolites to proteins depending on the linked aptamer’s specificity [5; 6].
Expanding the options for monitoring transcriptional and metabolic activities of living cells is important for advancing understanding of cell behavior, particularly when cells are in their natural environments (such as tissues or microbial communities) consisting of many cell types and signals that are not distributed equally through the population. With their abilities to faithfully fold and function inside cells, aptamers can uniquely fill this niche for intracellular quantitative biology. Aptamers have been applied as reporters of intracellular metabolite levels [7; 8] and to image transcription and RNA localization in real time and in living cells [3; 9; 10].
For imaging, the ratio of signal to noise of the aptamer-driven light-up signal should be high. Whereas signal/noise can be optimized in vitro by changing aptamer, ligand and buffer constituents, the inviolability of the cytoplasmic environment leaves only aptamer and its ligand to optimize for in vivo function. There are also additional important parameters for in vivo imaging, which include the ability of the aptamer ligand to enter the cells, no toxicity of the ligand and the faithfulness of correct aptamer folding in vivo.
Of the light-up aptamers with applications to intracellular signaling [1; 2; 3; 4; 10], the malachite green (MGA) and Spinach2 (SPN2A) aptamers have the highest signal/noise when assayed in vitro. This contrast is much higher than achieved with the MS2-based aptamer reporters that were developed for real-time monitoring of transcription using aptamers that recognize peptides fused to GFP [11; 12]. Although the MGA and SPN2A have similar affinities for their ligands, malachite green (MG) and many of its chemical variants are toxic to mammalian cells and yeast [13], DFHBI, the Spinach ligand is not toxic [3]. However, the in vivo signals received from Spinach [3; 9] and even the improved Spinach2 [10] are quite weak and not adequate for reporting on cellular mRNA levels [9]. The possibility of expanding the repertoire of aptamer reporters to enable the simultaneous tracking of more than one mRNA in a single cell motivated us to explore the use of light-up aptamers as a complement to IMAGEtags, from which a FRET signal is derived [9].
Here we have examined two aspects of light-up aptamers that are important for realtime in vivo imaging. The first is the aptamer ligand, its signal strength and lack of signal on non-specific interaction with cellular contents. The second is the impact of tandem aptamers on fluorescent output and aptamer affinity. The outcome of this work is 1) identification of a new higher affinity ligand for Spinach aptamers with higher fluorescent yield than DFHBI, 2) demonstration that, when applied to imaging cells, several light-up aptamers give a high background fluorescence that can be largely overcome by using FRET-based aptamer reporters such as IMAGEtags [9], and 3) the finding that Spinach aptamers and MGAs perform independently when present as multiple tandem copies in a single RNA molecule and they do not cooperate synergistically for higher affinity or unit fluorescent yield.
2. MATERIALS AND METHODS
2.1. Reagents
Buffer IC (13.5 mM NaCl, 150 mM KCl, 20 mM HEPES, 0.22 mM Na2HPO4, 0.44 mM KH2PO4, 0.12 mM MgCl2, 120 nM CaCl2, 0.1 mM MgSO4, pH 7.3) was formulated to approximate intracellular pH and ionic concentrations based on literature reports for these values [14; 15; 16; 17; 18; 19; 20]. Buffer IC+Mg differed from Buffer IC in containing 5 mM Mg2+ (13.5 mM NaCl, 150 mM KCl, 20 mM HEPES, 0.22 mM Na2HPO4, 0.44 mM KH2PO4, 5 mM MgCl2, 120 nM CaCl2, 0.1 mM MgSO4, pH 7.3). Buffer S was the buffer used in SELEX for selection of the Spinach aptamer (20 mM HEPES, 125 mM KCl, 5 mM MgCl2, pH 7.4). All buffers were prepared with deionized distilled water and the pHs were measured at 23–25°C. All chemicals used for buffers were from Fisher Scientific. Malachite green oxalate was purchased from Eastman Kodak (#1264).
Spinach2 aptamers and tandem malachite green aptamers were prepared by in vitro transcription with the AmpliScribe T7 flash in vitro transcription kit (Epicenter, Madison, WI) from templates created by oligonucleotide annealing and PCR amplification. The Spinach2 RNA sequence [10] included the additional two Gs at the 5’ end that are added by T7 polymerase during in vitro transcription. The single unit malachite green aptamer was purchased from Integrated DNA Technologies (IDT; Coralville, IA). Sequences of aptamers used in this study are found in the Supplementary materials.
2.2. Synthesis of imidazolone derivatives
The general procedure for the preparation of Spinach ligands was: To the azlactone (0.200 g, 0.711 mmol) in ethanol (10 mL) was added a solution of the primary amine (0.853 mmol) in ethanol (10 mL) followed by potassium carbonate (0.1474 g, 1.07 mmol). The reaction mixture was boiled for 12 h. After cooling to room temperature, solvent was removed by evaporation. Water (15 mL) was added and the pH was adjusted to 3 using 1 M HCl. The solution was left overnight at 4 °C and the resulting precipitated product was captured by filtration. The products were yellow solids. Additional purification of PFP-DFHBI was required after collecting the precipitate from water at pH = 3. PFP-DFHBI was purified by preparative TLC on silica gel with EtOAc - Hex, 1:1. Rf = 0.7 (EtOAc - Hex, 1:1). Each compound resolved as one spot on TLC. NMR and high resolution mass spectrometry data for DFHBI matched the data reported in the literature. The NMR and high resolution mass spectrometry data for PFP-DFHBI were consistent with the assigned structure: Yellow solid: 1H NMR (400 MHz, CD3OD) δ 7.82 (d, J = 9.6 Hz, 2H), 6.89 (s, 2H), 4.98 (s, 1H), 2.42 (s, 3H); HRMS ESI (m/z): calculated for C18H10F7N2O2 [M+H]+, 419.0625; found 419.0633.
2.3. Cell culture
The mink lung epithelial cell line, Mv1Lu (ATCC, CCL64, Manassas, VA), was obtained from R. W. Holley and maintained as monolayer cultures in Dulbecco-Vogt's medium (DMEM) containing 0.45 % glucose, 10 % calf serum, 10 units/ml penicillin and 10 µg/ml streptomycin. Chinese hamster ovary (CHO) cells were maintained as monolayer cultures in F12 medium, 10 % calf serum, 10 units/ml penicillin and 10 µg/ml streptomycin. Both cell lines were cultured at 37 °C, in a water-saturated atmosphere with 10 % CO2 in air.
Yeast cells were cultured at 30 °C in SD-uracil medium containing 2 % glucose for the growth period until they reach OD600 1.0 at which time they were incubated with the aptamer ligands (either with DFHBI or PFP-DFHBI) for 60 minutes prior to imaging. Yeast strains used in this study were: BY4735: MATα ade2Δ::hisG his3Δ200leu2Δ0met15Δ0trp1Δ63ura3Δ0::ppGAL1.MCS16.pYES2/JJR1.2, which expresses the control RNA, BY4735:MATαade2Δ::hisGhis3Δ200leu2Δ0met15Δ0trp1Δ63ura3Δ0[pp5S.5SrRNA.tRNA(Lys)aSpinachtRNA(Lys)Term.pYES2/JJR167.1], which expresses Spinach RNA in a tRNA cassette, BY4735:MATαade2Δ::hisGhis3Δ200leu2Δ0met15Δ0trp1Δ63ura3Δ0pp5S.5SrRNA.tRNA(Lys)a2xSpinach2-tRNA(Lys)Term. pYES2/JJR300.2, which expresses two tandem Spinach2 aptamers and BY4735:MATαade2Δ::hisGhis3Δ200leu2Δ0met15Δ0trp1Δ63ura3Δ0::pGAL1a6xPDC::pJJR1, which expresses the 6xPDC IMAGEtag [9].
2.4. Fluorescence measurements
Fluorescence intensities were measured by a Varian Cary Eclipse spectrofluorometer or a Biotek Synergy 2 plate reader. The dissociation constants (Kd) of the aptamer-ligand complexes were determined by measuring the increase in fluorescence with increasing ligand concentration while the aptamer concentration was held constant. RNA (1 µM) was incubated for 30 min at 23 °C with 10 µM DFHBI in S buffer with 5% DMSO. Fluorescence intensities were measured with λex = 447 nm and λem = 501 nm with 5 nm slit widths. The fluorescence data, normalized to the maximum value in each dataset, was fit in Microsoft Excel using the Solver Add-in (GRG Non-linear method) and the equation F = Fmin+(Fmax*Ln)/(Ln+ Kd n) [21], where F is fluorescence, L is the concentration of ligand, n is the Hill coefficient, and Kd is the binding constant. The fit was calculated by instructing the Solver to minimize the sum of the squares while allowing Fmin, Fmax, n, Kd to vary.
2.5. Live cell imaging
For light-up aptamer imaging, the yeast cells were preincubated for 60 min with aptamer ligands, DFHBI or PFP-DFHBI, placed on a poly L-lysine coated cover glass or a poly D-lysine coated glass bottom culture dish (MatTek) then observed with a Leica SP5X laser scanning confocal microscope through a 63X objective and immersion oil. Excitation was by a 470 nm white light laser (WLL, Leica) and the fluorescence images were taken using emission filters of 475–550 nm. Each field was selected randomly prior to collecting the fluorescence images, which were subsequently collected after focusing the field using DIC.
For FRET measurements, yeast cells were incubated with the Cy3- and Cy5-linked PDC ligands [9; 22] and FRET images by sensitized emission were collected using a Leica SP5X laser scanning confocal microscope with a 63X objective and immersion oil. Cells were imaged by a sequential scan, in the first sequence cells were excited by a 550 nm white light laser (WLL, Leica) and FRET images were taken to measure sensitized emission using emission filters for FRET at 660–754 nm and 560–626 nm for the Cy3 donor. In the second sequence cell were excited at 650 nm and Cy5 emission was recorded at 660–754 nm.
Mv1Lu cell transfection was mediated by Effectene (Qiagen, Cat# 301425). The plasmids for transfection expressed either a short control RNA or a tandem series of 4 malachite green aptamers (4xMGA) driven by the human U6 promoter (see Supplementary materials). The experiment was performed and images taken with the operator blinded to the nature of the plasmid used in each transfection. Nine hours after transfection, the medium over the cells was changed to HBSS (137 mM NaCl, 5.4 mM KCl, 0.33 mM Na2HPO4, 0.44 mM KH2PO4, 0.4 mM MgSO4, 1.3 mM CaCl2, 5.55 mM glucose, 0.49mM MgCl2, 0.006% w/v phenol red, pH 7.4) and the cells incubated with MG for varying periods as identified in the figures and legends. The cells were imaged using a Nikon Eclipse TE200 microscope with filter set Omega #XF110-2 (Cy5) and a 20X objective, λex = 630 nm; λem =695nm. Scale bars shown in one or more images of a set apply to all images in the entire set. Color versions of many images shown in figures 4 and 5 are found in the supplementary materials (Figs. S1–S7).
Figure 4. Imaging RNA in mammalian cells with MG, DFHBI and PFP-DFHBI.
A) Images of CHO cells transiently transfected with plasmids from which either 4xMGA or control RNA was expressed. The cells were imaged with MG 9 h after transfection, B) Time course of the increase in fluorescence background after the addition of 10 µM MG to the cell medium, C) Mv1Lu cells were transiently transfected with plasmids from which control or SPN1A RNA were expressed from the CMV promoter. All cells were cotransfected with a plasmid from which DsRed was expressed from the CMV promoter. DsRed was used as an internal control to verify transfection of the cells and transcription of RNA from the plasmids. Cells were imaged 24h after transfection.
Figure 5. Imaging RNA in yeast cells with Spinach and IMAGEtag reporters.
A) Yeast cells expressing control RNA, SPN1A or 2xSPN2A were incubated at 30°C with DFHBI or PFP-DFHBI and fluorescence and DIC images taken after 60 min, B) The fluorescence images in A, and others from the same experiment, were quantified for fluorescence per cell using ImageJ. The results are shown for each ligand normalized to the average fluorescence per cell of the control RNA-expressing cells. Images of field as higher resolution are shown in Fig. S1 and Fig. S2. C) Representative images of cells (63x objective with 3.5X zoom in) from the experiment in A to demonstrate that the high background signal is not due to vacuolar uptake of the ligands. D) Images of yeast cells expressing GAL1 promoter-driven 6XPDC IMAGEtags taken 99 min after the promoter was induced by the addition of 2% galactose to cells that had previously been grown for 12 h in 2% raffinose to reach 0.7 OD600. The images for Cy3-PDC only, Cy5-PDC only, or Cy3-PDC, Cy5-PDC are of cells that have been incubated with either one or the other PDC ligand or the ligand pair (Cy3-PDC, Cy5-PDC) that interacts in FRET. Images were taken in the Cy3, Cy5 or FRET channels. The DIC is of the field shown for the ligand pair and is representative of all fields in this experiment. E) FRET efficiencies from the experiment in D determined from images taken each 1 or 2 min after induction were quantified for 8 cells expressing control RNA and for 12 cells expressing 6xPDC IMAGEtags and averaged for each group. The average FRET/cell of the IMAGEtag expressing group was divided by the average FRET/cell of the control RNA expressing group and plotted as a function of time after the addition of galactose. F) The chemical structure of Cy3-PDC. Cy5-PDC is identical in structure with the exception that Cy5 replaces Cy3.
MuLv1 transfection was mediated by the K2 Transfection System (Biontex Laboratories GmbH). The plasmids for transfection expressed the Spinach aptamer (SPN1A) or dsRed from a CMV promoter. Cells were transfected with the dsRed-expressing plasmid with or without the SPN1A-expressing plasmid. Twenty four hours after transfection, either DFHBI or PFP-DFHBI was added to a final concentration of 20 µM, 0.1% DMSO and incubated at 37°C in 10% CO2 for 45 min prior to imaging using a Leica confocal microscope (Leica SP5 X MP confocal/multiphoton microscope system).
3. RESULTS
3.1. PFP-DFHBI, a DFHBI analog with higher affinity and fluorescent yield
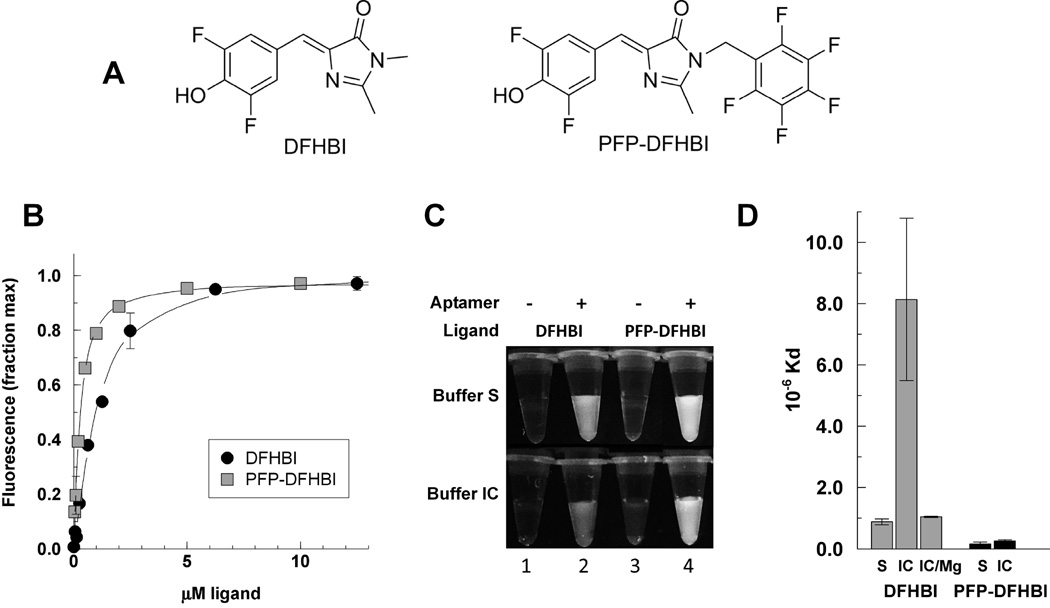
A limitation for using the Spinach and Spinach2 aptamers for live-cell imaging is the weak signal from DFHBI. To improve on this signal we prepared a new ligand PFP-DFHBI (Fig. 1A), which binds Spinach2 with a ~5-fold higher affinity than DFHBI (Fig. 1B). The Kd of Spinach2 for DFHBI is 0.88 ± 0.010 µM compared with a Kd of 0.16 ± 0.060 µM for Spinach2 and PFP-DFHBI. The fluorescent yield of the PFP-DFHBI-Spinach2 complex is also higher than for the DFHBI-Spinach complex (Fig. 1C). These measurements were made in the buffer in which the Spinach aptamer was selected (Buffer S). However, Buffer S contains 5 mM MgCl2, which is significantly higher than the free Mg2+ ion concentration in the eukaryotic cytoplasm [16; 20]. The dependence of Spinach2 on MgCl2 is evident when its affinity for DFHBI is measured in Buffer IC, which is formulated to resemble the eukaryotic cytoplasm salt concentrations (Fig. 1D). In Buffer IC, the affinity of Spinach2 for DFHBI plummets by 10-fold compared with in Buffer S. This reveals one reason for the weakness of the signal from intracellular Spinach aptamers and DFHBI. The effect of raising the Mg2+ concentration of Buffer IC to 5 mM is to increase the affinity to close to the Kd for DFHBI in Buffer S (Fig. 1D). By contrast, PFP-DFHBI binds equally well to Spinach2 in Buffers S and IC. Compared with DFHBI, the fluorescence yield of PFP-DFHBI is increased 3.0-fold in buffer S and 3.8-fold in buffer IC.
Figure 1. Affinities of Spinach2 for DFHBI and PFP-DFHBI and effect of Mg on the interactions.
A) Chemical structures. B) The ligands were measured for their affinity to the Spinach aptamer by the increase in fluorescence as a function of the ligand concentration in buffer S. C) Fluorescence of DFHBI versus PFP-DFHBI under UV illumination with 254nm UV lamp. D) Kds of DFHBI and PFP-DFHBI determined as in B in buffer S (black bars) or buffer IC (grey bars). Error bars are the standard deviations of two or more independent replicates under the same conditions.
3.2. Spinach2 and MGA Multimers show increased ligand binding capacities but the same affinities
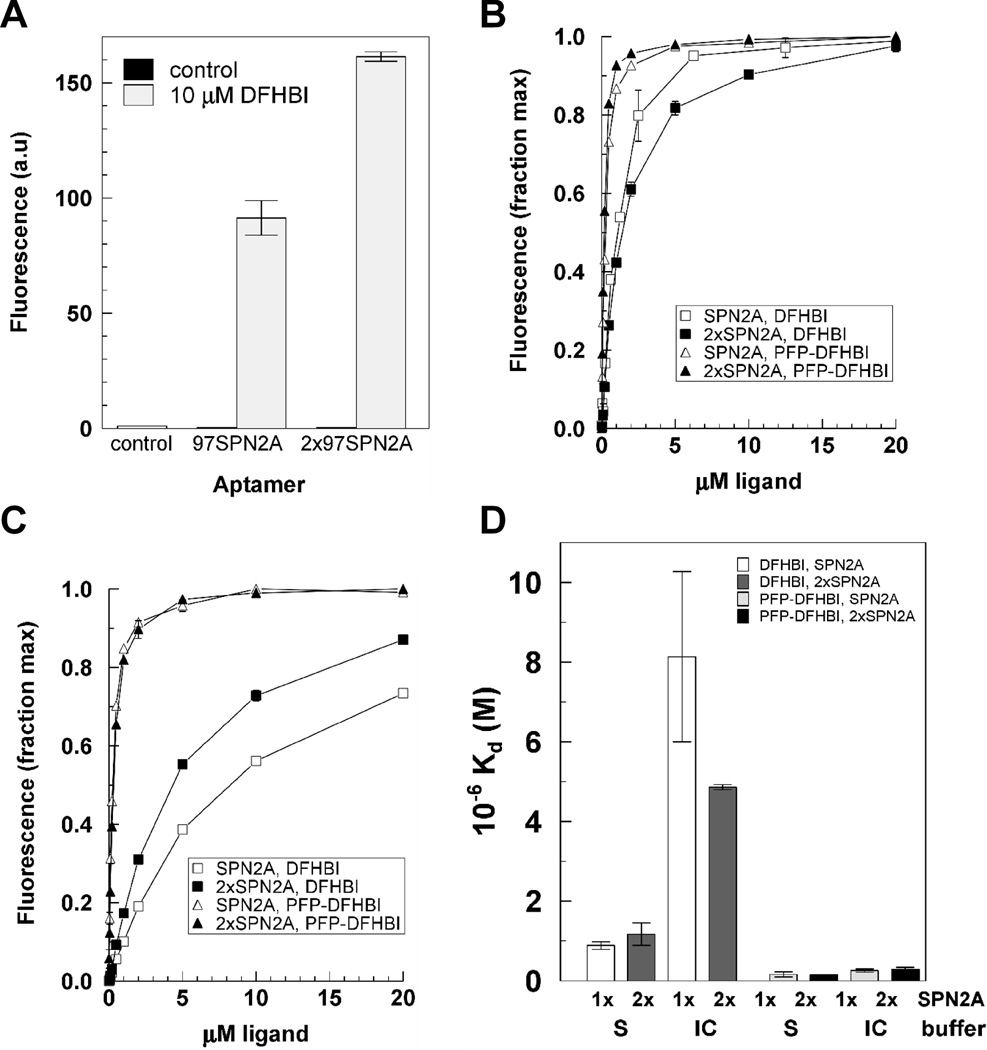
The strength of Spinach2 signal from an RNA reporter molecule can be increased if the reporter contains multiple tandem aptamers. If constructed to allow each aptamer to fold properly for ligand binding, the increase in brightness per RNA will be a linear function of the number of aptamers as observed for the Spinach2 aptamer (Fig. 2A). Affinity for ligand is also important because intracellular ligand concentrations are expected to reach levels much lower than outside the cell. Tandem aptamers have been reported to have higher affinities than single aptamers for their protein ligands [23; 24]. However, 2xSPN2A showed no increase in affinity for DFHBI or PFP-DFHBI either in Buffer S (Fig. 2B,D) or in Buffer IC (Fig. 2C,D).
Figure 2. Characteristics of ligand binding with tandem Spinach2 aptamers.
A) Relative fluorescence yield from SPN2A and 2xSPN2A on a molar basis, B) Binding curves for SPN2A and 2xSPN2A in Buffer S, C) Binding curves for SPN2A and 2xSPN2A in Buffer IC, D) Kds of DFHBI and PFP-DFHBI for SPN2A and 2xSPN2A in buffers IC and S.
We also tested tandem multimers of the MGA, another light-up aptamer for which the aptamer-ligand complex shows ~2360-fold increase in MG fluorescence [Fig. 3A, 1]). In the supplemental materials for a previous study, the data for multimers of one through five MGA aptamers showed an interesting step-wise increase in fluorescence intensity with increasing number of aptamers that is tightly defined by the small estimates of error that is better fit to a polynomial rather than a straight line [1]. Strings of 4 and 8 tandem MGA aptamers were compared with the single aptamer for their binding capacities at saturating ligand concentrations and their ligand affinities. As for Spinach2, the MGA multiaptamers (4xMGA and 8xMGA) bound MG in proportion to the number of tandem aptamers (Fig. 3B). Although we observed an apparent small increase in affinity with increasing numbers of tandem aptamers (Fig 3C,D), this increase was not statistically significant.
Figure 3. Characteristics of ligand binding with tandem malachite green aptamers.
A) Fluorescence spectrum of MGA upon binding 10 µM MG, λex = 630 nm. Dashed line, fluorescence of control RNA with 10 µM MG, B) Relative fluorescence yield from 10 µM MG interacting with MGA, 4xMGA and 8xMGA on a molar basis. Aptamer concentrations were 0.8, 0.2, and 0.08 µM respectively. C) Binding of MG to MGA, 4xMGA and 8xMGA (each normalized to the maximum fluorescence). D) Kds of MGA (1) 4xMGA (2) and 8xMGA (8) for MG.
3.3. Imaging RNA transcription in mammalian cells
The MGA was tested for its ability to report on RNA expression in mammalian cells. A control RNA or an RNA reporter containing four tandem MGA aptamers were expressed from the human U6 promoter in transiently transfected CHO cells. An increase in fluorescence was observed in cells that expressed the MGA (Fig. 4A). However, the effective imaging period was limited to less than 10 min because the background fluorescence increased rapidly with time (Fig. 4B), thereby decreasing signal/noise until the aptamer reporter signal could not be observed above the background.
The Spinach aptamer was tested for its ability to report on RNA expression from a CMV promoter in MuVL1 cells. The background from both DFHBI and PFP-DFHBI was undetectable. However, under our conditions, we were unable to observe a signal from the Spinach reporter from an RNA Pol II promoter (Fig. 4C) even though the majority of cells were transfected (as evidenced by the DsRed images).
3.4. Imaging RNA transcription in yeast
We tested the ability of Spinach and 2xSPN2A to report on RNA expression in yeast. The yeast 5S promoter was used to drive aptamer reporter or control RNA expression. As we have observed previously [9] the DFHBI signal was very weak and very few cells were fluorescent (Fig. 5A). By contrast, the PFP-DFHBI signal was quite strong. Whereas signal was observed in only 6% of the 955 cells examined that had been incubated with DFHBI, 56% of the 340 cells examined were visibly fluorescent that had been incubated with PFP-DFHBI. However, comparison of fluorescence intensity per cell showed that the signal was equally strong in cells that expressed or did not express 2xSpinach2 (Fig. 5A,B). Closer analysis of the cells showed that the high background with PFP-DFHBI was not due to accumulation of the compound in the vacuoles (Fig. 5D). However, staining over the cells was granular for both DFHBI and PFP-DFHBI, which suggested that these compounds were accumulating preferentially in certain cell compartments.
A high background of ligand retention such as observed with PFP-DFHBI and MG can be circumvented if tandem aptamer reporters are used in conjunction with ligand FRET pairs as shown for IMAGEtags [9]. Whereas the Cy3-PDC and Cy5-PDC ligands each give a significant background fluorescence (Fig. 5D), the FRET image, which requires that both ligands be held in close proximity by tandem aptamers, effectively reports on RNA expression from the GAL1 promoter (Fig. 5E).
4. DISCUSSION
Light-up aptamers have the capability of reporting on transcriptional activity and being monitored in real-time in living cells. However, to realize their promise, the sensitivity of aptamer detection in cells must be increased. For reporters such as Spinach2 and MGA, the sensitivity can be increased by increasing the length of tandem aptamers and by improving the ligand for cell permeability, aptamer affinity, and increased signal strength. We have tested two light-up aptamers and their ligands for their efficacy in reporting on RNA expression in yeast and mammalian cells and compared them to FRET-based IMAGEtags [9].
For increased signal strength from the Spinach aptamers, we identified a new ligand, PFP-DFHBI, that binds with a 40-fold higher affinity in intracellular conditions and for which binding is unaffected by the low intracellular free Mg2+ concentrations. The new ligand gives a brighter signal upon binding SPN2A with a 3-fold increase in fluorescence yield compared with the currently used Spinach aptamer ligand, DFHBI. The interaction of PFP-DFHBI with Spinach is independent of Mg2+, which is important for intracellular activity of the aptamer.
Increased signal strength can also be achieved with multiple aptamers per RNA reporter. A nonlinear increase in fluorescence emission with respect to aptamer numbers was shown in an early study in which it was suggested that each second aptamer might serve the function of a spacer and that increasing the spacer length could enable a linear increase in fluorescence intensity with number of aptamers [supplemental materials for ref. 1]. Whereas the previous constructs included 6 base spacers between aptamers, our multimers have 7 bases between most MGAs in the string with 19 bases between sets of 4 aptamers. Although the two results differ quantitatively, our results and the previously reported results are in agreement that the fluorescence intensity of multimers increases with the number of tandem aptamers.
Others have reported significant increases in affinities of aptamer that recognize proteins when the aptamers are linked in tandem [23; 24]. Ligand affinity was not evaluated for the previously reported MGA multimers [1] and we show here that the Spinach and the MGA tandem aptamers bind their ligands with the same affinity as the monomeric forms. These RNA aptamers recognize much smaller molecules than the aptamers for which affinity increases of tandem arrays have been reported. Based on the expectation that the off-rate of the aptamer-ligand equilibrium likely determines the affinity, we postulate that the basis of the observed increase in affinity with tandem aptamers is the ability of one aptamer to capture a ligand released by another. Molecular diffusion rates are an inverse function of molecular size and the more rapid movement of the small molecule ligands from the aptamers may disallow immediate recapture by an adjacent aptamer. However, these in vitro studies were performed in a buffer of low viscosity. Affinities of tandem aptamers may be higher than single aptamers in the more viscous intracellular environment.
An increase in signal was obtained in mammalian cells with the 4xMGA reporter for RNA expressed from the human U6 (Pol III) promoter. However, the rapidly increasing intracellular background prevents the use of the MGA for imaging long-term activities in mammalian cells. Also, MG is highly toxic to mammalian cells and yeast [13], which makes this light-up aptamer a poor choice as a reporter for RNA expression in living cells.
A critical weakness of the light-up aptamer approach for signaling RNA expression in living cells is that light-up ligands tend to fluoresce when they bind intracellular components and thereby decrease the signal/noise such that imaging is difficult or impossible. In cases where no background signal is produced by the light-up ligand (such as DFHBI), the reporter signal is also low and very few cells are labeled. With a brighter ligand such as PFP-DFHBI, it can clearly be seen that fluorescence per cell varies, with some cells being brighter than the others in the population. The cells were not always randomly distributed in a field and some areas contained a large number of cells with either higher or lower levels of fluorescence (Figs. S1, S2). However, in yeast, the same average fluorescence was observed per cell from the control, Spinach2 or tandem Spinach2 aptamer reporters. These results underline the need to examine large fields, rather than focusing on a few cells, and to follow good practices in analysis by microscopy, which include random selection of several fields and performing image collection and analysis by individuals blinded to the identities of the samples.
The Spinach aptamers have been used to detect 5S RNA and trinucleotide repeats in mammalian cells [3; 10; 21; 25]. These cellular RNA levels are very high, which may account for the observed signal from the reporters. By comparison, we were unable to detect RNA expressed from the CMV promoter, a pol II promoter, using the Spinach reporters. The ability to use light-up aptamer reporters may also depend on the cell type. A comparison of the background fluorescence from PFP-DFHBI for yeast and mammalian cells suggests that the light-up ligands may enter yeast cells more readily than mammalian cells. Whereas the light-up aptamer approach does not provide a signal for RNA expression in yeast, approaches that involve a change in the fluorescent properties of the aptamer ligand when it binds the aptamer, such as release of fluorescence quenching by turn-on aptamers [26; 27], or FRET as shown here with IMAGEtags [9] and by others with protein reporters [12; 28], are effective reporters of transcription even when the ligand adsorbs non-specifically to cellular materials.
5. CONCLUSION
From these studies we conclude that light-up aptamers will prove difficult to apply generally as in vivo reporters of RNA expression. The presence of high backgrounds that are not observed in vitro, necessitate that all studies with light-up aptamers include control conditions in which the ligand alone is present and that image collection and analysis is based on random selection of fields for analysis and performed by individuals blinded to the identities of the samples. Approaches that minimize background fluorescence, such as FRET-based aptamer reporters or turn-on reporters that require aptamer binding for fluorescence output, can be successfully applied to imaging RNA in living cells.
Supplementary Material
Highlights.
A new Spinach ligand with 40-fold higher affinity and 4-fold higher fluorescence
High background adsorption with light-up aptamer ligands
FRET-based IMAGEtags signal RNA expression in the presence of high backgrounds
Tandem arrays of light-up aptamers do not cooperate for increased binding affinity
ACKNOWLEDGEMENT
Financial support was provided by grants R01EB005075 and R21AI114283 to MN-H from the National Institutes of Health (chemistry and imaging) and funds from the U.S. Department of Energy, Office of Biological and Environmental Research through the Ames Laboratory (fluorescence measurements). The Ames Laboratory is operated for the U.S. Department of Energy by Iowa State University under Contract No. DE-AC02-07CH11358. We thank Jayeeta Banerjee and Lisa Cannistraci Patrin for preliminary work with the MGA and preparing some of the plasmids used in this study.
Abbreviations
- ddH2O
deionized distilled water
- MGA
malachite green aptamer
- MG
malachite green
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 2.Sando S, Narita A, Hayami M, Aoyama Y. Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: a blue fluorescent RNA. Chem Commun (Camb) 2008:3858–3860. doi: 10.1039/b808449a. [DOI] [PubMed] [Google Scholar]
- 3.Paige JS, Wu KY, Jaffrey SR. RNA Mimics of Green Fluorescent Protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dolgosheina EV, Jeng SC, Panchapakesan SS, Cojocaru R, Chen PS, Wilson PD, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 5.Kellenberger CA, Hammond MC. In vitro analysis of riboswitch-Spinach aptamer fusions as metabolite-sensing fluorescent biosensors. Methods Enzymol. 2015;550:147–172. doi: 10.1016/bs.mie.2014.10.045. [DOI] [PubMed] [Google Scholar]
- 6.Kellenberger CA, Chen C, Whiteley AT, Portnoy DA, Hammond MC. RNA-Based Fluorescent Biosensors for Live Cell Imaging of Second Messenger Cyclic di-AMP. J Am Chem Soc. 2015;137:6432–6435. doi: 10.1021/jacs.5b00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kellenberger CA, Hallberg ZF, Hammond MC. Live Cell Imaging Using Riboswitch-Spinach tRNA Fusions as Metabolite-Sensing Fluorescent Biosensors. Methods Mol Biol. 2015;1316:13187–13103. doi: 10.1007/978-1-4939-2730-2_8. [DOI] [PubMed] [Google Scholar]
- 8.Song RLSW, Jaffrey SR. Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria. Nature protocols. 2014;9:146–155. doi: 10.1038/nprot.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin I, Ray J, Gupta V, Ilgu M, Beasley J, Bendickson L, et al. Livecell imaging of Pol II promoter activity to monitor gene expression with RNA IMAGEtag reporters. Nucleic Acids Res. 2014;42:e90. doi: 10.1093/nar/gku297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nat Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA Particles in Living Yeast. Molecular Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 12.Wu B, Chen J, Singer RH. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Scientific Reports. 2014;4:3615. doi: 10.1038/srep03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraus GA, Jeon I, Nilsen-Hamilton M, Awad AM, Banerjee J, Parvin B. Fluorinated analogs of malachite green: synthesis and toxicity. Molecules. 2008;13:986–994. doi: 10.3390/molecules13040986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rorsman P, Berggren PO, Hellman B. Manganese accumulation in pancreatic beta-cells and its stimulation by glucose. Biochem J. 1982;202:435–444. doi: 10.1042/bj2020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ammann H, Noel J, Tejedor A, Boulanger Y, Gougoux A, Vinay P. Could cytoplasmic concentration gradients for sodium and ATP exist in intact renal cells? Can J Physiol Pharmacol. 1995;73:421–435. doi: 10.1139/y95-055. [DOI] [PubMed] [Google Scholar]
- 16.Grubbs RD. Intracellular magnesium and magnesium buffering. Biometals. 2002;15:251–259. doi: 10.1023/a:1016026831789. [DOI] [PubMed] [Google Scholar]
- 17.Auffinger P, Grover N, Westhof E. Metal ion binding to RNA. Met Ions Life Sci. 2011;9:1–35. [PubMed] [Google Scholar]
- 18.Christian JH, Waltho JA. The sodium and potassium content of nonhalophilic bacteria in relation to salt tolerance. J Gen Microbiol. 1961;25:97–102. doi: 10.1099/00221287-25-1-97. [DOI] [PubMed] [Google Scholar]
- 19.Csonka LN. Physiological and genetic responses of bacteria to osmotic stress. Microbiol Rev. 1989;53:121–147. doi: 10.1128/mr.53.1.121-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romani A. Regulation of magnesium homeostasis and transport in mammalian cells. Arch Biochem Biophys. 2007;458:90–102. doi: 10.1016/j.abb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, et al. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraus GA, Gupta V, Mokhtarian M, Mehanovic S, Nilsen-Hamilton M. New effective inhibitors of the Abelson kinase. Bioorg Med Chem. 2010;18:6316–6321. doi: 10.1016/j.bmc.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proceedings of the National Academy of Sciences. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonaka Y, Yoshida W, Abe K, Ferri S, Schulze H, Bachmann TT, et al. Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal Chem. 2013;85:1132–1137. doi: 10.1021/ac303023d. [DOI] [PubMed] [Google Scholar]
- 25.Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-Play Fluorophores Extend the Spectral Properties of Spinach. Journal of the American Chemical Society. 2014;136:1198–1201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunbul M, Jaschke A. Contact-mediated quenching for RNA imaging in bacteria with a fluorophore-binding aptamer. Angew Chem Int Ed Engl. 2013;52:13401–13404. doi: 10.1002/anie.201306622. [DOI] [PubMed] [Google Scholar]
- 27.Arora A, Sunbul M, Jaschke A. Dual-colour imaging of RNAs using quencher- and fluorophore-binding aptamers. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B, Miskolci V, Sato H, Tutucci E, Kenworthy CA, Donnelly SK, et al. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev. 2015;29:876–886. doi: 10.1101/gad.259358.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.