Abstract
Clustered, regularly interspaced, short palindromic repeats - CRISPR associated (CRISPR-Cas) systems are sequence specific RNA-directed endonuclease complexes that bind and cleave nucleic acids. These systems evolved within prokaryotes as adaptive immune defenses to target and degrade nucleic acids derived from bacteriophages and other foreign genetic elements. The antiviral function of these systems has now been exploited to combat eukaryotic viruses throughout the viral life cycle. Here we discuss current advances in CRISPR-Cas9 technology as a eukaryotic antiviral defense.
Keywords: Cas9, virus, antiviral, HIV, HBV, HCV
The Prokaryotic Immune System
Prokaryotic cells possess innate and adaptive immune systems that conceptually parallel those found in eukaryotic organisms [1]. Prokaryotic restriction endonucleases function as an ‘innate’ immune defense mechanism, recognizing conserved nucleotide sequences and subsequently cleaving foreign nucleic acids. These endonucleases have formed the platform for the generation of powerful tools such as recombinant DNA technology and genome sequencing. Alternatively, CRISPR-Cas systems serve as the ‘adaptive’ immune system of bacteria and archaea. By incorporating short sequences of bacteriophage-derived or other foreign nucleic acids into their own genome, prokaryotes use CRISPR-Cas systems to recognize new targets and degrade these invaders upon secondary encounter, analogous to a memory response in eukaryotic organisms [2].
Cas9-mediated Targeting of Bacteriophages
The clustered, repetitive sequences (CRISPR array) that form the central feature of CRISPR-Cas systems were first discovered in 1987 in Escherichia coli [3], although it was not until 2007 that the function of CRISPR sequences and their conserved, adjacent cas genes (CRISPR-associated genes) was first described. Barrangou and colleagues demonstrated that upon bacteriophage infection, Streptococcus thermophilus integrated phage genomic sequences into the CRISPR array, and that these sequences, in conjunction with the cas genes, provided protection from subsequent viral challenge [4]. CRISPR-Cas systems have now been identified in over 90% of sequenced archaea, as well as roughly 50% of bacterial species [5, 6]. The class II CRISPR-Cas9 system has been the most extensively studied and is discussed in detail below.
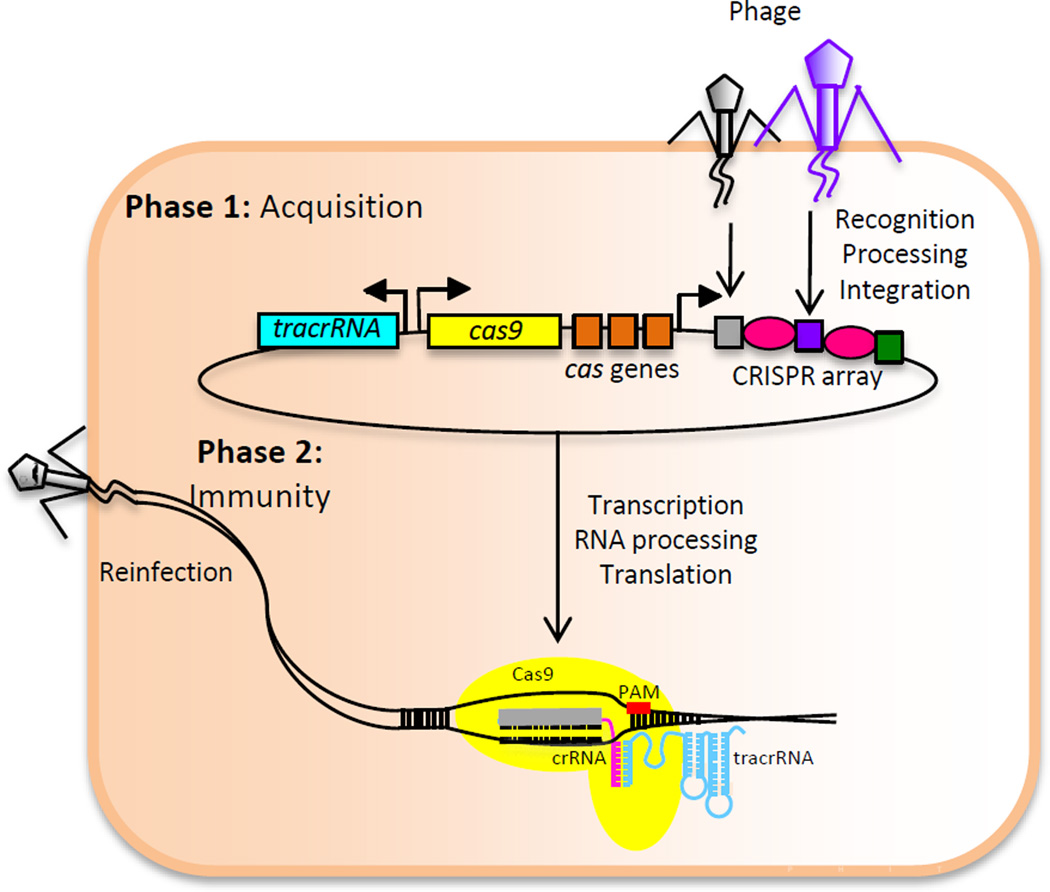
There are two distinct stages of Cas9-mediated immunity (Figure 1). During the acquisition phase, a bacterium encounters foreign nucleic acid, such as that of a phage genome. A portion of the phage genome is incorporated into the CRISPR array, and is termed a spacer [4, 7, 8]. The subsequent immunity phase occurs when the bacterium again encounters an identical foreign nucleic acid and proceeds to catalyze its cleavage. In this phase, Cas9 is guided by two small RNAs, the trans-activating RNA (tracrRNA) and the CRISPR RNA (crRNA) [9, 10]. When the associated crRNA, transcribed from the CRISPR array, has complementarity to the invading phage sequence, the two Cas9 endonuclease domains (RuvC and HNH) mediate cleavage of the targeted sequence. Endonuclease activity also requires that a short sequence on the foreign DNA adjacent to that bound by the crRNA, known as the protospacer adjacent motif (PAM), is recognized by Cas9 [10] (Figure 1).
Figure 1. Cas9-mediated Immunity in Type II CRISPR-Cas Systems.
Phase 1: Acquisition. Upon encounter with foreign nucleic acid, such as that of a bacteriophage genome, a short sequence of the phage DNA is recognized, processed into small portions, and integrated into the CRISPR array by Cas proteins. Each sequence, termed a spacer, (gray, purple, and green boxes) is incorporated between identical CRISPR repeats (pink ovals). Phase 2: Immunity. In this stage, the bacterium encounters an identical foreign nucleic acid. Two small RNAs, the crRNA and the tracrRNA, guide the Cas9 effector protein to the phage genome. If there is sufficient sequence complementarity between the crRNA and the phage genome and a protospacer adjacent motif (PAM) is present, Cas9 then cleaves both strands of the target.
In addition to its natural role within bacteria, the Cas9 system has been harnessed for diverse applications in eukaryotic cells. Cas9 can be programmed to target and cleave double-stranded DNA (dsDNA) sequences of interest by engineering single chimeric guide RNAs (gRNAs) composed of portions of the tracrRNA and crRNA [10] (Figure 2). Target specificity is achieved by simply modifying the short spacer region of the gRNA to a sequence complementary to the target. Co-expression of the gRNA with Cas9 in the cell of interest leads to target cleavage. Genomic alterations have already been performed in diverse cell types, including those from zebrafish [11, 12], mice [13–15], humans [10, 14, 16, 17], and an abundance of other organisms. Further, Cas9-based technologies have now been successfully exploited against eukaryotic viruses at different stages of their life cycle.
Figure 2. Cas9-mediated Cleavage of the HIV Provirus.
A synthetic guide RNA (gRNA), shown in blue, is composed of a duplex of the crRNA and tracrRNA and may be engineered to target virtually any sequence of interest. Cas9 targeting is dependent on sequence complementarity with the target, in this case the HIV provirus, and a protospacer adjacent motif (PAM), shown in red. Following DNA recognition and binding, two Cas9 endonuclease domains, the HNH and RuvC domains, cleave the complementary and non-complementary DNA strands, respectively.
Cas9-mediated Inhibition of Eukaryotic Viruses
While directly targeting viral nucleic acids is an obvious strategy to inhibit viral replication, Cas9 has also been targeted to disrupt host factors critical for the viral life cycle. Additional strategies include using Cas9-transcriptional fusion proteins to reactivate viruses and render them susceptible to killing by the immune system, or to induce transcription of antiviral genes. Together, these avenues may provide effective means to inhibit, or even clear, viral infections. In this review, we discuss examples of pathogenic eukaryotic viruses that have been targeted by Cas9, as well as current obstacles, future implications, and questions regarding this technology.
Human immunodeficiency virus
More than 35 million people worldwide are infected with human immunodeficiency virus (HIV) [18]. HIV infects cells using the primary receptor CD4 [19] and chemokine co-receptors CCR5 [20] or CXCR4 [21]. If left untreated, infection can lead to progressive impairment and depletion of immune cells, particularly CD4+ T cells, and acquired immunodeficiency syndrome (AIDS) [18]. Despite significant advances in antiretroviral regimens, treatment toxicity and viral resistance are limitations of current therapies. A vaccine remains elusive despite intense research effort [22–24].
Certain individuals possess a naturally occurring 32-nucleotide deletion within CCR5 that confers resistance to CCR5-tropic HIV-1 infection [25]. In the highly publicized case of the Berlin patient, Timothy Brown received a bone marrow transplant for myeloid leukemia from a donor who was homozygous for this 32-nucleotide deletion in the ccr5 gene. As a consequence, viremia was undetectable following the transplant, even after cessation of antiviral therapy [26, 27]. This fortuitous occurrence highlighted the potential for this CCR5 deletion to be harnessed therapeutically.
Since then, several gene editing strategies have been utilized to generate the CCR5 deletion, including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and Cas9 [28–31]. Briefly, ZFNs are recombinant proteins comprised of sequence-specific zinc finger DNA binding domains fused to the endonuclease domain of the bacterial FokI restriction enzyme. ZFNs are designed in pairs, and following DNA binding, FokI endonuclease domains dimerize to induce site-specific cleavage [32]. Like ZFNs, TALENs are proteins consisting of novel DNA binding domains fused to the endonuclease domain of FokI. Each TALE repeat confers specificity for a single nucleotide, so virtually any DNA sequence can be targeted. ZFNs and TALENs have been reviewed extensively elsewhere [33–35], but importantly, one of the major drawbacks of these technologies is that a new protein must be engineered for each target sequence, so protein design and synthesis remain significant obstacles. Consequently, the emerging Cas9 platform has quickly become the preferred approach for genome engineering due to its ease of use. Only an RNA needs to be altered to program the system for each new target.
Ye et al. recently demonstrated that deletion of the 32 bp region of the ccr5 gene with Cas9:gRNA complexes conferred resistance to CCR5 tropic HIV-1 in monocytes and macrophages differentiated from induced pluripotent stem cells [28]. Other groups have also reported CCR5 disruption in HIV-1 permissive cell lines [29], primary CD4+ T cells [30, 31], as well as hematopoietic stem and effector cells [31]. Together, these findings reveal the utility of Cas9 in generating CCR5 deletion mutations to inhibit HIV entry and infection.
As HIV-1 may also use CXCR4 as a co-receptor for cellular entry [21], Cas9 has been utilized to disrupt the cxcr4 gene in GHOST (a human osteosarcoma cell line expressing CXCR4) and Jurkat cell lines, as well as Rhesus macaque primary CD4+ T cells in vitro [36]. p24 capsid antigen levels were reduced by approximately four-fold, seven days post infection in the supernatants of primary CD4+ cells transfected with Cas9 and CXCR4-specific gRNA [36]. This concept of host gene modulation by Cas9 could potentially be applied to countless other viruses with life cycles that are dependent upon specific host proteins.
An alternative strategy to combat HIV is to directly target the HIV provirus by Cas9-mediated cleavage. The provirus is generated when virally encoded polymerase (Pol) protein reverse transcribes the RNA genome of the virion into a DNA form. RNase H, also virally encoded, degrades the RNA strand, thus allowing the DNA polymerase activity of Pol protein to synthesize the complementary DNA strand [37, 38]. This dsDNA form, when integrated into the host chromosome is termed the provirus, which serves as a fundamental hurdle to curing HIV infection [39–42]. An HIV provirus can be transcriptionally silent, though in the more common scenario, the provirus serves as a key transcriptional template for HIV RNAs [42]. Antiviral therapies target transcriptionally active proviruses, while transcriptionally inactive proviruses escape both therapeutics and immune recognition [42]. Cells containing the provirus maintain a persistent reservoir that poses a long-lasting health risk to infected individuals.
Several studies have demonstrated that the HIV provirus can be disrupted or excised from latently infected cell lines using Cas9 [43–45]. Ebina et al. utilized an HIV vector expressing GFP under the control of a long terminal repeat (LTR) promoter to mimic latent HIV infection. Transfections of plasmids encoding Cas9 or gRNA targeted to the LTR in Jurkat cells reduced GFP expression by more than half relative to cells transfected with a control vector [43]. Expression of Cas9 and gRNA targeted to the LTR region of an HIV-1 reporter virus in microglial cells also reduced the frequency of GFP expressing cells from 76% to 4% [44]. Others have shown that Cas9 treatment was efficacious in latently infected U1 monocytic cells, and that when HeLa cells stably expressing Cas9 and an LTR-specific gRNA were infected with a GFP reporter virus, the percentage of GFP+ cells was reduced to less than 10%, as compared to roughly 60% in cells expressing a control gRNA [44]. Further, Zhu and colleagues designed a panel of 10 gRNAs targeted to different regions of the HIV genome. Expression of three multiplexed gRNAs and Cas9 in Jurkat cells expressing a full length HIV-1 reporter virus reduced viral particle production by more than 20-fold relative to vector controls [45]. Liao et al. expanded on these studies to demonstrate that the viral cDNA may be disrupted prior to chromosomal integration [46]. More importantly, Cas9 and HIV-specific gRNA expression within primary CD4+ T cells and induced monocyte/macrophage cells reduced virus production by approximately three-fold and ten-fold, respectively [46].
An additional approach to inhibition of HIV has been based on Cas9-transcriptional activator proteins (fusion proteins of catalytically inactive Cas9 to a transcriptional activation domain from another protein), which were originally engineered to induce eukaryotic host gene expression [47, 48]. This approach has been applied to activate transcription of the antiviral restriction factor APOBEC3B, which can inhibit HIV-1 in cell culture [49]. Another strategy, termed ‘shock and kill’, relies on Cas9 to reactivate transcriptionally inactive virus from cellular reservoirs, which will then be cleared by host immune defenses and/or with conventional antiretroviral therapy. Several groups have engineered Cas9-transcriptional activator proteins that specifically target the HIV-1 LTR promoter sequence. These fusion proteins may provide enhanced specificity relative to traditional agents used for provirus reactivation [50–52]. Taken together, the aforementioned studies highlight the diverse ways in which Cas9 technology is being used to directly target the HIV provirus and/or the receptors important for its entry, reactivate latent virus, or stimulate antiviral gene induction. A critical question is how these technologies will translate to potential use in humans.
Although the aforementioned studies are promising, the efficient delivery of Cas9 and gRNAs in vivo poses a major challenge, not only due to the sheer number of intended recipient CD4+ T cells within the body, but also to the anatomic locations of viral reservoirs. Common viral vectors such as adeno-associated viruses (AAV) and lentiviruses are currently unsuitable for in vivo delivery for any of the approaches described above. For example, AAV does not readily transduce T cells [53], and lentiviruses are generally produced at titers of no greater than 109 transducing units/mL, even after extensive concentration, [54] which does not meet the necessary requirement for the in vivo transduction of each CD4+ T cell within the body, estimated to total >1011 cells [55]. Until these obstacles can be overcome, targeted modification of ccr5 or cxcr4 by Cas9 may be the most immediate approach. However, this approach has three obvious disadvantages. First, this technique requires ex vivo genomic alteration and cellular expansion, which will likely hinder its widespread use. Second, although lentiviruses are currently being utilized as ex vivo Cas9 delivery systems, transduction itself is a mutagenic event, which poses an inherent risk should the virus integrate at a site that could cause harm to the host cell. Finally, latent cellular reservoirs would still persist in spite of HIV co-receptor modification. Given the great potential of Cas9 in combating HIV, it will be interesting to see if and how these obstacles are overcome in the future.
Hepatitis B virus
Hepatitis B virus (HBV) is an enveloped partially dsDNA virus that primarily infects hepatocytes [56]. Despite the development of a highly effective HBV vaccine [57] as well as nucleoside and nucleotide analog therapeutics used in combination with interferon [56], HBV still affects approximately 250 million people worldwide [58], predisposing these individuals to complications such as liver fibrosis, cirrhosis, and hepatocellular carcinoma.
Following cellular entry, the relaxed circular, partially dsDNA HBV genome (rcDNA) is converted to covalently closed circular DNA (cccDNA), which serves as the transcription template for the generation of viral RNAs [56, 59]. Though antiviral therapies inhibit HBV replication, they do not eliminate cccDNAs, which stably persist in the nuclei of infected hepatocytes [60].
Several groups have demonstrated that both HBV rcDNA and cccDNA are susceptible to Cas9-mediated cleavage in cell culture [61–69] as well as in vivo [63–66, 69]. Numerous cell types and Cas9:gRNA delivery methods were used for the cell culture studies, but one feature common among several of the reports was a two to three fold reduction in HBV surface antigen (HBsAg) level in the lysates or supernatants of cells expressing HBV-specific gRNAs relative to cells expressing control gRNA [61, 63–66]. In vivo, NRG mice hydrodynamically injected with two different plasmids encoding HBV or Cas9 with a gRNA specific for the HBV polymerase-coding region displayed a four-fold decrease in viremia relative to mice injected with HBV and Cas9/control gRNA plasmids. The viral titer correlated with a mild reduction in serum HBV surface antigen (HBsAg) level [63]. Another group utilized the hydrodynamic injection system in which plasmids encoding HBV and plasmids encoding Cas9 with either control gRNA or gRNAs targeting the HBV surface (S) and X proteins, were injected into Balb/c nude mice [64]. Three days following injection, a 90% reduction in serum HBsAg level was observed [64]. Similar inhibition was retained up to ten days following injection, and Cas9-induced mutations were detected in up to 75% of total HBV genomes [64]. An additional study demonstrated at least a five-fold decrease in serum HBV DNA level and HBsAg four days post Cas9:gRNA treatment in Balb/c mice [69].
Perhaps the least efficacious report of the ability of Cas9 to inhibit HBV in vivo was performed in immunocompetent mice. C57BL/6 mice were hydrodynamically injected with two different plasmids encoding HBV and Cas9 with either control gRNA or gRNAs specific for the HBV P1 or X coding sequences [66]. Serum HBsAg levels were only slightly reduced in Cas9 expressing mice relative to those expressing the vector control, 24 and 72 hours post treatment, and Cas9-mediated insertions and deletions within the HBV sequence were only detected in four to five percent of total HBV genomes [66]. However, discrepancies in the level of HBV inhibition between this study and others could be due to multiple reasons other than the intact immune system, such as gRNA target region, injection time course, and/or ratio of injected HBV plasmid to Cas9 gRNA plasmid.
One caveat to the use of hydrodynamic injection of plasmids encoding the HBV genome is that cccDNAs are not efficiently generated, and therefore this system does not closely resemble the hepatocytes of chronically infected HBV patients which may harbor up to 40 copies of cccDNA per cell [70]. To overcome this obstacle, Dong and colleagues utilized an alternative in vivo model in which Balb/c mice were injected with plasmids that allowed for high expression of cccDNAs. Subsequently, hydrodynamic injection with Cas9/gRNA plasmids resulted in a two to threefold decrease in liver cccDNA, and a corresponding approximate fivefold decrease in HBeAg six days post injection [65]. Regardless of the mouse model and treatment length, Cas9/gRNA complexes did not facilitate 100% viral clearance in any of the aforementioned studies, highlighting the importance of effective delivery systems in which each infected cell receives Cas9 and gRNA.
In vivo delivery of Cas9 for therapeutic targeting of HBV and other hepatotropic viruses may be more readily achievable than for other eukaryotic viruses, as recombinant adeno-associated virus 8 (AAV8) is capable of efficiently transducing hepatocytes [71]. Recombinant AAV has gained popularity as a potential Cas9 delivery vector, and offers the advantages of high-titer production and continuous transgene expression [72]. However, recent observations that chromosomal insertions of AAV2 may activate proto-oncogenes in human hepatocellular carcinomas suggest a pathogenic role for this vector [73]. Further studies will need to address the potential oncogenic role for AAV8, a serious consideration given that viruses such as HBV are associated with hepatocellular carcinomas. An additional point to consider is that size constraints in many of these viral vectors may not allow for the full expression of Streptococcus pyogenes Cas9, gRNA, and their respective promoters. To overcome packaging restraints, split-enzyme Cas9 proteins have been generated. Each half of the enzyme can be packaged within a separate vector, and then the protein domains dimerize and form a functional endonuclease upon chemical induction or recruitment by gRNA [74, 75]. Further, smaller proteins, such as Cas9 from Staphylococcus aureus, are being investigated as alternatives [76, 77]. Despite the obstacles, Cas9 may have intriguing potential to combat hepatotropic viruses, specifically in the case of HBV and the ablation of cccDNA.
Papillomavirus
Human papillomavirus (HPV) encodes a circular dsDNA genome that may integrate into the host cell chromosome. HPV DNA is detected in greater than 99% of cervical cancers, and aberrant expression of HPV proteins promotes cervical cell malignant transformation and tumor expansion [78]. The HPV proteins E6 and E7 disrupt normal expression of the host cell cycle proteins p53 and retinoblastoma (Rb), respectively, enhancing cellular immortalization [79, 80]. Although the prophylactic vaccines, Gardasil ® (http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm111283.htm) and Cervarix ® (http://www.fda.gov/BiologicsBloodVaccines/Vaccines/ApprovedProducts/ucm186959.htm), have been developed, there is still a need for therapeutics that effectively control established HPV infection and associated tumors. Presumably, excision of these HPV oncogenes could restore expression and normal functioning of p53 and Rb tumor suppressors, leading to cell cycle arrest and inhibition of tumor/cancer progression.
Cas9 was used with cognate gRNAs to disrupt the HPV E6 and E7 coding sequences within the siHa cervical tumor cell line that retains integrated HPV-16, a subtype of HPV that has been associated with greater than 50% of cervical tumors [81]. Forty-eight hours following transfection of plasmids encoding Cas9 and gRNAs, viral transcript levels were reduced over 90% relative to cells transfected with control vectors [82]. The reduction in E6 and E7 transcript levels correlated with an increase in host p53 and p21 (a cyclin-dependent kinase inhibitor used as a readout for Rb) production, as well as a significant reduction in cell viability [82]. Kennedy et al. demonstrated similar findings in both siHa and HeLa cells, which retain integrated coding sequences from HPV-subtype 18 [83]. Further, expression of Cas9 and HPV targeting gRNAs impeded tumorigenesis of siHa cells in a xenograft mouse model [82]. No mice were completely free of tumors 45 days following transplantation, but tumor weights were reduced by three-fold relative to those from control treated mice [82]. However, in this model, siHa cells were transiently transfected with plasmids encoding Cas9 and HPV-targeting gRNAs, and subsequently implanted into immunodeficient Balb/c nude mice. Other models such as naturally occurring rabbit papillomavirus perhaps more closely resemble the course of infection and tumorigenesis of HPV, and it would be interesting to test the Cas9 system in these models.
It is plausible that AAV may be a potential delivery vehicle for Cas9:gRNAs that target HPV, since certain AAV serotypes can readily transduce HeLa cells [53], a cervical cancer cell line, and AAV sequences have been detected in primary cervical samples [84]. However, AAV tissue tropism is not limited to cervical cells [53]. Localized introduction of recombinant AAV in the female cervix could potentially minimize systemic cellular transduction. Similarly, the generation of mosaic capsid proteins to express peptide moieties or antibody light chains for a specific receptor may provide the specificity required for transducing a particular cell type [85]. Nonetheless, an underlying concern is again the potential oncogenic role of AAV. However, if delivery concerns can be overcome, Cas9 may hold considerable promise for the targeting HPV.
Epstein-Barr virus
Epstein-Barr virus (EBV) is a linear dsDNA herpes virus and causative agent of infectious mononucleosis. EBV infection can lead to cancers such as Burkitt’s lymphoma, Hodgkin’s lymphoma, and nasopharyngeal carcinoma, among others, when maintained as an episome [86]. The first report of Cas9 in the study of EBV was for promoter studies and the generation of an EBV reporter virus [87]. Later, Wang and colleagues utilized Raji cells, a B cell line derived from a patient with Burkitt’s lymphoma that harbors episomal copies of EBV [88], to demonstrate that Cas9 with gRNAs specific for EBV could excise the coding sequences of EBNA1 and EBNA3C, viral proteins involved in genome replication and host cell transformation, respectively [89]. Further, expression of Cas9 with seven multiplexed EBV-targeting gRNAs could arrest cellular proliferation and induce apoptosis [89]. Because it has been described that EBV proteins inhibit apoptotic processes in host cells, the induction of apoptosis may be due to the loss of these viral proteins [90]. Single cell quantification of EBV episomal copy number within Raji cells expressing both Cas9 and these multiplexed gRNAs indicated that the copy number was widely divergent from cell to cell. Of the 71-sorted cells expressing both Cas9 and gRNA, 19 had undetectable EBV genome levels, whereas 22 cells retained copy numbers identical to untreated cells. The remaining 30 retained varying EBV copy numbers [89]. Although this study demonstrates that Cas9 and targeting gRNAs can induce EBV genome disruption in Raji cells, the reasons why Cas9 and EBV gRNAs were only effective in reducing the EBV genome copy number in a portion of cells remain to be elucidated. To our knowledge, a delivery vector that specifically targets only B cells has yet to be developed, which would serve as a limitation in implementing Cas9 technology to combat EBV.
Geminiviruses
Geminiviruses are one of the largest families of plant viruses and are the causative agents of major crop losses in both the U.S. and around the world. These small, single stranded DNA viruses disrupt host machineries, often leading to plant developmental defects [91]. However, recent progress has been made in engineering plants that inhibit geminivirus replication. Ji and colleagues generated transgenic Nicotiana benthamiana and Arabidopsis thaliana plants expressing Cas9 and gRNA specific for beet severe curly top virus. Plants expressing high levels of Cas9 were protected from virus challenge, and no leaf curling was observed [92]. Similarly, Baltes et al. demonstrated that DNA loads of bean yellow dwarf viruses were reduced by approximately four-fold in transgenic N. benthamiana plants expressing Cas9 and gRNA [93], while others have demonstrated a reduction in tomato yellow leaf curl virus DNA accumulation in Cas9-transgenic plants [94]. These studies are extremely encouraging and may pave the way for the generation of other transgenic plants and animals that are resistant to viral pathogens through Cas9-mediated targeting.
Cas9 Targeting of RNA
In addition to their role in bacterial adaptive immunity, it has been demonstrated that type II CRISPR-Cas systems can regulate endogenous gene expression. In conjunction with a small, CRISPR-Cas-associated RNA (scaRNA) and tracrRNA, Francisella novicida Cas9 (FnCas9) can target an endogenous mRNA encoding a bacterial lipoprotein (BLP) [95]. Recognition of BLP by the host innate immune receptor Toll-like receptor 2 (TLR2) triggers an important pro-inflammatory cascade that contributes to host defense. By targeting BLP mRNA, Cas9 suppresses BLP expression and promotes evasion of TLR2-mediated host immune activation and F. novicida virulence [95, 96].
Further, O’Connell et al. described the ability of purified Cas9 from Streptococcus pyogenes (SpCas9) along with gRNA to bind and cleave single stranded RNA in the presence of an exogenous PAM oligonucleotide, termed the PAMmer. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA could also be isolated and precipitated from HeLa cell lysate using SpCas9:gRNA in the presence of a PAMmer [97]. Taken together, these studies highlight the ability of Cas9 to target RNA.
Given the success of Cas9 in targeting DNA for genome engineering in eukaryotic cells, it was theorized that the ability of Cas9 to target RNA might also be harnessed for use in eukaryotic cells. Such ability could be useful in targeting viruses that contain no DNA stage in their life cycle. Eukaryotic cells possess a mechanism of RNA interference (RNAi) in which small RNAs direct cellular machinery to mRNAs to dampen gene expression [98, 99]. RNAi has been harnessed to inhibit the RNA stage of numerous eukaryotic viruses, and does not require the presence of an exogenously expressed protein, as does the Cas9 platform. However, certain viruses have developed methods of circumventing RNAi [100, 101]. Eukaryotic viruses have not evolved in the presence of Cas9 and likely cannot inhibit it, thus raising a potential advantage of Cas9-based systems.
FnCas9 was recently used to target the genome of the positive sense single-stranded RNA (+ssRNA) virus, hepatitis C virus (HCV), as a proof of principle [102]. HCV is an important human pathogen that can cause liver fibrosis, cirrhosis, and may lead to hepatocellular carcinoma [103]. Transient expression of Cas9 and RNA-targeting guide RNA (rgRNA) complexes targeted to either the 5’ or 3’ untranslated regions of the genome prior to infection of a hepatoma cell line with HCV resulted in a 50–60% reduction in viral protein expression relative to cells transfected with vector controls. In a model of established infection, FnCas9-mediated inhibition was approximately 40%, once again highlighting the need for efficient delivery systems in which each cell receives the Cas9:rgRNA complexes. The endonuclease domains of FnCas9 were dispensable for FnCas9-mediated inhibition of HCV, as was the presence of a canonical PAM [102]. Collectively, these data suggest that inhibition was most likely due to a blockade of cellular translation and viral replication machineries that associate with the viral RNA, rather than HCV genome cleavage. This application of Cas9 technology more closely resembles CRISPR interference (CRISPRi), in which catalytically inactive Cas9 proteins are programmed to dampen gene expression, [104, 105] rather than canonical genomic editing. One potential obstacle to the use of Cas9:rgRNA complexes to directly target RNA viral genomes in vivo would likely be the requirement for continuous high-level Cas9:rgRNA expression during the course of viral infection. Nonetheless, these studies highlight the potential for using Cas9 to target RNA in diverse applications.
Obstacles to the Use of Cas9 as an Antiviral in Humans
Despite the advances demonstrating Cas9-mediated inhibition of eukaryotic viruses, significant obstacles must be overcome before Cas9 can be used for human therapeutics. One primary obstacle is the current lack of data addressing how the mammalian immune system responds to Cas9. It is currently unknown whether Cas9 elicits cross-reactive antibodies or T cell responses. This concern may be alleviated if delivery systems are developed that allow for transient expression of Cas9:gRNAs, as would likely be required for DNA cleavage. The introduction of a Cas9-destabilization domain or inducible promoter could also allow for transient Cas9 expression.
In contrast to using Cas9 to cleave DNA targets, prolonged Cas9 expression would likely be required for the transcriptional activation of antiviral effector proteins and latent viruses, as well as direct targeting of the RNA stage of virus infections. Sustained expression may not be feasible if the immune system is activated to kill cells expressing Cas9. It is plausible that truncated Cas9 proteins may be utilized to target the RNA stage of viral infections, which could potentially offer the advantages of a smaller packaging size for delivery systems such as AAV, as well as a decreased number of epitopes to be presented to the immune system, assuming a direct correlation between protein size and number of recognizable epitopes. As RNA virus inhibition may not be reliant on Cas9 endonuclease activity, shorter proteins could potentially protect the gRNA from nuclease digestion while still inhibiting translation and/or replication of viral nucleic acid. Further, it is possible that complete Cas9-mediated viral clearance may not be required for viruses that do not establish latency, as a Cas9-mediated decrease in viral load may synergize with other antiviral therapeutics and/or the immune system to fight infection, thus lessening the length of time Cas9 would need to be expressed.
A second major obstacle, as discussed in several sections above, is the development of a highly effective Cas9 delivery vehicle. Ensuring that the Cas9:gRNA machinery is expressed in the correct anatomic locations and proper cell types is absolutely necessary to translate this technology from the bench to the bedside. While transgenic plants and livestock may be readily produced to bypass delivery issues, the potential for human therapeutics is limited at this time. Nevertheless, numerous viral vectors such as adenoviruses, AAV, and lentiviruses or retroviruses are potential approaches for the delivery of the antiviral Cas9 machinery. Other approaches include non-viral delivery methods such as lipopeptide nanoparticles, cell-penetrating peptides conjugated to Cas9:gRNA complexes, and DNA nanoclews. Each of these approaches offers both advantages and drawbacks that have been discussed in detail elsewhere [72, 85, 106–109], but underlying concerns include cell target specificity, proper Cas9 expression levels, and host safety. An additional concern is the immunogenicity of the delivery vehicle itself. For viral vectors, untoward immune responses, such as the production of neutralizing antibodies, could prevent effective vector entry and transgene expression in the cell type of interest, which is especially concerning if multiple rounds of treatment are warranted. Even if delivery methods are established so that every infected cell receives Cas9:gRNAs, the necessary ratio of the number of Cas9:gRNA molecules per virus genome or genomic cleavage site will still need to be established.
Finally, Cas9 is a powerful tool that could wreak havoc on the host should cleavage occur at genomic off-target sites. Generating Cas9 proteins with altered PAM specificities or enhanced target recognition are approaches being pursued as methods of reducing off-target effects [110, 111]. Other methods include utilizing truncated gRNAs, paired Cas9 nickases, and high-fidelity Cas9 nucleases in order to alleviate this hurdle [15, 112, 113].
Concluding Remarks
Overall, significant advances have been made in employing the Cas9:gRNA machinery to inhibit pathogenic eukaryotic viruses, though questions still remain (see Outstanding Questions). gRNA multiplexing could facilitate resistance to a panoply of different viruses in transgenic plants and animals, and may also circumvent viral escape. Targeting host factors necessary for viral infection and replication represents yet another route of viral inhibition. However, the possibility of off target effects on the host remains a major concern. Further research will need to address the feasibility of co-administration of currently available antiviral therapeutics and Cas9:gRNA, as well as potentially damaging immunological responses generated toward the foreign Cas9 protein. Despite its limitations, the use of the prokaryotic adaptive immune system as a eukaryotic antiviral defense holds considerable promise. With continued progress and engineering feats, Cas9-based technologies may represent the next generation of antiviral prophylactics and therapeutics.
Presently, however, the majority of studies have been performed in cell culture, with a small number of in vivo experiments. Surmounting the existing challenges will likely require intensive collaborative efforts from experts in the fields of immunology, molecular biology, bacteriology, and biomedical engineering. Undoubtedly huge strides will be made in the near future, but the use of Cas9 in humans should be weighed very carefully given the numerous potential risks.
Outstanding questions.
Can multiplexing gRNAs in transgenic plants and animals simultaneously facilitate resistance to multiple viruses, as occurs naturally with crRNAs in CRISPR arrays in prokaryotes? In addition, can multiplexed gRNAs targeting the same virus prevent viral escape?
Can current antiviral therapies synergize with Cas9:gRNA to promote viral clearance?
Are there off target effects on the host when Cas9 technology is used in vivo? How can such off-target effects be limited? Tissue-restricted expression of Cas9 and gRNA, as well as continued engineering to generate increased Cas9:gRNA specificity are possibilities.
How does the mammalian immune system respond to expression of the foreign Cas9 protein? The large Cas9 proteins from Streptococcus pyogenes, Francisella novicida, and other organisms likely contain numerous B and T cell epitopes. Cas9 proteins may be immunogenic and result in clearance of the protein:RNA complex. Additional studies are also needed to address whether these epitopes are cross-reactive to host epitopes and induce aberrant autoimmune responses.
Trends.
Cas9 technology has been utilized to inhibit pathogenic DNA and RNA viruses in cell culture. This has been accomplished by modulating expression of host factors required for viral entry, directly targeting virus genomes, transcriptionally activating antiviral genes, and cleaving the DNA stage of viruses that integrate into the host cell chromosome.
The Cas9 machinery has been shown to be efficacious in restricting pathogenic viruses in small rodent models.
Transgenic plants have recently been developed that inhibit the replication stage of geminiviruses, a major cause of crop losses worldwide.
Cas9 technology is a promising tool for the generation of virus-resistant transgenic plants and animals, as well as antiviral therapeutics.
Acknowledgments
We would like to express our gratitude to Dr. Timothy Sampson and Hannah Ratner for their outstanding comments and critical reading of the manuscript. We sincerely apologize to our colleagues for the omission of other key literature citations due to space limitations and the rapidly expanding Cas9 field. We would like to acknowledge support from the NIH grants R01AI070101, R01AI124680 and 1R21AI118337 to A.G, and ORIP/OD P51OD011132 (formerly NCRR P51RR000165) to the Yerkes National Primate Research Center. DSW is supported by NIH grant R01-AI110701 and a Burroughs Welcome Fund Investigator in the Pathogenesis of Infectious Disease award. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldberg GW, Marraffini LA. Resistance and tolerance to foreign elements by prokaryotic immune systems - curating the genome. Nature reviews. Immunology. 2015;15:717–724. doi: 10.1038/nri3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. doi: 10.1038/nature15386. [DOI] [PubMed] [Google Scholar]
- 3.Ishino Y, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of bacteriology. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Makarova KS, et al. Comparative genomics of defense systems in archaea and bacteria. Nucleic acids research. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberger AD, et al. Viral diversity threshold for adaptive immunity in prokaryotes. mBio. 2012;3:e00456–e00412. doi: 10.1128/mBio.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath P, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus . Journal of bacteriology. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus . Journal of bacteriology. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang N, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell research. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho SW, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 17.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(UNAIDS), J.U.N.P.o.H.A. Global Report: UNAIDS report on the global AIDS epidemic. 2013 [Google Scholar]
- 19.Dalgleish AG, et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 20.Alkhatib G, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 21.Bleul CC, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 22.Carr A. Toxicity of antiretroviral therapy and implications for drug development. Nature reviews. Drug discovery. 2003;2:624–634. doi: 10.1038/nrd1151. [DOI] [PubMed] [Google Scholar]
- 23.Palella FJ, Jr, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. The New England journal of medicine. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 24.Barre-Sinoussi F, et al. Past, present and future: 30 years of HIV research. Nature reviews. Microbiology. 2013;11:877–883. doi: 10.1038/nrmicro3132. [DOI] [PubMed] [Google Scholar]
- 25.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 26.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. The New England journal of medicine. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 27.Allers K, et al. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- 28.Ye L, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W, et al. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PloS one. 2014;9:e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. The Journal of general virology. 2015;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 31.Mandal PK, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell stem cell. 2014;15:643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YG, et al. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta RM, Musunuru K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. The Journal of clinical investigation. 2014;124:4154–4161. doi: 10.1172/JCI72992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaj T, et al. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou P, et al. Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Scientific reports. 2015;5:15577. doi: 10.1038/srep15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panganiban AT. Retroviral DNA integration. Cell. 1985;42:5–6. doi: 10.1016/s0092-8674(85)80092-1. [DOI] [PubMed] [Google Scholar]
- 38.Rabson AB, Martin MA. Molecular organization of the AIDS retrovirus. Cell. 1985;40:477–480. doi: 10.1016/0092-8674(85)90189-8. [DOI] [PubMed] [Google Scholar]
- 39.Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nature medicine. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 40.Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 41.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 42.Archin NM, et al. Eradicating HIV-1 infection: seeking to clear a persistent pathogen. Nature reviews. Microbiology. 2014;12:750–764. doi: 10.1038/nrmicro3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebina H, et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Scientific reports. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11461–11466. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu W, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao HK, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nature communications. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 47.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogerd HP, et al. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E7249–E7256. doi: 10.1073/pnas.1516305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Scientific reports. 2015;5:16277. doi: 10.1038/srep16277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saayman SM, et al. Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/dCas9 Activator Complex. Molecular therapy : the journal of the American Society of Gene Therapy. 2015 doi: 10.1038/mt.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Limsirichai P, et al. CRISPR-mediated Activation of Latent HIV-1 Expression. Molecular therapy : the journal of the American Society of Gene Therapy. 2015 doi: 10.1038/mt.2015.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellis BL, et al. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virology journal. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Segura MM, et al. New developments in lentiviral vector design, production and purification. Expert opinion on biological therapy. 2013;13:987–1011. doi: 10.1517/14712598.2013.779249. [DOI] [PubMed] [Google Scholar]
- 55.Ganusov VV, De Boer RJ. Do most lymphocytes in humans really reside in the gut? Trends in immunology. 2007;28:514–518. doi: 10.1016/j.it.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 56.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 57.Chang MH, et al. Decreased incidence of hepatocellular carcinoma in hepatitis B vaccinees: a 20-year follow-up study. Journal of the National Cancer Institute. 2009;101:1348–1355. doi: 10.1093/jnci/djp288. [DOI] [PubMed] [Google Scholar]
- 58.Ott JJ, et al. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 59.Kennedy EM, et al. Targeting hepatitis B virus cccDNA using CRISPR/Cas9. Antiviral research. 2015;123:188–192. doi: 10.1016/j.antiviral.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. Journal of virology. 2007;81:6164–6174. doi: 10.1128/JVI.02721-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kennedy EM, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Molecular therapy. Nucleic acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramanan V, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Scientific reports. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhen S, et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene therapy. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 65.Dong C, et al. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral research. 2015;118:110–117. doi: 10.1016/j.antiviral.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 66.Lin SR, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Molecular therapy. Nucleic acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, et al. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World journal of gastroenterology. 2015;21:9554–9565. doi: 10.3748/wjg.v21.i32.9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karimova M, et al. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Scientific reports. 2015;5:13734. doi: 10.1038/srep13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, et al. Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. The Journal of general virology. 2015;96:2252–2261. doi: 10.1099/vir.0.000159. [DOI] [PubMed] [Google Scholar]
- 70.Laras A, et al. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology. 2006;44:694–702. doi: 10.1002/hep.21299. [DOI] [PubMed] [Google Scholar]
- 71.Lisowski L, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kotterman MA, Schaffer DV. Engineering adeno-associated viruses for clinical gene therapy. Nature reviews. Genetics. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nault JC, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nature genetics. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 74.Zetsche B, et al. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature biotechnology. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright AV, et al. Rational design of a split-Cas9 enzyme complex. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kennedy EM, et al. Optimization of a multiplex CRISPR/Cas system for use as an antiviral therapeutic. Methods. 2015;91:82–86. doi: 10.1016/j.ymeth.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walboomers JMM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. Journal of Pathology. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 79.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nature reviews. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 80.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nature reviews. Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 81.Bosch FX, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. Journal of the National Cancer Institute. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 82.Zhen S, et al. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochemical and biophysical research communications. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 83.Kennedy EM, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. Journal of virology. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Venturoli S, et al. Detection of adeno-associated virus DNA in female genital samples by PCR-ELISA. Journal of medical virology. 2001;64:577–582. doi: 10.1002/jmv.1088. [DOI] [PubMed] [Google Scholar]
- 85.Buchholz CJ, et al. Surface-Engineered Viral Vectors for Selective and Cell Type-Specific Gene Delivery. Trends in biotechnology. 2015;33:777–790. doi: 10.1016/j.tibtech.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 86.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nature reviews. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 87.Yuen KS, et al. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. The Journal of general virology. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 88.Adams A, Lindahl T. Epstein-Barr virus genomes with properties of circular DNA molecules in carrier cells. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruf IK, et al. Epstein-Barr virus regulates c-MYC, apoptosis, and tumorigenicity in Burkitt lymphoma. Molecular and cellular biology. 1999;19:1651–1660. doi: 10.1128/mcb.19.3.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hanley-Bowdoin L, et al. Geminiviruses: masters at redirecting and reprogramming plant processes. Nature reviews. Microbiology. 2013;11:777–788. doi: 10.1038/nrmicro3117. [DOI] [PubMed] [Google Scholar]
- 92.Ji X, et al. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nature Plants. 2015;1:15144. doi: 10.1038/nplants.2015.144. [DOI] [PubMed] [Google Scholar]
- 93.Baltes NJ, et al. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nature Plants. 2015;1:15145. doi: 10.1038/nplants.2015.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ali Z, et al. CRISPR/Cas9-mediated viral interference in plants. Genome biology. 2015;16:238. doi: 10.1186/s13059-015-0799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sampson TR, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sampson TR, et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11163–11168. doi: 10.1073/pnas.1323025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Connell MR, et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 99.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 100.Misasi J, Sullivan NJ. Camouflage and misdirection: the full-on assault of ebola virus disease. Cell. 2014;159:477–486. doi: 10.1016/j.cell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fabozzi G, et al. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. Journal of virology. 2011;85:2512–2523. doi: 10.1128/JVI.01160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Price AA, et al. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6164–6169. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nature medicine. 2013;19:837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt F, Grimm D. CRISPR genome engineering and viral gene delivery: a case of mutual attraction. Biotechnology journal. 2015;10:258–272. doi: 10.1002/biot.201400529. [DOI] [PubMed] [Google Scholar]
- 107.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome research. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sun W, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angewandte Chemie. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen X, Goncalves MA. Engineered Viruses as Genome Editing Devices. Molecular therapy : the journal of the American Society of Gene Therapy. 2015 doi: 10.1038/mt.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fu Y, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature biotechnology. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016 doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]