Introduction
Wuchereria bancrofti (Wb) is a parasitic nematode of the order Spirurida and the main cause of lymphatic filariasis (LF) in humans—the others being Brugia malayi and Brugia timori. Presently, more than 90 million people in 73 countries are infected with Wb and an estimated 789 million people at risk by living in Wb endemic areas (Hooper et al. 2014; Ramaiah & Ottesen 2014).
The life cycle of Wb includes both a human and mosquito host. In the human host, Wb reproduces sexually giving birth to thousands of offspring—termed microfilaria (MF). The MF then infects a mosquito host while it is taking a blood meal from the Wb infected human host. Over the next 8-12 days the MF mature to L3 stage larvae in the mosquito host. The now infective L3 larvae passes back to a human host during the next blood meal by the infected mosquito. Now back in the human host the L3 migrate to the lymphatics and mature to reproductive adults over the next 8-14 months (Farrar et al. 2013). It is the adult stage, residing in the lymphatics, that causes most of the clinical symptoms of LF such as elephantiasis and hydrocele.
The history of Wb is likely the history of human migration. Wb is specific to Homo sapiens with only a single species, W. kalimantani, discovered to infect silver leaf monkeys (Presbytis cristatus) (Palmieri et al. 1980). Symptoms of LF are documented in medical records and illustrations dating as far back as 3000 BC (Laurence 1989). From these sources, LF causing nematodes are proposed to have spread from Africa to the Caribbean Islands and Southern United States (last documented in Charleston, South Carolina in 1920) by way of the trans-Atlantic slave trade (Laurence 1989).
Our current knowledge on the distribution and diversity of Wb is limited to clinical and historical records. Wb is also more difficult to culture than other filarial nematodes like B. malayi. In turn this limits our understanding of infection dynamics, directly affecting how we treat and understand Wb infections.
To deal with Wb, the Global Programme to Eliminate Lymphatic Filariasis (GPELF) was created in year 2000 with the goal to halt Wb transmission by year 2020. In the first 13 years over 4.4 billion treatments have been distributed in 56 endemic countries reducing the risk of infection by 46% from previous estimates in the year 2000 (Hooper et al. 2014; Ramaiah & Ottesen 2014). However, the program has identified some outliers regions where prevalence is rebounding after an initial reduction (Won et al. 2009; Simonsen et al. 2013; Lau et al. 2014; Rebollo et al. 2015).
In other parasites the addition of population genetics have improved upon our understanding of infection dynamics (i.e., transmission patterns) and epidemiology (Steinauer et al. 2010; Volkman et al. 2012). For example, in Senegal population genomic data was used to infer transmission rate in Plasmodium falciparum (Daniels et al. 2015). Parasite transmission rapidly declined following an intensive elimination program as inferred from the population genomic data and supported by incidence data collected at the same time (Daniels et al. 2015). There exists an opportunity to integrate population genetics into Wb elimination programs. Thus far, studies on population genetics in Wb have revealed high haplotype diversity within infections (Small et al. 2013; Souza et al. 2014) and a striking lack of population stratification at the local scales, possibly explained by extensive human travel between rural and densely populated areas (Thangadurai et al. 2006; Hoti et al. 2008).
Population genetics studies of filarial nematodes are made difficult by host DNA contamination and the presence of an entire population of infecting worms. Typically, Wb is collected via a blood sample from an infected human host. The infected blood sample contains a small quantity of parasite DNA but many times more abundant host DNA. The second problem is that infections contain a population of parasites, due to multiple infection events and the necessary sexual reproduction. Multiple parasites complicate population genetic analysis because infections contain a mixture of genetically heterogeneous genomes of an unknown number (Small et al. 2013).
Here we circumvented the two main problems associated with studying population genetics of filarial worms by sequencing from L3 stage worms isolated from mosquitoes. By sequencing L3 stage worms we are able to sample single genomes from the same infected host generating data on within infection diversity and relatedness. We use these genome sequences to improve upon our knowledge of Wb biology and infection dynamics.
Material and Methods
Materials acquisition
In 2012, adults were recruited from Tau, Papua New Guinea (PNG). Individuals consenting to participate in the study were screened for the presence of Wb using BinaxNow© Filariasis rapid card tests (Alere Inc.). On consent, antigen positive study participants were asked to provide a venous blood sample. Microfilaremia was confirmed via microscopy using 20ml of blood in a 2% formalin wet mount (Erickson et al. 2013). All studies were conducted under IRB approved by Case Western Reserve University and Papua New Guinea – Institute for Medical Research for both donors and feeding assays.
De Novo Genome Assembly: Infrapopulation Sample Preparation and Sequencing
A single participant, pt0022, with high MF density (>6,000 mf/ml) was selected for initial sequencing and analysis of the Wb genome. Details of the sample preparation, data generation and analysis are presented in Supporting Information—De novo Genome assembly.
Wb L3 Genomes: Strategic Individual L3 Sample Preparation and Sequencing
Blood from three MF positive individuals originating from either Tau, PNG or Bogia, PNG (pt17 [Tau], pt48 [Tau], pt74 [Bogia]) was used for feeding mosquitoes of Anopheles punctulatus or An. farauti s.s. via water-jacketed membrane feeders fitted with parafilm (Erickson et al. 2013). After exposure mosquitoes were euthanized and dissected to recover individual L3 larvae (Erickson et al. 2013) (Fig. S1, Supporting Information). Recovered parasites were immediately placed into a 0.25 mL microcentrifuge tube containing 200uL of RNAlater® (Life Technologies) and stored at −20°C. A sequencing library was then prepared from each L3 larva and sequenced on an Illumina HiSeq 2000 as described in Supporting Information – Wb L3 Genomes.
Mapping and Variant calling for Wb L3
Sequencing reads generated from each Wb L3 library were processed using the bioinformatics pipeline as described in Supporting Information – Wb L3 Genomes. Briefly, reads were pre-processed to check quality using FastQC (http://www.bioinformatics.babraham.ac.uk/index.html) and adapters were trimmed using TrimGalore. Next, reads were filtered by aligning to the mitochondrial genome and the Wolbachia genome using Bowtie2 (Langmead & Salzberg 2012). Finally, remaining reads were aligned to the de novo Wb pt0022 genome assembly (De novo genome assembly, Supporting Information). The resulting alignments were processed using a custom script to remove incorrectly paired reads as well as reads mapping to more than one location in the genome.
Prior to variant calling, repeated sequences and putative paralogous genomic regions were masked. Repetitive elements in the Wb pt0022 assembly were identified using RepeatMasker (Tarailo-Graovac & Chen 2009). RepeatMasker was run with default parameters using both a RepeatModeler dataset built from the Wb assembly and a pre-built dataset from the previously assembled B. malayi genome (Ghedin et al. 2007). Next, potential paralogous genomic regions were identified by mapping the pt0022 library reads back to the Wb de novo assembly (De novo genome assembly, Supporting Information) and then calculating a sliding window of sequencing coverage following a correction for GC-bias (broadinstitute.github.io/picard). Finally, regions of the genome identified as having coverage greater than 95% of the contigs, were masked before calling variants. Variants were called using Freebayes v0.9.20 following the recommended settings for a diploid genome and calculating genotype-qualities (Garrison & Marth 2012). The resulting variant call format file (VCF file) was filtered using a series of custom python scripts and bcftools v1.20 (samtools.github.io/bcftools/). In brief, any variants were removed if they did not meet all of the following criteria: read coverage > 20.0, Phred-scaled base quality > 30.0, allele balance > 0.1, minor allele frequency > 0.20, Phred-scaled mapping quality > 25.0, Phred-scaled strand probability < 60.0, Phred-scaled genotype quality > 30.0. Sites with more than 20% missing data were excluded from further population analysis.
Relatedness
The SNP-based pairwise kinship coefficients (ΦSNP), which denote the relationship between two genomes, were estimated using the program KING (Manichaikul et al. 2010) for each pair of Wb L3 genomes. The input file was prepared using VCFtools v1.12 (Danecek et al. 2011) by exporting data in PLINK-binary format. KING was run using the kinship and ibs option on sites with no missing data.
Population Data Analysis
Summary statistics were calculated using custom python scripts. Summary statistics included: number of heterozygous sites per genome, number of sites fixed for the alternative allele, average coverage per genotype, and percent of genotypes missing. PopGenome (Pfeifer et al. 2014) was used to calculate θ (the population mutation rate) (Watterson 1975) and π (the pairwise nucleotide diversity) (Nei 1987) per 10,000 base pair (10kb) sliding window using contigs > 10,000bp in length. To determine the ancestral allele for the unfolded SFS, B. malayi (PRJNA10729), Loa loa (PRJNA60051), and Wb genomes were aligned using the program Mugsy (Angiuoli & Salzberg 2011). A custom python script was then used to transform the data to the correct format. The empirical SFS was calculated using ANGSD (Korneliussen et al. 2014) this was then compared to the SFS from a simulated population of constant size. Briefly, θ was calculated for each 10kb window and used to simulate a population of constant size in the software MaCS (Chen et al. 2009). The SFS was then calculated using mssfs (available from github.com/rossibarra/msstats). The average distance of linkage disequilibrium (LD) was calculated using VCFtools v1.12 (Danecek et al. 2011). A background value of LD was calculated by selecting among variants from different contigs using a custom python script and the program covld (Rogers & Huff 2009). Population level relationships were visualized using phylogenetic trees. Trees were reconstructed using an unrooted maximum likelihood approach as implemented in SNPhylo (Lee et al. 2014). For comparison to Wb from PNG, read data generated from one Wb genome from Mali, Africa (Desjardins et al. 2013) and one from Jakarta, Indonesia (PRJEB536) were mapped onto the de novo pt0022 assembly and included in the analysis. Using SNPhylo, variable positions were thinned to account for linkage, which resulted in ~19,000 informative positions remaining. The resulting tree was bootstrapped using phangorn (Schliep 2011). To test whether sequence coverage could influence the results of this analysis, each Wb L3 genome was randomly down-sampled to 30x coverage, the minimum genotype coverage for L3-4851. SNPs were recalled following the criteria described above. The second phylogenetic tree was then constructed using the subsampled data and the same method as detailed above.
For each Wb L3, the filtered mitochondrial genome reads were aligned against the published Wb mitochondrial genome (JF775522) (Ramesh et al. 2012). Sequencing reads made available from Wb in Jakarta (PRJEB536) were also aligned against the published Wb mitochondrial genome (JF775522) (Ramesh et al. 2012). The mitochondrial genome for Wb in Mali, Africa (JN367461) (Ramesh et al. 2012) was downloaded and included in the analysis. The mitochondrial genome phylogeny was reconstructed using MrBayes (Huelsenbeck & Ronquist 2001) as made available in the program Geneious v7.1.8 (Biomatters).
Demography
The pairwise sequential markovian coalescent (PSMC) (Li & Durbin 2011) model was used to reconstruct past demography. The PSMC model uses a single diploid genome to infer past variations in the effective population size (Ne). The PSMC input file was created using a custom script to modify the Wb assembled genome by replacing heterozygous sites with the appropriate IUPAC letter and then masking all sites covered at less than 20X. The programs available with the PSMC distribution were then used to construct the final input file. PSMC was run on contigs greater than 50kb in length for Wb L3 genomes with an average genotype coverage >50x: pt17 (L3-17A, L3-17E, L3-17D), pt48 (L3-4853, L3-4873, L3-48B), and pt74 (L3-74E). Each model was run for a total of 25 iterations with the following tuned parameters: θ/ρ = 4, tmax = 10, and atomic time intervals “3*4+7*2+6”. Bootstrap replicates were run using scripts provided with the PSMC distribution and an average chunk size of 50kb. All results were plotted in R (R Development Core Team 2009) using the ggplot2 package (Wickham 2009).
PSMC utilizes two chromosomes and is therefore limited in its ability to infer the recent past. MSMC (Schiffels & Durbin 2014), a variation on the same concept, allows for simultaneous analysis of multiple individual genomes and thereby offers increased resolution of events in the recent past. Phasing was carried out using Shapeit2 (Delaneau et al. 2013) and 10 of the 13 highest coverage genomes (excluding L3-17B, L3-74A, L3-74F). Kinship information (see above) between pairs of individuals was included as a prior as were any phase-informative sequencing reads (sequencing reads spanning a variant). Previously identified paralogous sequences and repeat locations were masked. MSMC was run using six haplotypes from three unrelated Wb worms exhibiting the highest coverage: L3-4853, L3-4873, and L3-17E. The input file was checked for anomalies, such as excessive consanguinity, using getStats.d script available from msmc-tools repository (github.com/stschiff/msmc-tools). MSMC was run with the default parameters and a fixed recombination rate for 20 EM-iterations. Results were plotted in R (R Development Core Team 2009) using package ggplot2 (Wickham 2009). Wb L3 genome L3-4853 was run as both phased and unphased data to check for the affect of phasing on demographic inference.
Selection
The DH test statistic, a compound test of neutrality that combines Tajima’s D, Fay and Wu’s H (Zeng et al. 2006), was calculated for each 10kb non-overlapping windows of the assembly using PopGenome (Pfeifer et al. 2014) excluding full-sib relationships. The empirical distribution of DH test statistic was calculated by sorting the normalized test statistics and then multiplying by the inverse of the ranks (Esteve-Codina et al. 2013). The 2.5% most extreme negative and positive windows were retained for blast annotation to the genome of B. malayi (PRJNA10729). For identified protein-coding genes the rate of adaptive substitutions was evaluated using the approximate McDonald-Kreitman test (MKT) (McDonald & Kreitman 1991) as implemented in PopGenome (Pfeifer et al. 2014).
Multi-locus Hudson-Kreitman-Aguadé (HKA) (Hudson et al. 1987) tests were calculated using the HKAdirect algorithm (Esteve-Codina et al., 2013) on 10kb windows across the genome. The input file was prepared using a custom python script. A correction for multiple tests was performed in R (R Development Core Team 2009) using the qvalue package and a 1% false discovery rate.
A final test was done to search for local adaptation. Briefly, the two genomes from Jakarta and PNG were compared to identify regions with an excess of fixed differences compared to neutral expectation using a custom python script. The numbers of fixed differences between the PNG and Jakarta genomes were counted for each 10kb sliding windows and normalized for variation in mutation rate using the diversity observed in the PNG genomes. Significance was determined at a false discovery rate of 10% using the qvalue package in R (R Development Core Team 2009). Significant genomic regions identified by DH and fixation tests were blasted against the NCBI nucleotide database using blastn and tblastn. The top 10 hits for each region based on E-value and percent identity were retained.
Results
Infrapopulation Genome Sequence Generation and Analysis
We de novo assembled a Wb genome using DNA from a single patient infection (De novo genome assembly, Supporting Information). Prior to DNA extraction, we reduced the proportion of human contaminating leukocytes by using two rounds of Histopaque gradients. Following Histopaque gradients, we selected the infection that displayed the most favorable ratio of Wb/Human DNA (qPCR Ct ratio 28:34 Wb:human) — patient 0022 (pt0022). Sequencing of the prepared library yielded a total of 199,921,394 paired-end reads. 60% of the reads consisted of human or contamination DNA (E. coli, PhiX); the remaining 40% of reads were used for de novo assembly (Table 1). The final Wb genome assembly consisted of 6,412 contigs with a N50 of 52kb, a maximum contig size of 689kb, and a total assembly length of 91.7 Mb – comparable to the published genomes of other filarial nematodes (Scott & Ghedin 2009; Desjardins et al. 2013). The assembly contained 680 (649 complete and 31 partial) out of the total 843 BUSCOs (Benchmarking sets of Universal Single-Copy Orthologs) (Simão et al. 2015) in the metazoan dataset (busco.ezlab.org). The complete assembly, as well as raw reads used to construct the assembly, is available at NCBI under bioproject PRJNA275548.
Table 1.
Wuchereria bancrofti de novo assembly statistics
| Assembly Statistic | pt22 |
|---|---|
| Total Reads (millions) | 199,912,394 |
| % mapping Human | 36% |
| % mapping contaminants | 23% |
| % Wb mtGenome | 0.017% |
| Number of contigs (>500bp) | 6412 |
| Assembly size (kb) | 91753 |
| N50 contig size (kb) | 52.417 |
| Max contig size (kb) | 689 |
| Position covered >20x | 83% |
| Predicted Genes | 13247 |
Wb L3 Sequence Generation and Analysis
13 individual Wb L3 worms derived from three separate patient infections (patient = pt): pt17, pt48 and pt74, yielded sufficient DNA for library preparation. Barcoded libraries were pooled (5 libraries per lane) and sequencing yielded an average of 30 million reads per sample genome (range: 7 to 158 million reads) (Table 2). Paired-end reads mapping to our de novo Wb genome assembly resulted in an average of 80% mapping to the Wb nuclear genome and 10% mapping to the mitochondrial genome. The raw reads for all 13 individuals are available through NCBI under bioproject PRJNA278287.
Table 2.
Wuchereria bancrofti L3-stage Genome Statistics
| Individual Infection | L3-Sample | Mosquito* | Reads (millions) | %mtGenome | %Nuclear | Average Coverage | Total Sites | Homs | Hets |
|---|---|---|---|---|---|---|---|---|---|
| Pt17 | L3-17A | Af ss | 35.1 | 13 | 74 | 74.3 | 49987 | 6289 | 15801 |
| L3-17B | Af ss | 13.4 | 14 | 72 | 40.8 | 22256 | 2486 | 6534 | |
| L3-17C | Af ss | 14.6 | 4 | 79 | 33.5 | 14279 | 1474 | 3996 | |
| L3-17D | Af ss | 44.2 | 15 | 73 | 81.8 | 49465 | 6234 | 16366 | |
| L3-17E | Af ss | 46.6 | 17 | 72 | 79.6 | 53677 | 6916 | 17542 | |
| Pt48 | L3-48A | Af ss | 28.7 | 8 | 63 | 36.1 | 18791 | 2409 | 4435 |
| L3-48B | Af ss | 37.1 | 15 | 70 | 60.7 | 48014 | 7412 | 12296 | |
| L3-4851 | Ah | 7.0 | 0.7 | 82 | 30.4 | 22652 | 2969 | 5491 | |
| L3-4853 | Af ss | 158.4 | 2 | 82 | 136.4 | 72774 | 11902 | 25289 | |
| L3-4873 | Ah | 17.0 | 7 | 83 | 64.9 | 60462 | 7625 | 21641 | |
| Pt74 | L3-74A | Ap | 39.8 | 0.1 | 29 | 46.6 | 12307 | 1603 | 2985 |
| L3-74B | Ap | 33.4 | 3 | 50 | 62.7 | 38039 | 2126 | 19916 | |
| L3-74E | Ap | 30.7 | 0.2 | 79 | 56.3 | 29264 | 4377 | 7491 |
A total of 73,922 SNPs were identified among the 13 Wb L3 genomes. Table 2 highlights the results of variant calling on the Wb L3 genomes. For each sample, we identified the total number of sites that were heterozygous (Hets) as well as the total number sites for which the derived allele was homozygous relative to the ancestral state (Homs) (Table 2). The ancestral state was determined by alignment to B. malayi (PRJNA10729). We also identified two samples from pt74 (L3-74A and L3-74B) where analysis of the reference allele frequency indicated peaks at 25%, 50%, and 75% rather than the single expected peak at 50% for a diploid genome. We believe this indicates the presence of multiple genomes within the sample (i.e., contamination with free DNA from other worms present in the same mosquito) and therefore exclude these samples from further population genetic analysis.
Population Genetic Diversity
Using 11 Wb L3 genomes, we estimated the population mutation rate, θ, as 2.18 × 10−4 (1 mutation expected per 4.5kb) and nucleotide diversity, π, as 2.47 × 10−4. The population recombination rate, ρ, is 4.12 × 10−5. We calculated that pt48 had on average 1.4-fold higher θ than pt17, 2.37 × 10-4 and 1.43 × 10−4, respectively. Nucleotide diversity between the infections pt17 and pt48 was 3.17 × 10−4. Given the nucleotide diversity within each infection, the ratio of within infection to between infection is 0.58 for pt17 and 1.2 for pt48.
The SFS was similar to the simulated SFS except with a slight excess of intermediate frequency alleles (Fig. S2, Supporting Information). Linkage disequilibrium, calculated as r2 against the expected physical distance, decayed to a value of r2 = 0.3 for SNPs separated by at least 4,700 bases (Fig. S3, Supporting Information). The r2 among contigs was 0.1267.
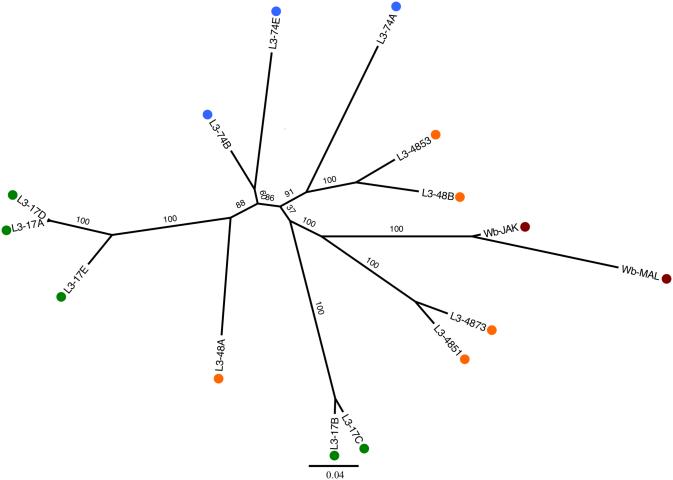
We constructed a phylogenetic tree from 19,000 SNPs distributed throughout the genome to examine the relationship among the Wb L3 worms (Figure 1). Typically, the genome of one Wb L3 larva is most closely related to an L3 genome from the same patient infection. However, there are exceptions, for example, the genomes of the closely related larvae L3-17B and L3-17C are more similar to the genomes of worms originating from infection pt48 (i.e., L3-4873) than they are to the genomes of other larvae from pt17 (L3-17A, L3-17D, and L3-17E). Moreover, Wb from Mali appeared more distant to PNG than to Jakarta, although this may be an artifact of incomplete sampling. The down-sampled phylogenetic tree supported the same relationships between worms and infections (Fig. S4, Supporting Information). The mitochondrial genome tree reconstructed using only DNA polymorphisms located on the mitochondrial genome (and illustrating the maternal relationships among the Wb L3 worms) supported within infection relationships as observed in the nuclear phylogeny (Fig. S5, Supporting Information).
Figure 1. Phylogeny.
Maximum Likelihood phylogeny with bootstrap support using all Wb L3 worms as well as sequences from Jakarta, Indonesia and Mali, Africa. The scale bar is in units of substitutions. Colored circles represent the infection where each worm originated: Patient 17 (green), Patient 48 (orange), Patient 74 (blue), Mali/Jakarta (red).
Relatedness
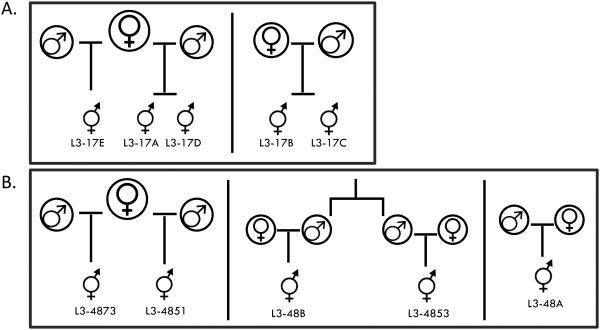
The kinship coefficient is the probability that two alleles sampled at random are identical by descent (Manichaikul et al. 2010). Kinship coefficients, ΦSNP, denoting 1st-degree relationships range from 0.177 to 0.354, while 2nd and 3rd degree relationships correspond to ranges of 0.0884 - 0.177 and 0.0442 - 0.0884, respectively. Using this metric we reconstructed the pedigree of the L3 worms stemming from each infection. The worms sampled from infection pt17 were most simply explained by two groups (Figure 2A), where L3-17A and L3-17D are 1st-degree relatives (ΦSNP =0.30) as are L3-17B and L3-17C (ΦSNP =0.26). The relationship between L3-17E to the pair 17A/17D is uncertain, but is consistent with them sharing a maternal ancestor based on the mitochondrial genome phylogeny (ΦSNP =0.039-0.098) (Figure 2A). For worms sampled from infection pt48 the pedigree is more complex (Figure 2B). Worms L3-4873 and L3-4851 are 2nd degree relatives (ΦSNP =0.14), likely ½ -sibs as based on the mitochondrial genome they share a maternal relative. There is also a weak 2nd-3rd degree relationship between L3-4853 and L3-48B, possibly equivalent to ‘2nd cousins’, although there are many other paths that lead to the same coefficient (ΦSNP =0.086).
Figure 2. Kinship Analysis.
Example pedigree for Wb L3 from within infection A) Patient 17 (pt17) and B) Patient 48 (pt48). The pedigree was constructed using the Kinship coefficients calculated in the program KING (Manichaikul et al. 2010). Shared maternal lineages were calculated using the mitochondrial genome phylogeny (Fig. S5, Supporting Information).
Demography
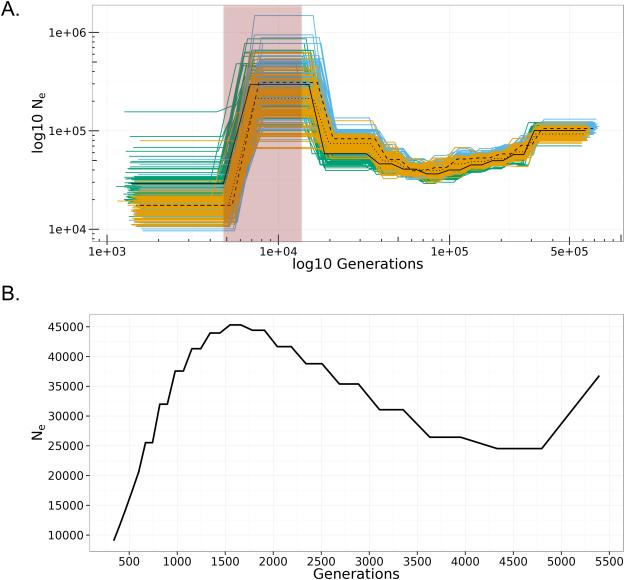
We reconstructed past demography of the Wb population in PNG using two whole genome methods where each provided a different temporal resolution. First, we used PSMC on single genomes to reconstruct the distant past of the Wb population, then MSMC on pooled haplotypes to reconstruct the more recent past. For PSMC analysis we included one Wb L3 worm from each patient infection, but since all the Wb L3 were from the same location in PNG (East Sepik Province, Tau) they all likely share the same population history and therefore we only presented three of the resulting traces (Figure 3A). The PSMC traces were scaled into Ne and generations using a mutation rate of 2.9 × 10−9 (Denver et al. 2009) (unscaled in Fig. S6, Supporting Information).
Figure 3. Reconstruction of Historical Demography.
A) Pairwise sequential Markovian coalescent (PSMC) plots of individual Wb L3 worm genomes: L3-17E (green/solid), L3-4873 (orange/dotted), L3-74E (blue/dash). The horizontal axis represents time in Log10 generations, while the vertical axis represents the Log10 of Ne (effective population size). Clusters of like-colored lines indicate bootstrapping support for each trajectory, while heavy colored lines indicate mean values. The shaded region (red box) corresponds to the peak effective population size of Anopheles farauti no 4. from Logue et al. (2015). B) Multiple sequential Markovian coalescent (MSMC) of 6 haplotypes from three unrelated Wb L3 worms: L3-17E, L3-4873, and L3-4853. The horizontal axis represents time in generations, while the vertical axis represents Ne, the effective population size.
Following the trace, at approximately 500,000 generations in the past, the Wb effective population size was predicted to have been approximately 100,000. The size increased to ~300,000 approximately 7,000-10,000 generations ago, then underwent a rapid decline to reach a size of 20,000-30,000 circa 3,000-4,000 generations in the past.
For the MSMC analysis we combined three Wb L3 worms for a total of six haplotypes. With MSMC we were able to resolve the demographic history from 5,000 to as recent as 400 generations in the past (Figure 3B). At 5,000 generations in the past the population size was 30,000-40,000. The Wb population increased in size and reached a peak size at 1,500 generations in the past of ~45,000. The size then declined until the last time epoch detectable, ~400 generations, with a final size of ~8,000.
Selection
For eight of the 11 L3 genomes (excluding full-sib pairs), we calculated the DH statistic in 10kb windows. We identified 16 regions throughout the genome that displayed a pattern consistent with selection. 12 windows (0.18% of the total genomic windows) were significant for directional selection with five also significant by the HKA test (Table S1, Supporting Information). Four windows (0.06% of the total genomic windows) were significant for balancing selection with three also significant by the HKA test (Table S1, Supporting Information). A MK test performed on the protein coding sequences found within the 16 windows returned no significant results. To test for local adaptation, we compared the genomes of L3 from PNG with a genome from Jakarta. After correcting for multiple comparisons, there were no remaining regions suggestive of local adaptation. The full results and accession numbers associated with blast annotation are available in Table S1 (Supporting Information).
Discussion
Little is known about the biology Much of the biology of W. bancrofti (Wb) because adult stages are difficult to access and we are not able to maintain a lab culture. Here our goal was to improve upon our knowledge of Wb biology through whole genome sequencing multiple individual worms. We choose to focus on Wb populations in the East Sepik Province of Papua New Guinea (PNG) because of the high prevalence infection (20 - 90% of the population positive for MF) (Bockarie & Kazura 2003; Cano et al. 2014) and study site in Tau, PNG that was not included in earlier elimination programs.
Genetic Diversity of Wuchereria bancrofti Infections
Complexity of infection (COI) is the presence of multiple parasites from the same species present in the same infection. COI complicates population genetic analysis because a variable position cannot be assigned to a single individual nor can we determine the number of individual parasites within an infection. Estimating the number of adult worms present in a patient infection or the relative change in adult worms (by sampling the larval cohort) is necessary if we wish to test whether elimination programs (either by reducing transmission or drugs) are effective. Other methods to quantify adult worm populations, i.e., ultrasonography of the lymphatics of Wb infected patients, indicate that adult worms exist in a “nest” structure (Amaral et al. 1994) and are thus difficult to correctly quantify via this method alone.
Our population genomic data provides a way to evaluate COI and track changes in the adult worm population providing a method to evaluate relative change in adult worm number. First, summary statistics on genetic diversity give us a baseline for COI. Then we compare how COI changes over time, where we predict that COI will gradually decrease while continuing treatment. However, to correctly capture the genetic diversity and track its change, it is necessary to sample MF from the same patient infection at multiple time points combined with sampling multiple patient infections from the same area. The genetic diversity from our two sample infections, pt17 as 1.43 × 10−4 and pt48 as 2.37 × 10−4, provide us with a first estimate of genome wide genetic diversity in a Wb infection. However, with our limited sample size it is difficult to distinguish whether the higher diversity in pt48 is due to differences in transmission, i.e., more infection events, or a larger, more distantly related breeding population which would produce more diverse offspring. Future studies will be needed to define the range of genetic diversity in Wb infections. The mitochondrial and nuclear genomes include information on how different worms are related and provides another way we can estimate COI. The pedigree structure of infections indicates the minimum number of adults contributing to the sampled larval cohort. Compared across multiple time points this is a method to characterize reproductive adults in an infection. From our relatedness data we hypothesize that there are more adult worms in pt48 than in pt17. The data also indicates multiple paternities where two worms share a single mitochondrial haplotype although they are not full-sibs. This may support a polyandrous mating system in Wb and while polyandry has been observed in other species of nematodes (Redman et al. 2008) this is the first time it has been demonstrated to occur in Wb. The addition of relatedness data from individual genomes supports a hypothesis of more breeding adult worms in pt48 than pt17. However, without further time points from these infections we cannot exclude alternative hypotheses such as asynchronous breeding or multiple infections as drivers of genetic diversity.
Another critical aspect of Wb elimination programs is quantifying transmission. Wb worms travel between infections through an intermediate host mosquito vector. The maturation from MF to an infective L3 can take upwards of 9 days over which time the mosquito may have dispersed up to 2km (Bockarie et al. 1996). Closely related worms found in different infections may indicate a direct transmission event between two patients or a common origin of infective worms. Through sampling from multiple infections within the same area we can use haplotype networks to reconstruct transmission dynamics. Our phylogenetic analysis provides some preliminary information on transmission events by combing information from nuclear and mitochondrial genomes. We observe that individual worms did not always cluster according to the patient infection of origin a potential sign of transmission. For example, worms sampled from the pt17 infection formed two distinct clusters suggesting that sampled worms are more closely related to worms from other infections, i.e., pt48 infection, than worms residing in the same infection. This may indicate a direct transmission event or alternatively adult worms with a shared history through multiple infection events. A better estimate of transmission networks would require a larger sample size incorporating more patient infections. If treatment is successful on reducing transmission then we would observe a progressively more clustered phylogeny at advancing time points. The goal then would be to observe a progressively “pruned” phylogeny as transmission is reduced during treatment. All three of these analyses combined provide an overview of the infection dynamics of the Wb population and will be informative for measuring the effectiveness of elimination programs.
Demographic History of Wuchereria bancrofti Populations in PNG
The different Wb L3 genomes (Figure 3A) display the same historical pattern of Ne which is likely representative of the entire history of Wb in PNG. We focus our interpretation on three distinctive events in the demographic history: i) the initial decline and stabilization ii) the population peak at 10,000 years, and iii) the population decline at 1,500 years. If the history of Wb is indeed the history of human migration (Laurence 1989), then the deeper demographic fluctuations observed in the PSMC analysis may be linked to human migration. If we assume a Wb generation time of 1-1.2 generations per year, as implied by stage specific maturation times (Farrar et al. 2013), then the population size fluctuations began around 240,000 years ago and stabilized 50,000 years ago. The continuous bottlenecks associated with human migration out of Africa might explain the reduction in Wb effective size over this time period, eventually leading to stable population size when dispersing humans settled in PNG 50,000 years ago—although the history for different endemic regions may determine a different stability point. Sampling of more endemic populations would give a better picture of Wb dispersal routes and verify a hypothesis of human mediated dispersal.
The peak Wb population size of 300,000 around 7,000-10,000 years ago, may have been influenced by a change in vector population size and vector species composition in PNG’s past. In PNG, multiple members of the Anopheles punctulatus group are known to transmit Wb (Bockarie et al. 1996). The different species have different competencies for transmitting Wb, where A. punctulatus is a less efficient vector of Wb when compared to A. farauti s.s and A. hinesorum (Erickson et al. 2013). If A. punctulatus were to replace A. farauti then transmission of Wb may be reduced. Logue et al. (2015) used PSMC to reconstruct the demographic history of mosquito species from the Anopheles punctulatus group in PNG. They observed a divergent history for A. punctulatus and A. farauti no.4 approximate to the timing of human arrival in PNG. A. punctulatus readily utilizes disturbed habitats, wheel-ruts and ditches, associated with human habitation and has been suggested to competitively displace more competent vectors in some locations of the Southwest Pacific (Beebe & Cooper 2002). Interestingly, a secondary peak population size in A. farauti no.4 corresponds with the observed peak population size in Wb, approximately 8,000 years ago. Following the peak size, A. farauti no.4 experienced a dramatic bottleneck in contrast to A. punctulatus which has been increasing (Logue et al. 2015). Future studies on Wb should include data on vector species composition that could then be tested for a correlation with genetic diversity of Wb infections, specifically the COI.
The final event in Wb history is the population crash which began 1,500 years ago (500 A.D.) and continued to ~300 years before the present day, resulting in the smallest observed population size in Wb history of 8,000-10,000.
The initial cause of Wb decline in PNG beginning 1,500 years ago is uncertain, we do know that the population size ~300 years ago is the smallest in the reconstructed history. To estimate contemporary Ne, future studies will need to incorporate temporal sampling of MF, ideally performed before, during, and after drug treatments, as well as larger samples sizes. However we postulate that the ongoing Wb elimination programs as well as other factors such as heavy malaria pressure in the north coastal lowlands (Peters & Christian 1960a) and malaria elimination programs using bed-nets and insecticides (Peters & Christian 1960b; Parkinson 1974; Spencer et al. 1974) has driven the Ne of Wb in PNG to be even lower than the most recent estimate obtained from MSMC.
Regions of the Genome Possibly Influenced by Natural Selection
We identified 12 regions as being consistent with directional selection, however only five were also significant by HKA test. The potentially most relevant gene is Innexin UNC-7, which has been implemented in modulating the sensitivity to ivermectin in C. elegans (Dent et al. 2000). Other identified genes were related to the nervous system of nematodes. For example, genes such as TwiK, a family of potassium channels most often expressed in the nervous system and ubiquitin carboxyl-terminal hydrolase which is highly specific to neurons and neuroendocrine system. Last we identified NOL-1, an ortholog to the human NOP2 (nucleolar protein), which in nematodes may be associated with larval development and reproduction.
We identified four regions of the genome that harbor diversity patterns consistent with the action of balancing selection: 1,4-alpha-glucan involved in glycogen storage, KH domain involved in RNA binding, ribose 5-phosphate isomerase involved in the Calvin cycle, and a ribosomal protein (Table S1, Supporting Information). Paralogous sequences as misalignments can falsely present as high diversity and be misidentified as balancing selection. To guard against misalignments we took steps to exclude paralogous regions (see Methods). Demography (e.g., population structure) can also confound the detection of balancing selection. We chose to use the DH test, a combination of the Tajima’s D and Fay and Wu’s H statistics, because it has been demonstrated to be less affected by demography than other frequency spectrum tests alone (Zeng et al. 2007). As a further test of balancing selection we examined the haplotype distribution for genes in the top regions. We found multiple haplotypes at three of the four regions. 1,4-alpha-glucan contained two distinct haplotype groups (45%, 55%), ribose 5-phosphate had three distinct haplotype groups (25%, 42%, 33%), and KH domain had two distinct haplotype groups (35%, 65%). The ribosomal protein region was a mixture of intermediate frequency alleles with no clear haplotype distinction (Fig. S7, Supporting Information). Multiple alleles coding for 1,4-alpha-glucan may indicate functional differences in glycogen storage or rate of glycogen conversion. Glycogen storage may be important in L3 as during host invasion the L3 worm becomes highly motile as it must migrate from the site of inoculation to the human lymphatics. Biological benefits of multiple alleles at ribose 5-phosphate isomerase is less obvious, however the gene has been investigated as a drug target in treating Trypanosoma brucei, the cause of African sleeping sickness (Loureiro et al. 2015).
Applications to monitoring and elimination programs
Wb elimination programs focus on distributing drugs that primarily kill MF but have limited efficacy against adult worms. The goal is to suppress transmission long enough for the adult worms to senesce. However, we have no reliable method of counting the number of adult worms in an infection. If we possessed this information we could make regional specific evaluations on when to stop drug distribution, potentially saving money and time. Thus, we need a way to count adult worms before, during, and after elimination programs. Currently this is not possible since adult worms reproduce in breeding “nests” which make it difficult to quantify the number of worms (Amaral et al. 1994). We propose a population genetic approach to monitor Wb populations by characterizing genetic diversity at different time points. For example, we can use genetic diversity indirectly to monitor the change in the complexity of an infection. As elimination programs progress we would predict that genetic diversity, coupled with complexity, should decrease especially if there is no ongoing transmission. A more accurate estimate of the adult worm population could be achieved using whole genome data, i.e., 79,000 SNPs identified here, to reconstruct the pedigree of the MF cohort and estimate the minimum number of reproductive adult worms needed to produce the sampled cohort. To evaluate elimination programs using population genetics, first, genetic material should be collected from numerous time points of reference, before, during and after drug administration. Next, genotype the samples using methods outlined here to create a catalogue of parasite genotypes from different time points (~30 individual to detect an allele present as low as 5% frequency). By comparing among time points we could then predict the decline in the number of adult worms with a slope that could then be used as a metric for elimination progress. In the event that elimination fails to progress or does not proceed as expected, data would now be available to test hypotheses of biological idiosyncrasies of the problematic Wb population against those for which elimination has been deemed successful.
Conclusions
Wuchereria bancrofti (Wb) is a widespread nematode parasite responsible for 90% of cases of lymphatic filariasis (LF). In this manuscript we examined the genome wide genetic diversity in 13 Wb L3 larvae isolated from mosquitoes experimentally fed on blood from patient infections. Genome wide data provides information on variable positions, demographic history, and genes influenced by selection. Future surveys will require more samples from infection patients to place into context our observed patterns of demography and natural selection in the Wb genome data. Our results and SNPs identified here are the first population data for Wb on a whole genome level and will provide useful tools for future studies of this important human parasite.
Supplementary Material
Data Accessibility.
- DNA sequencing reads for Bioproject PRJNA275548 deposited at NCBI SRA: SRP056161
- DNA sequencing reads for Bioproject PRJNA278287 deposited at NCBI SRA: SRP056210
- This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LAQH00000000. The version described in this paper is version LAQH01000000.
- Custom python scripts used in this manuscript: github.com/stsmall/Wb_Genome_L3
- Sampling location is Tau, Papua New Guinea: −3.666163, 142.766774
Acknowledgements
We would like to acknowledge: Matt Berriman for access to Wuchereria bancrofti reads from Jakarta and all the volunteers and collaborators from Papua New Guinea IMR. We would also like to thank two anonymous reviewers and editors at Molecular Ecology whose comments improved the final manuscript. Financial support for this study was provided through grants from the National Institutes of Health (AI103263) and the Clinical and Translational Science Collaborative of Cleveland (UL1TR000439). STS received support through a T32 Postdoctoral Fellowship in Geographic Medicine and Infectious Disease (AI007024).
Footnotes
Contributions:
S.T.S, L.J.R, and D.S designed the research; L.J.R, B.M.C, C.K, D.J.T, D.S and J.W.K provided materials and reagents; S.T.S and D.S analyzed the data; S.T.S, P.A.Z, and D.S wrote the manuscript.
Supporting Information
Methods: Wb de novo assembly
Table S1: Genes under Natural Selection
Fig. S1: Sampling Protocol
Fig. S2: Site Frequency Spectrum (SFS)
Fig. S3: Linkage disequilibrium decay over genomic distance
Fig. S4: Down-Sampled Phylogeny
Fig. S5: Mitochondrial Genome Phylogeny
Fig. S6: PSMC Analysis scaled in θ and pairwise sequence divergence
Fig. S7: Haplotype Distribution for balancing selection
References
- Amaral F, Dreyer G, Figueredo-Silva J, et al. Live adult worms detected by ultrasonography in human Bancroftian filariasis. Am J Trop Med Hyg. 1994;50:753–7. doi: 10.4269/ajtmh.1994.50.753. [DOI] [PubMed] [Google Scholar]
- Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–42. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe NW, Cooper RD. Distribution and evolution of the Anopheles punctulatus group (Diptera: Culicidae) in Australia and Papua New Guinea. International journal for parasitology. 2002;32:563–74. doi: 10.1016/s0020-7519(01)00359-9. [DOI] [PubMed] [Google Scholar]
- Bockarie M, Kazura J. Lymphatic filariasis in Papua New Guinea: prospects for elimination. Medical Microbiology and Immunology. 2003;192:9–14. doi: 10.1007/s00430-002-0153-y. [DOI] [PubMed] [Google Scholar]
- Bockarie M, Kazura J, Alexander N, et al. Transmission dynamics of Wuchereria bancrofti in East Sepik Province, Papua New Guinea. Am J Trop Med Hyg. 1996;54:577–81. doi: 10.4269/ajtmh.1996.54.577. [DOI] [PubMed] [Google Scholar]
- Cano J, Rebollo MP, Golding N, et al. The global distribution and transmission limits of lymphatic filariasis: past and present. Parasites & vectors. 2014;7:466. doi: 10.1186/s13071-014-0466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GK, Marjoram P, Wall JD. Fast and flexible simulation of DNA sequence data. Genome research. 2009;19:136–42. doi: 10.1101/gr.083634.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RF, Schaffner SF, Wenger EA, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proceedings of the National Academy of Sciences. 2015;112:7067–7072. doi: 10.1073/pnas.1505691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Howie B, Cox AJ, Zagury J-F, Marchini J. Haplotype estimation using sequencing reads. American journal of human genetics. 2013;93:687–96. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent JA, Smith MM, Vassilatis DK, Avery L. The genetics of ivermectin resistance in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2000;97:2674–2679. doi: 10.1073/pnas.97.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Dolan PC, Wilhelm LJ, et al. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proceedings of the National Academy of Sciences. 2009;106:16310–4. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins CA, Cerqueira GC, Goldberg JM, et al. Genomics of Loa loa, a Wolbachia-free filarial parasite of humans. Nature Genetics. 2013;45:495–500. doi: 10.1038/ng.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson SM, Thomsen EK, Keven JB, et al. Mosquito-parasite interactions can shape filariasis transmission dynamics and impact elimination programs. PLoS neglected tropical diseases. 2013;7:e2433. doi: 10.1371/journal.pntd.0002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Codina A, Paudel Y, Ferretti L, et al. Dissecting structural and nucleotide genome-wide variation in inbred Iberian pigs. BMC Genomics. 2013;14:148. doi: 10.1186/1471-2164-14-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar J, Hotez PJ, Junghanss T, et al. Manson’s tropical diseases. Elsevier; 2013. [Google Scholar]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012. arXiv:1207.3907.
- Ghedin E, Wang S, Spiro D, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–60. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper PJ, Chu BK, Mikhailov A, Ottesen EA, Bradley M. Mukoko D, editor. Assessing progress in reducing the at-risk population after 13 years of the global programme to eliminate lymphatic filariasis. PLoS neglected tropical diseases. 2014;8:e3333. doi: 10.1371/journal.pntd.0003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoti SL, Thangadurai R, Dhamodharan R, Das PK. Genetic heterogeneity of Wuchereria bancrofti populations at spatially hierarchical levels in Pondicherry and surrounding areas, south India. Infection, Genetics and Evolution. 2008;8:644–52. doi: 10.1016/j.meegid.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–9. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: Analysis of Next Generation Sequencing Data. BMC bioinformatics. 2014;15:356. doi: 10.1186/s12859-014-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–360. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CL, Won KY, Becker L, et al. Gyapong JO, editor. Seroprevalence and spatial epidemiology of lymphatic filariasis in American Samoa after successful mass drug administration. PLoS neglected tropical diseases. 2014;8:e3297. doi: 10.1371/journal.pntd.0003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence BR. The global dispersal of Bancroftian filariasis. Parasitology today (Personal ed.) 1989;5:260–4. doi: 10.1016/0169-4758(89)90260-3. [DOI] [PubMed] [Google Scholar]
- Lee T-H, Guo H, Wang X, Kim C, Paterson AH. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC genomics. 2014;15:162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–6. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue K, Small ST, Chan ER, et al. Whole genome sequencing reveals absence of recent gene-flow and separate demographic histories for Anopheles punctulatus mosquitoes in Papua New Guinea. Molecular Ecology. 2015;24:1263–74. doi: 10.1111/mec.13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro I, Faria J, Clayton C, et al. Ribose 5-phosphate isomerase B knockdown compromises Trypanosoma brucei bloodstream form infectivity. PLoS neglected tropical diseases. 2015;9:e3430. doi: 10.1371/journal.pntd.0003430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, et al. Robust relationship inference in genome-wide association studies. Bioinformatics (Oxford, England) 2010;26:2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–4. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University press; 1987. [Google Scholar]
- Palmieri JR, Purnomo, Dennis DT, Marwoto HA. Filarid parasites of South Kalimantan (Borneo) Indonesia. Wuchereria kalimantani sp. n. (Nematoda: Filarioidea) from the silvered leaf monkey, Presbytis cristatus Eschscholtz 1921. The Journal of parasitology. 1980;66:645–651. [PubMed] [Google Scholar]
- Parkinson AD. Malaria in Papua New Guinea 1973. PNG Med J. 1974;17:8–16. [Google Scholar]
- Peters W, Christian SH. Studies on the epidemiology of malaria in New Guinea. IV. Unstable highland malaria-the clinical picture. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1960a;54:529–536. doi: 10.1016/0035-9203(60)90027-4. [DOI] [PubMed] [Google Scholar]
- Peters W, Christian SH. Studies on the epidemiology of malaria in New Guinea. V. Unstable highland malaria-the entomological picture. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1960b;54:537–542. doi: 10.1016/0035-9203(60)90028-6. [DOI] [PubMed] [Google Scholar]
- Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. PopGenome: An Efficient Swiss Army Knife for Population Genomic Analyses in R. Molecular biology and evolution. 2014;31:1929–36. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. 2009 [Google Scholar]
- Ramaiah KD, Ottesen EA. Bockarie M, editor. Progress and impact of 13 years of the global programme to eliminate lymphatic filariasis on reducing the burden of filarial disease. PLoS neglected tropical diseases. 2014;8:e3319. doi: 10.1371/journal.pntd.0003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh A, Small ST, Kloos ZA, et al. The complete mitochondrial genome sequence of the filarial nematode Wuchereria bancrofti from three geographic isolates provides evidence of complex demographic history. Molecular Biochemical Parasitology. 2012;183:32–41. doi: 10.1016/j.molbiopara.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo MP, Mohammed KA, Thomas B, et al. Cessation of mass drug administration for lymphatic filariasis in Zanzibar in 2006: was transmission interrupted? PLoS neglected tropical diseases. 2015;9:e0003669. doi: 10.1371/journal.pntd.0003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman E, Grillo V, Saunders G, et al. Genetics of mating and sex determination in the parasitic nematode Haemonchus contortus. Genetics. 2008;180:1877–87. doi: 10.1534/genetics.108.094623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AR, Huff C. Linkage disequilibrium between loci with unknown phase. Genetics. 2009;182:839–44. doi: 10.1534/genetics.108.093153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nature genetics. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AL, Ghedin E. The genome of Brugia malayi - all worms are not created equal. Parasitology international. 2009;58:6–11. doi: 10.1016/j.parint.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Simonsen PE, Derua Y a, Kisinza WN, et al. Lymphatic filariasis control in Tanzania: effect of six rounds of mass drug administration with ivermectin and albendazole on infection and transmission. BMC infectious diseases. 2013;13:335. doi: 10.1186/1471-2334-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small ST, Ramesh A, Bun K, et al. Jex AR, editor. Population genetics of the filarial worm Wuchereria bancrofti in a post-treatment region of Papua New Guinea: insights into diversity and life history. PLoS neglected tropical diseases. 2013;7:e2308. doi: 10.1371/journal.pntd.0002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza DK, Osei-Poku J, Blum J, et al. The epidemiology of lymphatic filariasis in Ghana, explained by the possible existence of two strains of Wuchereria bancrofti. Pan Afr Med J. 2014;17 doi: 10.11604/pamj.2014.17.133.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer T, Spencer M, Venters D. Malaria vectors in Papua New Guinea. PNG Med J. 1974;17:22. [Google Scholar]
- Steinauer ML, Blouin MS, Criscione CD. Applying evolutionary genetics to schistosome epidemiology. Infection, Genetics and Evolution. 2010;10:433–43. doi: 10.1016/j.meegid.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] 2009 doi: 10.1002/0471250953.bi0410s25. Chapter 4, Unit 4.10. [DOI] [PubMed] [Google Scholar]
- Thangadurai R, Hoti SL, Kumar NP, Das PK. Phylogeography of human lymphatic filarial parasite, Wuchereria bancrofti in India. Acta Tropica. 2006;98:297–304. doi: 10.1016/j.actatropica.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Neafsey DE, Schaffner SF, Park DJ, Wirth DF. Harnessing genomics and genome biology to understand malaria biology. Nature reviews. Genetics. 2012;13:315–328. doi: 10.1038/nrg3187. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theoretical Population Biology. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- Won KY, Beau de Rochars M, Kyelem D, Streit TG, Lammie PJ. Assessing the impact of a missed mass drug administration in Haiti. PLoS neglected tropical diseases. 2009;3:e443. doi: 10.1371/journal.pntd.0000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Fu YX, Shi S, Wu CI. Statistical tests for detecting positive selection by utilizing high-frequency variants. Genetics. 2006;174:1431–1439. doi: 10.1534/genetics.106.061432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng K, Shi S, Wu C-I. Compound tests for the detection of hitchhiking under positive selection. Molecular biology and evolution. 2007;24:1898–908. doi: 10.1093/molbev/msm119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.