SUMMARY
Animals generate movement by engaging spinal circuits that direct precise sequences of muscle contraction, but the identity and organizational logic of local interneurons that lie at the core of these circuits remain unresolved. Here we show that V1 interneurons, a major inhibitory population that controls motor output, fractionate into highly diverse subsets on the basis of the expression of nineteen transcription factors. Transcriptionally defined V1 subsets exhibit distinct physiological signatures and highly structured spatial distributions with mediolateral and dorsoventral positional biases. These positional distinctions constrain patterns of input from sensory and motor neurons, arguing that interneuron position is a determinant of microcircuit organization. Moreover, V1 diversity indicates that different inhibitory microcircuits exist for motor pools controlling hip, ankle, and foot muscles, revealing a variable circuit architecture for interneurons that control limb movement.
INTRODUCTION
Animals interact with the world through movement, translating intent into action through the transformation of neural activity into the orderly contraction of muscles. The spinal circuits assigned to the control of limb movement take advantage of interneurons that drive motor neurons in precisely timed sequences, and that serve as relays for sensory feedback and descending command pathways (Arber, 2012; Windhorst, 2007). Defining the circuit logic through which local interneurons direct motor output is therefore a central issue in the neural control of movement.
Inhibitory interneurons have major roles in shaping spinal motor output, through the formation of local networks with highly selective synaptic input and output connectivity (Goulding et al., 2014). Electrophysiological studies have provided insight into the organizational logic of mammalian inhibitory microcircuits, notably those mediating feedforward, reciprocal, and feedback inhibition (Jankowska, 1992). GABAergic interneurons mediate presynaptic inhibition of sensory input, glycinergic interneurons control the reciprocal inhibition of flexor/extensor antagonists, and glycinergic and/or GABAergic Renshaw interneurons mediate the recurrent inhibition of motor neurons (Baldissera et al., 1981). The neuronal diversity implied by these physiological descriptions has been complemented by the molecular delineation of four cardinal classes of ventral spinal interneurons, termed V0 to V3 interneurons, each possessing a distinctive transcription factor character and different patterns of local connectivity (Briscoe et al., 2000).
Despite advances in elucidating the wiring of spinal motor systems, the organization of local circuit interneurons remains obscure. At its core, defining the organizing principles of spinal motor circuitry depends on determining the diversity of discrete interneuron subtypes that nucleate circuits for limb movement. In much the same way that limb-innervating motor neurons acquire diverse pool identities, cardinal interneuron classes defined by transcriptional character may fragment into multiple subtypes. Moreover, different physiological subtypes of interneurons appear to occupy stereotypic settling positions within the intermediate and ventral spinal cord (Alvarez and Fyffe, 2007; Hultborn et al., 1971). Nevertheless, it remains unclear whether the construction of interneuronal circuits takes advantage of distinctions in neuronal position when establishing local connectivity, a precedent set by the role of motor neuron position in establishing stereotypic patterns of proprioceptive input (Sürmeli et al., 2011). It is also uncertain whether the local circuits that control motor output adhere to a canonical wiring diagram, reiterated for each motor pool.
To gain insight into the organization of inhibitory circuits for motor control we have analyzed diversity and connectivity within the V1 interneuron population. V1 interneurons express the homeodomain transcription factor En1, and use GABA and/or glycine as inhibitory neurotransmitters (Benito-Gonzalez and Alvarez, 2012; Saueressig et al., 1999). This set constitutes over one third of all inhibitory interneurons in the ventral spinal cord, and includes all Renshaw and many reciprocal inhibitory interneurons (Sapir et al., 2004; Zhang et al., 2014). Genetic ablation of V1 neurons slows the speed of rhythmic locomotor output and perturbs flexor/extensor alternation (Gosgnach et al., 2006; Zhang et al., 2014). But in the absence of information about the diversity and connectivity of component V1 subtypes, it has not been possible to resolve the circuit logic that underlies aberrant motor behavior.
In this study we use molecular genetic approaches, and in an accompanying paper statistical analysis (Gabitto et al., 2016), to delineate the extent and consequences of diversity within the V1 interneuron population. This combined analysis reveals that V1 interneurons can be fragmented into fifty transcriptionally defined subtypes. V1 subpopulations exhibit distinct physiological signatures and settle in highly localized positions along both the mediolateral and dorsoventral axes of the neonatal spinal cord, a feature that constrains patterns of sensory and motor input. Moreover, the wiring diagram of inhibitory interneuron microcircuits differs for motor pools innervating hip, ankle, and foot muscles, a feature likely to reflect differences in the biomechanical properties of individual muscles.
RESULTS
Transcriptional Diversity of V1 Inhibitory Interneurons
To explore the diversity of spinal inhibitory interneurons we compared gene expression profiles for En1+ V1 and Ptf1a+ dI4/dILA interneurons (Glasgow et al., 2005; Sapir et al., 2004) (Figure 1A). We reasoned that a comparison of these two inhibitory populations, which settle at different dorsoventral positions and contact distinct postsynaptic targets, could reveal genes that fractionate the parental V1 and dI4/dILA populations, while excluding generic inhibitory markers. We focused this analysis on transcription factors (TFs), given their roles in specifying neuronal subtype identity (Dalla Torre di Sanguinetto et al., 2008).
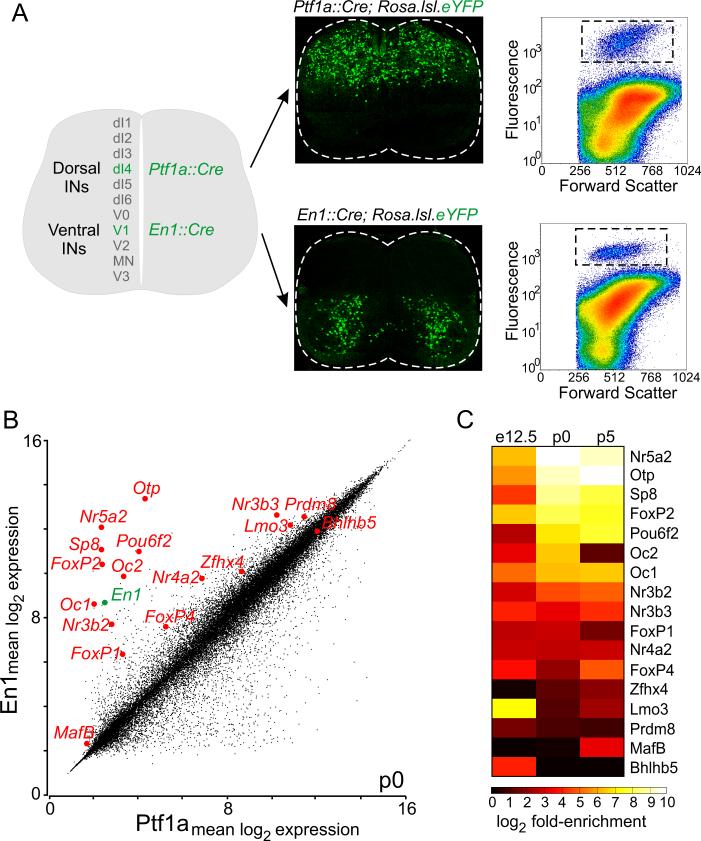
Figure 1. Transcription Factors Enriched in V1 Interneurons.
(A) Isolation of V1 and dI4/dILA interneurons. Left, interneuron populations. Middle, En1::Cre (V1) and Ptf1a::Cre (dI4/dILA) lineage-traced interneurons in p0 lumbar spinal cord. Right, FACS-isolated eYFP+ interneurons for microarray analysis.
(B) Scatter plot of expression levels of transcription factors (TFs, red) enriched in V1 interneurons from p0 mice.
(C) TFs with > 3-fold enrichment (p ≤ 0.02, one-way ANOVA) at one or more developmental ages. See also Figure S1.
V1 and dI4/dILA interneurons were isolated by fluorescence-activated cell sorting, from spinal cords dissociated from En1::Cre; Rosa.lsl.eYFP and Ptf1a::Cre; Rosa.lsl.eYFP mice respectively. Microarrays were performed at e12.5, p0, and p5 - a developmental window that covers the emergence of interneuron identity and the formation of synaptic connections (Figures 1B,C and S1A-C). Comparative microarray analysis identified 56 genes that encoded TFs with a >3-fold enrichment in V1 interneurons (mean V1:dI4 enrichment: 74.5; range: 3.1 to 930-fold; p ≤ 0.02, one-way ANOVA) at one or more developmental stages, and 160 TF genes with a >3-fold enrichment in dI4/dILA interneurons (mean dI4:V1 enrichment: 38.5; range: 3.1 to 784-fold; p ≤ 0.02, one-way ANOVA). In this study, we focus on diversity within the parental V1 interneuron population.
Analysis of gene expression databases revealed that 32 of the 56 V1 TFs exhibited mosaic expression in the embryonic (e11.5-e15.5) or neonatal (p4) ventral spinal cord (Figure S1A). Two additional genes, MafA and Prox1, exhibit scattered expression in the ventral spinal cord, and were included in subsequent analyses. From these 34 candidates, we focused on 19 TFs for which we were able to generate (FoxP1, FoxP2, FoxP4, Lmo3, MafA, Nr3b2, Nr4a2, Nr5a2, Otp, Pou6f2, Prdm8, and Sp8) or obtain (Bhlhb5, MafB, Nr3b3, Oc1, Oc2, Prox1, and Zfhx4) antibodies (Figure S1D-O; see Supplemental Information for description of antibodies).
To evaluate the prevalence of these 19 TFs within the parental V1 population we marked V1 interneurons by LacZ expression in En1::Cre; Tau.lsl.nLacZ (En1.nLacZ) mice, and performed dual immunohistochemical analysis with antibodies directed against LacZ and each TF individually. V1 interneurons arise from the p1 progenitor domain between e9.5 - e12.5 and undergo migration to their final settling position during embryogenesis (Matise and Joyner, 1997). We therefore focused our analysis on p0 lumbar spinal cord, when initial specification and migration is complete and before extinction of expression of many developmentally regulated TFs (Benito-Gonzalez and Alvarez, 2012). All 19 TFs were expressed in subsets of V1 interneurons (Figures 2A and S2A), with the incidence of expression ranging from 5% (MafA) to 74% (Lmo3) of the parental V1 population (Figure 2B). Conjoint exposure to antibodies directed against 14 of these TFs marked > 90% of V1 interneurons in p0 lumbar spinal segments, indicative of near-complete coverage of the parental V1 population (Figure 2C). In addition, 12 of the 19 V1 TFs were expressed within subsets of V2a interneurons (Figure S2C; threshold: 3%; range: 3-62% of parental V2a interneurons), indicating that this set of TFs reveals diversity in both V1 inhibitory and V2a excitatory interneurons.
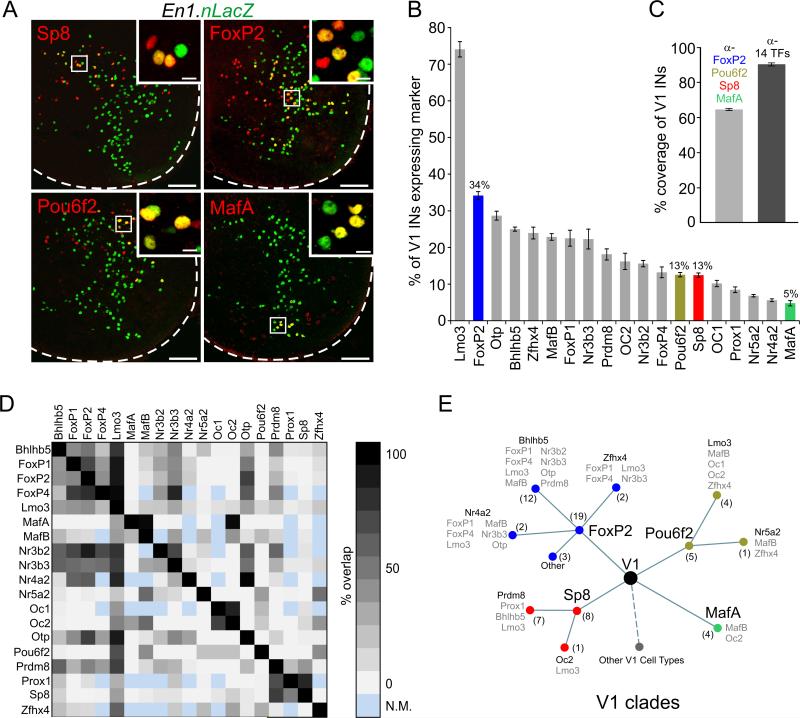
Figure 2. V1 Transcriptional Diversity and Cladistic Analysis.
(A) TFs (red) label subsets of V1 interneurons (green) in L3-L5 spinal segments from p0 En1.nLacZ mice. Scale bars = 100 μm or 10 μm (inset).
(B) V1 interneurons expressing TFs at p0 L3-L5 spinal segments (n ≥ 3 animals, mean ± SEM). See Figure S2B,C for characterization of TFs in dI4/dILA and V2a interneurons.
(C) Coverage of V1 interneurons in p0 L3-L5 spinal cord. Anti-FoxP2, MafA, Pou6f2, and Sp8 antibodies label 64.2 ± 0.6% of V1 interneurons (n = 3). Application of 14 antibodies - Bhlhb5, FoxP1, FoxP2, MafB, Nr3b2, Nr4a2, Nr5a2, Oc1, Oc2, Otp, Pou6f2, Prdm8, Prox1, and Sp8 - labels 90.4 ± 0.8% of V1 interneurons (n = 3). Mean ± SEM.
(D) Matrix of pairwise overlap for 148 out of 171 comparisons. N.M. = not measured, due to antibody incompatibility.
(E) V1 interneurons segregate into four clades defined by mutually exclusive expression of FoxP2, MafA, Pou6f2, and Sp8 (< 1% overlap in each pairwise comparison). Clades are further subdivided by distinct TFs (black). Dotted line represents additional V1 cell types. The number of V1 cell types is indicated in parenthesis. See also Figures S2D and S3C.
We estimated the number of neuronal types within the parental V1 population through application of a Bayesian sparse linear regression algorithm that assigns V1 interneuron cell types on the basis of TF expression and settling position (Gabitto et al, 2016). The extent of co-expression was determined for binary TF combinations, with each pairing gated to LacZ+ neurons in p0 En1.LacZ mice (Figure 2D). This analysis assigns 50 ± 2 V1 interneuron cell types, some defined by as many as 9 TFs (Gabitto et al., 2016). Four TFs - FoxP2, MafA, Pou6f2, and Sp8 - delineate non-overlapping clades that comprise 64% of the parental V1 population at p0, with each clade containing multiple cell types (Figures 2C,E and S2D). This combination of experimental observation and statistical inference therefore defines the extent of V1 diversity and its hierarchical organization.
Spatial Organization of V1 Interneuron Subpopulations
We considered whether V1 cell types are clustered in stereotypic settling patterns. Since individual V1 cell types are defined by as many as 9 TFs (Gabitto et al, 2016), it is not practical to delineate them on the basis of their complete transcriptional profile. We therefore assessed the spatial distributions defined by each of the 19 V1 TFs individually (Figure S3A) or in a few cases by the superimposition of two TFs (Figure S3B). These larger sets of V1 interneurons, termed “composite groups”, are predicted to contain multiple V1 cell types (Figure S3C). Any spatial restriction of these larger V1 interneuron groups would imply clustering of individual V1 types.
We first examined the single instance in which an inferred V1 cell type can be delineated by the co-expression of just two TFs, Nr5a2 and Pou6f2. With respect to the parental V1 population, which extends ~400 μm along the dorsoventral (D/V) and mediolateral (M/L) axes in p0 lumbar spinal cord, the V1Nr5a2/Pou6f2 cell type revealed a markedly confined settling position (Figure 3A,B). Each of the V1 composite groups also occupied a domain more restricted than that of the parental V1 distribution profile (Table S1, p < 0.001 by one-tailed Monte Carlo test; single TF-gated fractional area (Fa) range: 0.217 to 0.855, dual TF-gated Fa range: 0.085 to 0.365). By extension, it follows that the individual V1 cell types contained within these composite groups will also be clustered.
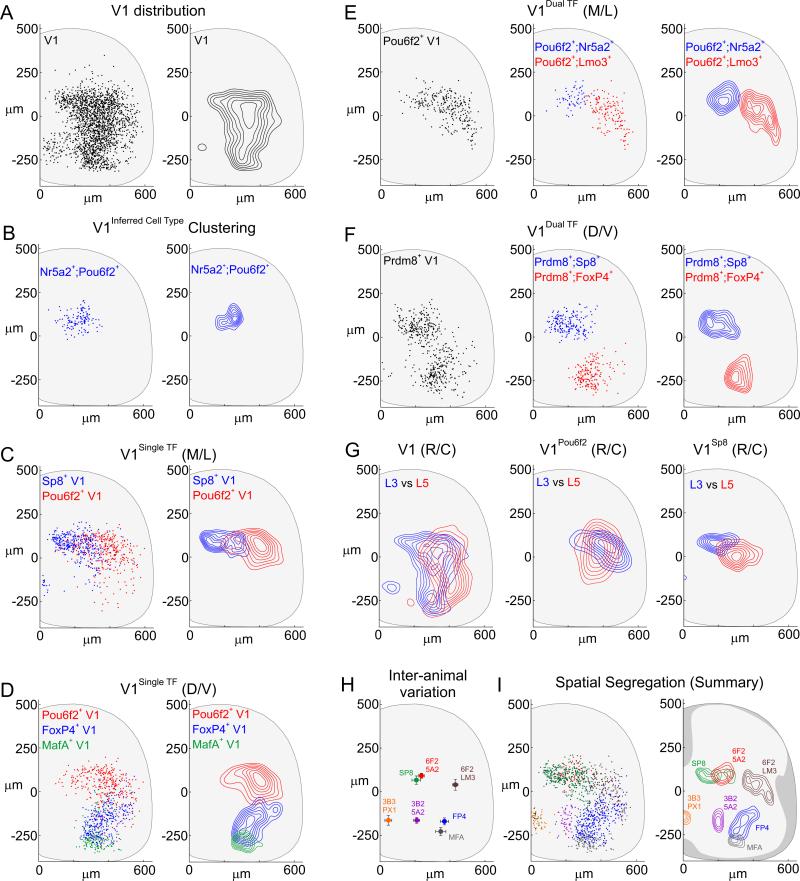
Figure 3. Spatial Segregation of V1 Interneuron Subpopulations.
(A) V1 interneurons in p0 L3-L5 segments of En1.nLacZ mice. D/V axis range: 132 to −265 μm; M/L axis range: 127 to 487 μm, 5th-95th percentiles from central canal. Contours represent density at the 30th-90th percentiles.
(B) Spatial clustering of V1Pou6f2/Nr5a2 interneurons (blue, Fa = 0.236) (p < 0.00001, one-tailed Monte Carlo test compared to parental V1).
(C) M/L biases in distributions of V1Sp8 (Xepicenter = 162 μm) and V1Pou6f2 (Xepi = 403 μm) interneurons. p < 1 × 10−20, Wilcoxon Rank Sum test in x-axis, V1Sp8 or V1Pou6f2 vs V1Parental, and V1Sp8 vs V1Pou6f2.
(D) D/V biases in distributions of V1Pou6f2 (Yepi = 66 μm), V1FoxP4 (Yepi = −158 μm), and V1MafA (Yepi = −277 μm) interneurons. V1Sp8 interneurons (Yepi = 72 μm) also occupy a dorsal position. p < 1 × 10−20, Wilcoxon Rank Sum test in y-axis for V1Pou6f2, V1FoxP4, V1MafA, or V1Sp8 vs V1Parental.
E) Subdivision of V1Pou6f2 interneurons into medial (Nr5a2+, blue) and lateral (Lmo3+, red) subsets in p0 L3-L4 spinal segments.
(F) V1Prdm8 interneurons fractionate into dorsal Sp8+ (blue) and ventral FoxP4+ (red) composite groups.
(G) V1, V1Pou6f2, and V1Sp8 settling position at L3 (blue) or L5 (red) in p0 mice. p < 0.0001 for L3 vs L5, 2D KS test.
(H) Constancy of x,y position (mean ± SD) for V1 interneurons expressing Sp8 (n = 7), Pou6f2/Nr5a2 (n = 8), Pou6f2/Lmo3 (n = 4), FoxP4 (n = 7), Nr3b2/Nr5a2 (n = 8), Nr3b3/Prox1 (n = 6), and MafA (n = 7 animals).
(I) Spatial distributions of seven V1 subsets. Contours represent 60th-90th percentile densities. See also Figures S3 and S4, and Table S1.
We examined the degree to which subsets of V1 interneurons settle at distinct positions in the ventral spinal cord, focusing initially on the four primary V1 clades. The V1MafA, V1Pou6f2, and V1Sp8 clades showed discrete distributions along the D/V and M/L axes (Figure 3C,D and Table S1), whereas the FoxP2 clade occupies a broader domain (Figure S3A, Fa = 0.855). V1Pou6f2 interneurons fractionated into medial Nr5a2+ and lateral Lmo3+ interneurons, with 93% mutual exclusion, demonstrating segregation within the members of a single clade (Figure 3E). Similarly, the V1Prdm8 composite group fractionated into dorsal Sp8+ and ventral FoxP4+ subsets (Figure 3F). The mean position of V1 cell types and composite groups at L3-L5 levels of lumbar spinal cord was constant between animals (Figure 3H). Moreover, V1 settling positions are stable from e15.5 to p20, an indication that V1 neuronal clustering is not a transient reflection of developmental stage (Figure S4A-B).
The ventral location of the V1MafA clade prompted us to examine its relationship to Renshaw (V1R) interneurons. This V1 subtype occupies the most ventral region of the parental V1 distribution, expresses the transcription factors Oc1, Oc2, and MafB and the calcium binding protein calbindin, and mediates recurrent inhibition of motor neurons (Alvarez and Fyffe, 2007; Stam et al., 2012). Analysis of V1R interneurons at e13.5, shortly after their initial specification and migration within the ventral spinal cord, demonstrated the existence of MafA+ and MafA− V1R subsets, with 61.6 ± 5.8% of V1R interneurons expressing MafA (Figure S2E,F). A similar molecular diversity is evident at p0, potentially corresponding to descriptions of morphological heterogeneity amongst V1R interneurons (Fyffe, 1990). Conversely, ~20% of V1MafA interneurons are located > 200 μm from the ventral edge of the spinal cord, a dorsal position that effectively excludes them as V1R interneurons. Thus, V1R and V1MafA interneurons are not co-extensive - while many V1R neurons are contained within the V1MafA clade, others comprise ventral V1 interneurons distinct from the V1MafA clade as well as more dorsal V1FoxP2, V1Pou6f2, or V1Sp8 clades.
V0c interneurons exhibit rostrocaudal variation in transmitter phenotype and connectivity (Zagoraiou et al., 2009), prompting us to determine whether neurons in V1Pou6f2 and V1Sp8 clades exhibit rostrocaudal distinctions in settling position. The overall positional bias of V1Pou6f2 and V1Sp8 interneurons was maintained at single segmental levels between L3 and L5 (Figure S4C-E). Nevertheless, minor differences in the settling position of V1Pou6f2 and V1Sp8 clades were evident, with a ventromedial shift for V1Pou6f2 and a ventrolateral shift for V1Sp8 at progressively more caudal lumbar levels (Figures 3G and S4D,E; see also Figure 5C,E). Thus, V1 interneuron settling position exhibits overall constancy, albeit with subtle differences along the lumbar rostrocaudal axis.
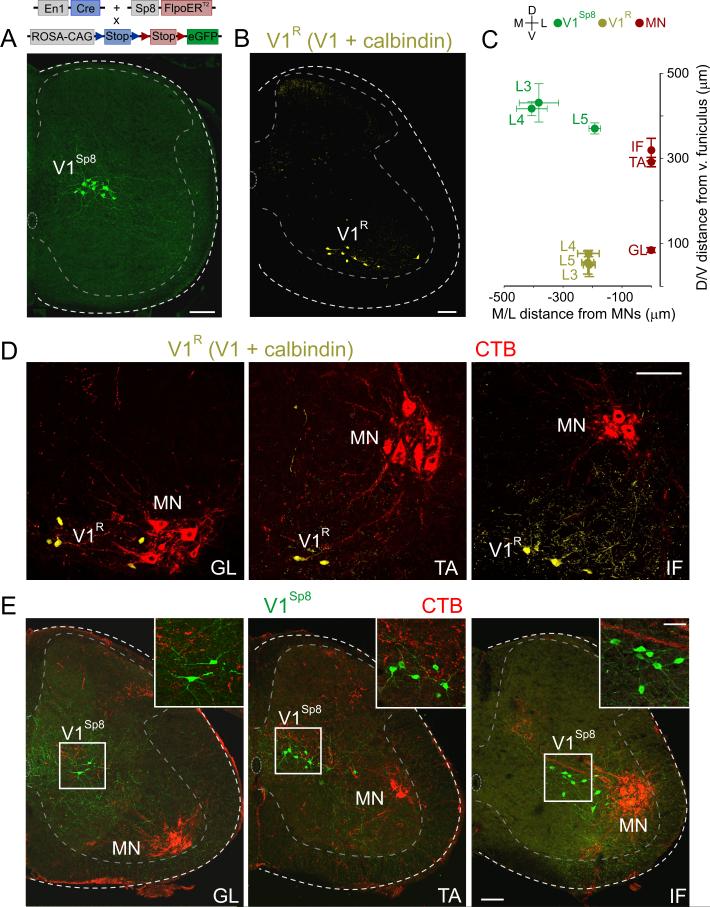
Figure 5. Relative Position of V1R and V1Sp8 Interneurons to Motor Pools.
(A) V1Sp8 interneurons (green), in p12 lumbar spinal cord of En1::Cre; Sp8::FlpoERT2; RCE.dual.GFP mice.
(B) V1R interneurons (yellow, colocalization mask of eGFP and calbindin immunoreactivity) in ~p21 En1::Cre; RCE.lsl.GFP lumbar spinal cord.
(C) V1R and V1Sp8 position with respect to GL, TA, and IF motor pools in ~p21 mice. Motor pool D/V positions: GL: 84 ± 3 μm, TA: 291 ± 6 μm, IF: 321 ± 15 μm, from dorsal border of ventral funiculus.
(D) D/V position of V1R interneurons (yellow) with respect to CTB-backfilled GL, TA, and IF motor pools (MN, red) in ~p21 lumbar spinal cord. D/V distances: V1R ventral to GL, TA, and IF motor neurons by 8 ± 3 μm, 242 ± 14 μm, and 264 ± 13 μm, respectively. p < 0.0001, one-way ANOVA; Bonferroni post-hoc test: p < 0.001, TA or IF vs GL. M/L distances were not significantly different (p = 0.99, one-way ANOVA).
(E) D/V position of V1Sp8 interneurons (green) with respect to CTB-backfilled GL, TA, and IF motor pools (red) in ~p21 lumbar spinal cord. V1Sp8 dorsal to GL, TA, and IF by 332 ± 8 μm, 139 ± 23 μm, and 50 ± 8 μm, respectively (p < 0.0001, one-way ANOVA; Bonferroni post-hoc test: p < 0.001, TA or IF vs GL; p < 0.05, TA vs IF). In the M/L axis, V1Sp8 interneurons were significantly closer to IF than to GL or TA (192 ± 11 μm versus 406 ± 26 μm or 382 ± 33 μm, respectively; p < 0.01, one-way ANOVA; Bonferroni post-hoc test; p < 0.01, IF vs GL or TA). Values are mean ± SEM, n ≥ 3 animals per condition. Scale bars = 100 μm or 50 μm (inset). See also Figure S6.
In total, this analysis of V1 identity and settling position identifies numerous spatially discrete V1 subpopulations, seven of which are illustrated (Figure 3I, see also Gabitto et al, 2016). These seven clusters represent ~44% of the parental V1 population and comprise 39 inferred cell types. The extent of V1 interneuron diversity goes beyond that recognized previously for interneuron populations in the mammalian CNS (Sanes and Masland, 2015).
V1 Clades Contain Physiologically Distinct Interneuron Subsets
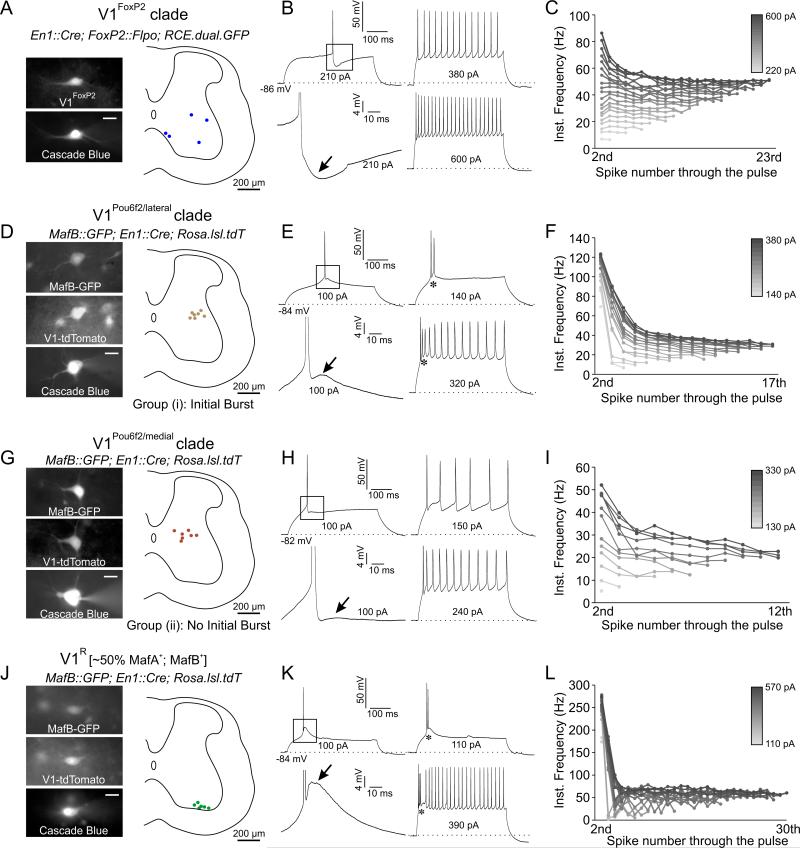
To determine whether molecular distinctions in V1 identity reflect differences in interneuron physiology, we analyzed the electrophysiological properties of neurons in V1FoxP2 and V1Pou6f2 clades, as well as V1R interneurons, in a spinal cord slice preparation at p10-p14. To label neurons in the V1FoxP2 clade, we generated FoxP2::Flpo transgenic mice, and used an intersectional genetic strategy in which En1::Cre; FoxP2::Flpo; RCE.dual.GFP mice selectively express GFP in V1FoxP2 interneurons (Figure S5A-C). To identify both the V1Pou6f2 clade and V1R interneurons, we used MafB::GFP; En1::Cre; Rosa.lsl.tdT mice, in which two distinct GFP+/tdT+ V1 subsets could be distinguished: a dorsal subset fully contained within the V1Pou6f2 clade, and a ventral subset corresponding to V1R interneurons (Figure S5D). Approximately half of all V1R interneurons express MafA, and they serve as a proxy for the V1MafA clade (Figure S2E,F).
We found that V1FoxP2, V1Pou6f2, and V1R subsets could be distinguished by their passive and active membrane properties (Figure 4 and S5E-I). At hyperpolarized (< −80 mV) membrane potentials, distinctive active properties included: (i) the prominence of spike after-hyperpolarization (AHP) and early transient low-threshold depolarizations, (ii) the extent of initial spike bursting, and (iii) the degree of spike-frequency adaptation (SFA) during steady-state firing.
Figure 4. Electrophysiological Characterization of V1 Clades.
(A-C) Physiology of V1FoxP2 interneurons. (A) V1FoxP2 interneurons (n = 5) targeted for recording and filled with Cascade Blue in En1::Cre; FoxP2::Flpo; RCE.dual.GFP mice. Scale bar = 20 μm. (B) Firing properties of V1FoxP2 cells show a prominent after-hyperpolarization (AHP, arrow), a non-bursting phenotype, and an absence of spike frequency adaptation (SFA). (C) Instantaneous firing (IF) frequency for each action potential (dot) through pulses of increasing current amplitudes (20 pA steps). Little or no SFA is observed below 460 pA.
(D-F) Physiology of V1Pou6f2/lateral interneurons. (D) Position of V1Pou6f2/lateral interneurons (n = 7) in MafB::GFP; En1::Cre; Rosa.lsl.tdT mice. (E) Transient low-threshold depolarization (arrow), with an initial burst (asterisks), and the presence of SFA throughout the pulse. (F) SFA, indicated by the decreasing instantaneous frequency of successive action potentials.
(G-I) Physiology of V1Pou6f2/medial interneurons. (G) Position of V1Pou6f2/medial interneurons (n = 7). (H) Neurons exhibit a non-burst phenotype and a weak low-threshold depolarization (arrow, H). (I) IF plot showing SFA.
(J-L) Physiology of V1R interneurons, representing the V1MafA clade. (J) Position of V1R interneurons (n = 6) in En1::Cre; Rosa.lsl.tdT; MafB::GFP mice. (K) Neurons show prominent low-threshold depolarization (arrow), and burst firing (asterisks). (L) IF plot shows absence of SFA. See also Figure S5.
Analysis of V1FoxP2 interneurons using whole-cell current-clamp recording revealed action potentials with a large and fast-rising AHP, no transient low-threshold depolarizations, no initial spike bursting, and little or no SFA (Figure 4A-C). V1Pou6f2 interneurons segregated into a lateral bursting subset with a large low-threshold depolarization (Figure 4D, E) and a medial non-bursting subset with a much smaller transient depolarization (Figure 4G, H). At a molecular level these physiological distinctions likely correspond to the mediolateral positional segregation of V1Pou6f2/Nr5a2 and V1Pou6f2/Lmo3 interneurons (Figures 3E). Both V1Pou6f2 subsets exhibited SFA, likely resulting from the buildup of long duration AHPs during successive spikes (Figure 4F, I). V1R interneurons exhibited a large low-threshold depolarization and short AHPs, resulting in a strong bursting phenotype with no evident SFA during steady state firing (Figure 4J-L). Thus, V1FoxP2, V1Pou6f2, and V1R interneurons can be distinguished by their biophysical properties, consistent with their molecular and positional segregation into distinct V1 clades.
Mapping the Relative Position of V1 Subpopulations and Motor Pools
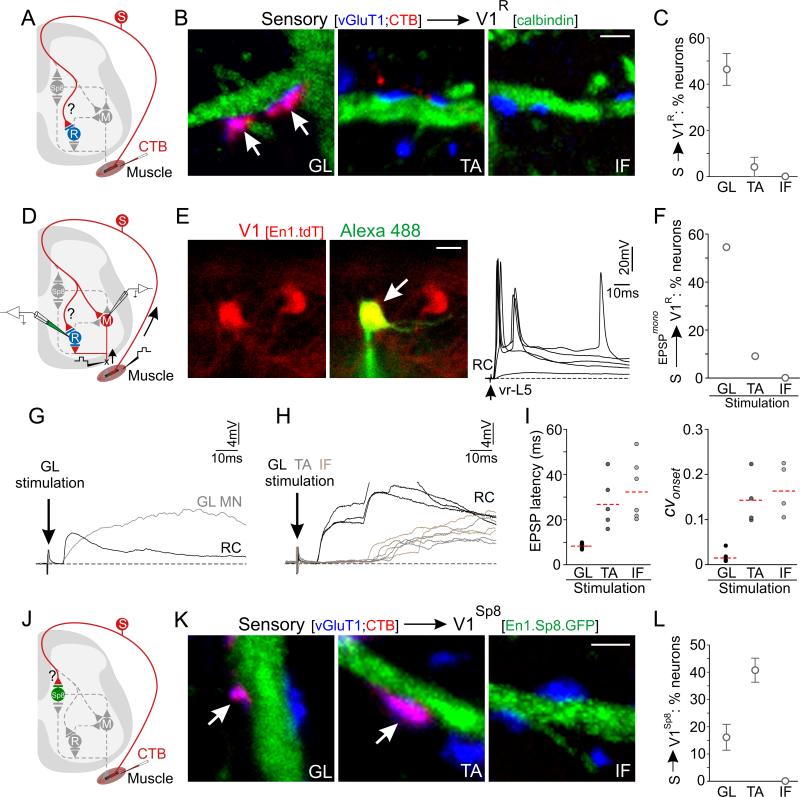
The stereotypic nature of V1 neuronal position led us to examine whether the spinal motor system takes advantage of spatial segregation in the construction of inhibitory microcircuits. We focused on the connectivity of V1 subpopulations that settle at dorsal and ventral extremes of the parental V1 domain. Ventrally, we examined V1R interneurons, and as a dorsal comparator population we examined neurons in the V1Sp8 clade (Figure 2E).
We employed intersectional and inducible genetic strategies to mark V1Sp8 interneurons, generating a Sp8::FlpoERT2 transgenic mouse line and administering tamoxifen at p0, to evade the early expression of Sp8 in neuronal progenitors (Figure S6). The use of Sp8::FlpoERT2 and En1::Cre driver lines crossed to a RCE.dual.GFP reporter allele resulted in selective labeling of a cluster of V1Sp8 interneurons in the dorsomedial region of the parental V1 domain (Figure 5A). To mark V1R interneurons we took advantage of their expression of calbindin (Figure 5B).
To probe the organizational features of inhibitory microcircuits that control limb musculature, we focused on three hindlimb motor pools. Gluteus (GL) motor neurons innervate hip extensor muscles and occupy an extreme ventral position in the LMC. Tibialis anterior (TA) and instrinsic foot (IF) motor neurons innervate ankle flexor and foot plantar-flexor muscles respectively, and occupy a similar dorsal position within the LMC (McHanwell and Biscoe, 1981). Assessment of V1 subsets with respect to identified GL, TA, and IF motor pools in p21 En1::Cre; Rosa.lsl.FP (tdT or eGFP) mice indicated that V1R interneurons occupy a position close to that of GL motor neurons, whereas V1Sp8 interneurons occupy a position close to that of TA and IF motor neurons (Figure 5C-E).
The dorsal and ventral positioning of the V1R and V1Sp8 populations provided a spatial reference point for assessing three elements of spinal motor microcircuitry: (i) the nature of proprioceptive sensory input to V1 interneuron subsets, (ii) the nature of motor collateral input to V1 interneuron subsets and (iii) the organization of V1 interneuron output onto discrete motor neuron pools.
Positional Selectivity of Proprioceptive Sensory Inputs to V1R and V1Sp8 Interneurons
We examined first whether V1R and V1Sp8 interneurons receive sensory input, and if so, whether their settling position is predictive of proprioceptive input. Proprioceptive afferents target distinct D/V domains in a motor neuron-independent manner, as a function of their distal-to-proximal muscle origins in the limb (Sürmeli et al., 2011). On this basis we reasoned that the ventral restriction of V1R interneurons may constrain their ability to receive sensory input from dorsally-projecting proprioceptive afferents. In contrast, V1Sp8 interneurons might receive input both from dorsally-projecting sensory afferents supplying more distal muscles, as well as from the collaterals of ventrally-projecting afferents supplying proximal musculature (Brown, 1981).
To determine the pattern of proprioceptive input to V1R and V1Sp8 interneurons, we monitored the origin of proprioceptive terminals through transganglionic sensory transport and terminal accumulation of CTB after injection into individual limb muscles (Figure 6A,J). We found in ~p21 En1::Cre; Rosa.lsl.tdT mice that 47% (14/30) of V1R interneurons received input from CTB+; vGluT1+ GL sensory afferents. In contrast, only 5% (2/38) of V1R interneurons received input from TA afferents, and none (0/32) received input from IF afferents (Figure 6B,C). Differential innervation was not the result of preferential labeling of GL sensory afferents, in that we detected a similar incidence of homonymous CTB-labeled sensory terminals onto GL, TA, and IF motor neurons (Figure S7A,B).
Figure 6. Specificity of Sensory-Interneuron Connectivity at Individual Joints.
(A) Assay of proprioceptive input to V1R interneurons.
(B-C) CTB+; vGluT1+ proprioceptive input to V1R interneurons. (C) Percent of V1R interneurons with pool-specific input. p = 0.0007, one-way ANOVA; Bonferroni post-hoc test: p < 0.01, GL vs TA or IF, n = 3 animals. Innervation density (inputs/100 μm dendrite) for neurons receiving sensory input: GL, 5.1 ± 1.0; TA, 2.8 ± 1.9. All data are mean ± SEM. Scale bar = 2 μm.
(D) Assay of monosynaptic input from pool-specific Ia afferents onto V1R interneurons.
(E) Left, V1R interneurons (red, labeled in En1::Cre; Rosa.lsl.tdT mice) targeted for intracellular recording, filled with Alexa-488 hydrazide (green). Right, stimulation of L5 ventral root (vr-L5) at different intensities evoked graded short-latency synaptic potentials. Scale bar = 10 μm.
(F) V1R interneurons (GL: 6/11 cells; TA: 1/11 cells; IF: 0/11 cells) receiving monosynaptically-evoked EPSPs from peripheral muscles.
(G) Monosynaptic EPSPs in a V1R interneuron (black) or GL motor neuron (gray) following GL muscle stimulation.
(H) Short-latency EPSPs evoked in V1R interneurons after GL stimulation (3 superimposed responses evoked at 0.1 Hz stimulation frequency, black). Stimulation from TA (gray) or IF (brown) muscle sensory fibers resulted in long (>20 msec), variable latencies, indicative of polysynaptic activation. Arrow, stimulation artifact.
(I) Left, latencies from synaptically evoked responses in V1R interneurons after stimulation of GL, TA, and IF muscles in p4 to p5 mice. The latency from GL muscle stimulation was significantly shorter than from TA or IF (p < 0.05, one-way ANOVA). Right, coefficient of variation (CVonset) of synaptic response latency. Red line = mean.
(J) Assay of proprioceptive sensory input onto V1Sp8 interneurons.
(K-L) CTB+; vGluT1+ proprioceptive input to V1Sp8 interneurons. (L) V1Sp8 interneurons with pool-specific input. p = 0.004, one-way ANOVA; Bonferroni post hoc test: p < 0.01, GL vs TA; p < 0.001, TA vs IF; n.s., GL vs IF, n ≥ 3 animals. Average CTB+; vGluT1+ input density/100 μm of V1Sp8 dendrite: GL: 0.92 ± 0.23; TA: 2.20 ± 0.14; IF: 0 ± 0; p < 0.0001, oneway ANOVA; Bonferroni post hoc test: p < 0.01, GL vs TA; p < 0.05, GL vs IF; p < 0.001, TA vs IF. Similar labeling efficiency was observed for GL, TA, and IF sensory afferents (Figure S7C). Scale bar = 2 μm. See also Figure S7.
To determine whether the profile of sensory contacts with V1R neurons correlates with functional input we monitored monosynaptic connections between proprioceptive afferents and V1R interneurons in an isolated spinal cord-hindlimb preparation (Figures 6D and S7E). In p4-p5 En1::Cre; Rosa.lsl.tdT mice, ventrally-positioned tdTomato+ neurons were identified as V1R interneurons by virtue of their short-latency monosynaptic responses after antidromic stimulation of L4 or L5 ventral roots (Figure 6E). Individual muscle nerves were stimulated to activate defined classes of proprioceptive afferents. We found that 6 of 11 V1R interneurons responded to stimulation of the GL muscle nerve with EPSP latencies similar to those recorded from homonymous motor neurons (8.1 ± 0.3 ms and 8.4 ± 0.4 ms in V1R and motor neurons, respectively; Figure 6F,G). Moreover, repetitive stimulation of GL afferents at 0.1 or 1 Hz elicited EPSPs of low onset jitter in V1R interneurons (CVonset < 0.05, Figures 6H,I and S7F), an indication of the monosynaptic nature of sensory input (Vrieseling and Arber, 2006).
TA and IF stimulation, in contrast, evoked long-latency input with variable onset to the vast majority of V1R interneurons, with only one TA neuron exhibiting a short latency, low jitter response (Figures 6F,H-I and S7F-H). Thus, physiological studies indicate selective input to V1R interneurons from proprioceptors innervating proximal (GL) muscles. Moreover, the strength of connections between GL proprioceptors and V1R interneurons indicates the existence of a selective group Ia-mediated feed-forward disynaptic inhibitory influence on motor output.
We next compared the pattern of proprioceptive sensory connections with V1Sp8 interneurons. Analysis of ~p20 En1::Cre; Sp8::FlpoERT2; RCE.dual.GFP mice revealed that ~15% of V1Sp8 interneurons (7/44 cells) received CTB+; vGluT1+ contacts from GL afferents. TA sensory afferents formed synaptic contacts with ~40% of V1Sp8 interneurons (23/58 cells) (Figure 6K,L; fraction of cells innervated by TA vs GL: p = 0.004 by Fisher's exact test). In addition, TA afferents established a >2-fold higher innervation density than GL afferents onto the proximal dendrites of V1Sp8 interneurons. This increased incidence of TA connectivity suggests that dorsally-positioned V1Sp8 interneurons receive a broader spectrum of proprioceptor input that ventrally-positioned V1R interneurons. In contrast, IF sensory afferents failed to contact V1Sp8 interneurons (Figure 6K,L), an indication that proximity alone is not sufficient to explain all aspects of proprioceptive input to V1 interneurons. Despite the lack of IF sensory contacts, these findings support the idea that positional constraints imposed by the D/V matching of interneuron settling position and sensory afferent arborization domain explain many features of the pattern of selectivity of sensory input to V1R and V1Sp8 interneurons.
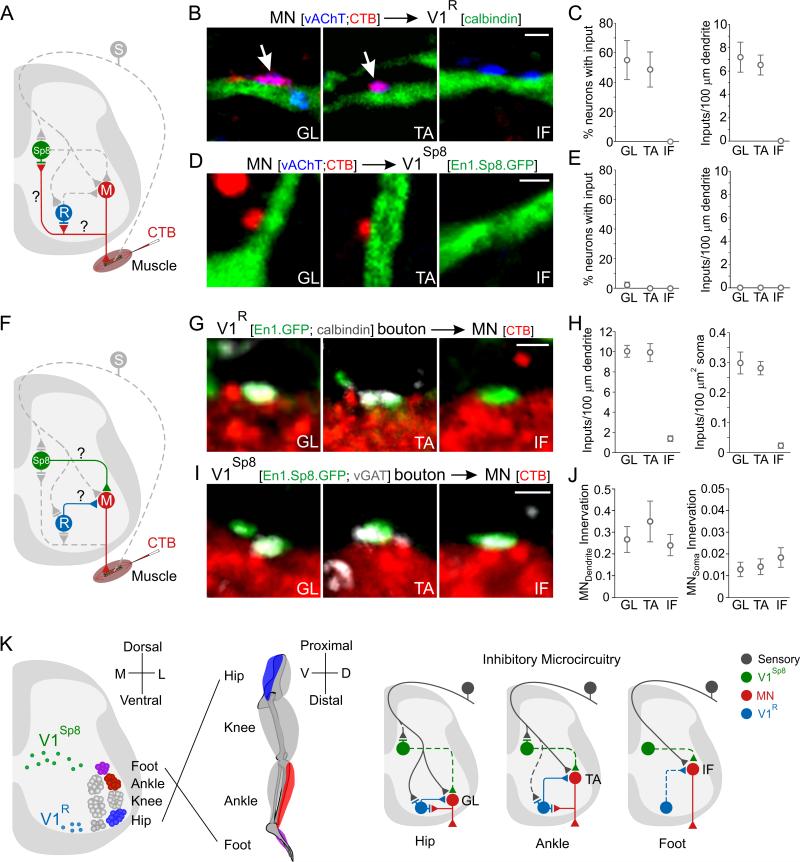
Positional Constraints on V1 Interneuron-Motor Neuron Connections
We next considered whether the settling position of V1R and V1Sp8 interneurons predicts their interconnectivity with motor neurons, analyzing first the extent to which V1R and V1Sp8 interneurons receive motor neuron collateral input (Figure 7A). Analysis of the location of CTB+; vAChT+ motor axon collateral terminals indicated that GL and TA, but not IF motor axons form dense collateral arbors that are confined to a ventral domain overlapping with V1R interneurons, and ventral to V1Sp8 interneurons (Figure S7D), in agreement with studies performed in cat (McCurdy and Hamm, 1992). By implication, V1R but not V1Sp8 interneurons are positioned to receive synaptic input from GL and TA, but not IF, motor neurons.
Figure 7. Specificity of Interneuron-Motor Neuron Interconnectivity at Individual Joints.
(A) Assay of pool-specific motor input to interneurons.
(B-C) V1R interneurons receive CTB+; vAChT+ input from GL and TA (arrows) but not IF motor neurons (MN). Scale bar = 2 μm. (C) Left, V1R interneurons with input from GL, TA, or IF MNs. p = 0.02, one-way ANOVA; Bonferroni post-hoc test: p < 0.05, GL or TA vs IF. Right, CTB+ MN inputs/100 μm of V1R dendrite length. p = 0.002, one-way ANOVA; Bonferroni post-hoc test: p < 0.01, GL or TA vs IF; p > 0.5, GL vs TA, n ≥ 3 animals, and 23 (GL), 24 (TA), or 15 (IF) cells.
(D-E) Absence of MN input to V1Sp8 interneurons. GL, n = 4 animals, 43 cells; TA, n = 2 animals, 52 cells; IF, n = 3 animals, 43 cells.
(F) Assay of interneuron input onto motor pools.
(G-H) V1R interneurons preferentially innervate GL and TA relative to IF motor pools, on proximal MN dendrites (H, left) or soma (H, right). p < 0.0001, one-way ANOVA; Bonferroni post hoc test: p < 0.001, GL or TA vs IF, n = 4 animals, and 31 (GL), 21 (TA), or 27 (IF) cells.
(I-J) V1Sp8 interneurons sparsely and uniformly innervate motor pools acting on different joints. Number of V1Sp8 inputs/100 μm MN dendrite or 100 μm2 of soma area, normalized to V1Sp8 interneuron number. p = 0.53 or 0.65 for dendrites and soma, respectively, one-way ANOVA, n ≥ 3 animals, 35 (GL), 42 (TA), or 59 (IF) cells. Scale bars = 2 μm. All data are mean ± SEM.
(K) V1R and V1Sp8 microcircuits operating on hip, ankle, and foot motor neurons. Solid and dotted lines represent prevalent and sparse synaptic connectivity. See also Figure S7.
To assess motor neuron input to V1R or V1Sp8 interneurons, CTB was injected into individual muscles and motor axon collateral labeling analyzed in p21 En1::Cre; RCE.lsl.GFP or En1::Cre; Sp8::FlpoERT2; RCE.dual.GFP tamoxifen-treated mice. The density of CTB-labeled, vAChT+ boutons was determined on the soma and proximal dendrites of V1R and V1Sp8 interneurons (Figure 7B-E). Both GL and TA motor neurons provide synaptic input to V1R interneurons, with each innervating about half of all V1R interneurons at comparable CTB+; vAChT+ bouton densities (Figure 7C). Nevertheless, V1R interneurons did not receive collateral input from IF motor neurons (Figure 7C). V1Sp8 interneurons received no motor axon collateral contacts (Figure 7D,E), indicating that motor neuron collateral innervation is restricted to V1R interneurons, presumably a function of their ventral position. Thus, the settling position of V1R and V1Sp8 interneurons predicts the status of motor as well as sensory neuron input.
We next examined whether subsets of V1 interneurons provide differential input to motor pools in accord with their settling position, focusing on the connections of V1R or V1Sp8 interneurons with GL, TA, and IF motor neurons (Figure 7F). V1-derived, calbindin+ terminals from V1R interneurons contacted GL and TA motor neurons at similar densities in ~p21 En1::Cre; Rosa.lsl.tdT or En1::Cre; RCE.lsl.GFP mice (Figure 7G,H; p = 0.94 or 0.84 for dendrites and soma, respectively, by two-tailed Student's t-test). Thus, ventrally located V1R interneurons target motor pools at different D/V locations with similar efficacy. In contrast, IF motor neurons received only sparse input from V1R neurons (Figure 7G,H), despite the similar D/V settling position of TA and IF motor pools (Figure 5C). The incidence of V1R innervation was ~7-fold greater on TA proximal dendrites and ~14-fold greater on TA soma, by comparison with IF motor neurons (Figure 7H). Thus the ventral position of V1R interneurons does not appear to constrain their ability to innervate motor neurons at different D/V positions, with the exception of a reduced innervation of the IF pool.
We also explored whether the dorsal settling position of V1Sp8 interneurons limits their connectivity with motor neurons. In En1::Cre; Sp8::FlpoERT2; RCE.dual.GFP mice, V1Sp8 interneurons provided sparse and uniform contacts with GL, TA, and IF motor neurons (Figure 7I,J). The innervation density of V1Sp8 inputs was <20% that of V1R inputs onto GL or TA motor neurons - although this value is likely to underestimate the actual incidence of motor neuron innervation, because only ~30% of V1Sp8 interneurons are labeled after tamoxifen exposure (Figure S6E). These observations indicate that the dorsal position of V1Sp8 interneurons does not limit their ability to innervate motor pools. Together, these data provide evidence that in spinal motor circuits the settling position of inhibitory interneurons is a determinant of input but not output connectivity.
DISCUSSION
Spinal interneurons shape motor activity and limb movement, but the organizational logic of their encoded microcircuits has remained obscure. Through a focus on V1 interneurons we identified nineteen transcription factors whose combinatorial expression documents extensive diversity within this inhibitory set. Different V1 subsets have distinct physiological characteristics and occupy stereotypic clustered positions in the ventral spinal cord, establishing a spatial plan with predictive relevance for inhibitory microcircuit organization. Indeed, variant V1 microcircuits are used to control motor pools that innervate muscles at different limb joints, documenting the absence of a fixed circuit architecture for interneurons that control limb movement (Figure 7K).
Interneuron Diversity and its Implications for Motor Control
V1 interneurons comprise a highly diverse set of transcriptionally distinct neuronal types, posing questions about the purpose of such heterogeneity. We have found that the transcriptional diversity of V1 clades matches physiologically distinct excitable properties. Diversity may also reflect the demand that interneurons receive varied inputs from numerous sources. The activity of motor neurons is regulated by over a dozen supraspinal neuronal systems (Lemon, 2008), many of which engage only a restricted set of all possible motor pools: thus rubrospinal input is restricted to motor pools controlling distal muscles, and vestibulospinal input to motor pools innervating extensor muscles (Grillner and Hongo, 1972; McCurdy et al., 1987). These descending systems presumably engage interneurons with a selectivity that matches the specificity of motor neuron recruitment. Distinct subsets of V1 interneurons may therefore be recruited by different descending systems so as to link sensory input with intermediary descending control pathways. The high degree of V1 transcriptional diversity could provide a means of establishing distinctions in settling position or molecular recognition cues that facilitate the integration of multiple input systems and output modules.
The heterogeneity exhibited by V1 interneurons is likely to extend to other spinal interneuron populations. Small subsets of spinal V0 interneurons have been delineated on the basis of selective profiles of transcription factor expression, best exemplified by a compact cluster of Pitx2+ V0c interneurons that represent the source of cholinergic C-bouton inputs to motor neurons (Zagoraiou et al., 2009). Moreover, many of the transcription factors that delineate V1 subsets are expressed by small subsets of inhibitory V2b and excitatory V2a interneurons, raising the possibility that conserved elements of input and output wiring specificity are encoded by a common set of transcription factors within different excitatory and inhibitory interneuron sets. If the extent of diversity of V1 interneurons extends to each cardinal (V0, V2a/b, and V3) interneuron population (Francius et al., 2013), the fidelity of motor output could depend on the coordinated activity of > 200 subsets of ventral interneurons.
It remains unclear whether the diversity evident in V1 interneurons has predictive relevance for other CNS circuits. The spinal motor system could require a greater degree of interneuron diversification than the brain, because of the last-order and non-redundant nature of motor neuron output and the behavioral imperative to confer precise patterns of muscle activation. Nevertheless, the predictive view may be nearer the mark. Single-cell transcriptional profiling from interneurons in primary somatosensory cortex and CA1 hippocampus have revealed at least sixteen different subsets, with the potential for yet greater diversity (Zeisel et al., 2015). In addition, many of the transcription factors that delineate subsets of V1 interneurons are expressed by subsets of cortical interneurons (Tasic et al., 2016). Thus, it is likely that principles of spinal interneuron heterogeneity and function have relevance for circuit organization and function in the brain.
Position as a Determinant in the Organization of Inhibitory Microcircuits
The relevance of neuronal settling position in spinal connectivity has emerged from studies on the synaptic organization of sensory connections with motor neurons. Proprioceptive afferents target distinct dorsoventral domains of the ventral spinal cord in a manner independent of motor neuron character (Sürmeli et al., 2011), and thus the stereotypy of settling position is needed for the formation of selective sensory connections. Similarly, V1R interneurons receive input from ventrally projecting hip afferents, whereas dorsal V1Sp8 interneurons receive input both from dorsally-directed ankle afferents as well as from hip afferents. Thus, V1 positional stereotypy has implications for motor microcircuit organization in the realm of input selectivity.
The finding that V1R interneurons receive selective input from hip muscle afferents sheds light on a long-standing uncertainty about the status of sensory input to V1R interneurons. Classical studies in cat focused on sensory feedback from knee and ankle muscles, and argued for the absence of functional monosynaptic sensory connectivity with V1R interneurons (Ryall and Piercey, 1971). Later studies in rodent spinal cord, however, provided physiological evidence for direct sensory input to V1R interneurons during early postnatal development (Mentis et al., 2006). These divergent conclusions can be reconciled through an appreciation of the dominance of proprioceptive input from hip afferents, an afferent source not examined in cat. Nevertheless, the extent to which this circuit functions at later developmental stages is unclear because the strength of sensory inputs to V1R interneurons decreases in the adult (Mentis et al., 2006).
The density of hip afferent inputs to V1R interneurons presumably forms a disynaptic feedforward inhibitory pathway to motor neurons, in addition to the role of V1R interneurons in recurrent inhibition. Sensory-evoked feedforward inhibition could modulate the temporal features and dynamic range of excitatory responses of hip motor neurons, as with inhibitory interneurons in hippocampal and cortical circuits (Pouille et al., 2009). An inhibitory signal dependent on hip position could also modulate flexion/extension transitions during the step cycle (McVea et al., 2005) and/or reflex actions at the ankle joint (Knikou and Rymer, 2002).
The link between interneuron settling position and microcircuit wiring is so far largely correlative. Nevertheless our data, combined with previous findings on the relevance of motor neuron positioning (Sürmeli et al., 2011), supports the view that the precision of interneuron location constrains circuit wiring. The role of neuronal settling position in organizing interneuron circuits appears restricted to input connectivity. V1 interneuron position is not predictive of motor pool target connections, reminiscent of observations that motor neuron settling position is not required for the innervation of specific limb muscles (Demireva et al., 2011).
Positional constraints are likely to act in conjunction with molecular recognition systems in defining final connectivity profiles. Precedent for such recognition systems has emerged from analysis of repellant sema3e-plexinD1 signaling in sensory-motor connectivity (Fukuhara et al., 2013; Pecho-Vrieseling et al., 2009). The existence of repellent cues could explain how the dorsal termination zone of IF sensory afferents is not associated with direct synaptic contact with V1Sp8 interneurons, despite the proximity of presynaptic axons and V1Sp8 dendrites. In addition, the extent of interneuron dendritic arborization could relieve constraints on input connectivity imposed by somatic clustering. Although the dendritic arbors of V1R interneurons are largely confined to the ventral spinal cord (Lagerback and Kellerth, 1985), V1Sp8 interneurons exhibit larger dendritic arbors (J. Bikoff, unpublished observation), potentially expanding synaptic input. Thus it seems unlikely that position alone directs input connectivity with V1 subsets.
Variant Microcircuits for Joints and Muscles
The variant circuit architectures exhibited by V1 interneurons may be a general feature of spinal motor microcircuits. The anatomical and physiological characterization of the circuitry of V1R interneurons is consistent with physiological descriptions in cat of a reduced degree of recurrent inhibition for motor neurons that innervate distal compared to proximal limb musculature (Illert and Wietelmann, 1989; McCurdy and Hamm, 1992). Thus, recurrent inhibition is not implemented uniformly across motor pools. Other classically-defined inhibitory motifs also exhibit variant circuit architecture: Group Ib feedforward inhibition is strongly activated by extensor afferents, but only weakly by flexor afferents (Eccles et al., 1957). Moreover, reciprocal inhibition is prominent for flexor/extensor antagonist pairs, but absent for abductor/adductor antagonists (Eccles and Lundberg, 1958). Local motor microcircuits are therefore differentially tailored to the workings of individual muscles.
Finally, our studies raise the issue of the functional consequences of variation in V1 interneuron microcircuitry. Evolutionary adaptation for dexterous hand movements has fine-tuned the ability of distal limb muscles to control digits independently. Yet recurrent inhibition functions broadly to silence motor pools (Eccles et al., 1961), suggesting that the absence of recurrent inhibition in distal musculature contributes to the independence of neural control for individual digit muscles. From an evolutionary perspective, the diversity exhibited by mammalian V1 interneuron circuits may be needed to accommodate the complex control features of multi-joint limbs.
EXPERIMENTAL PROCEDURES
Mouse Genetics
Mouse lines are described in Supplemental Experimental Procedures. All experiments and procedures were performed according to NIH guidelines and approved by the Institutional Animal Care and Use Committee of Columbia University.
Microarray Screen
Fluorescence activated cell sorting, RNA processing, and microarray analyses are described in Supplemental Experimental Procedures. Microarray data are available at the NCBI Gene Expression Omnibus, under accession number GSE69560.
Immunohistochemistry
Immunohistochemistry was performed as described (Betley et al., 2009). Confocal images of cryostat or vibratome-sectioned tissue were obtained on a LSM 710 Meta Confocal microscope (Carl Zeiss). Antibodies, immunohistochemical methods, and detection of transcription factor co-expression are documented in Supplemental Experimental Procedures.
Analysis of Interneuron Spatial Distributions
V1 interneuron positional analysis is described in Supplemental Experimental Procedures.
Retrograde Labeling of Motor and Sensory Neurons
Motor and sensory neurons were retrogradely labeled in vivo at p14-p16 by intramuscular injection of 1% unconjugated cholera toxin B subunit (CTB, List Biologicals). Animals were analyzed after 5-7 days (Sürmeli et al., 2011). Details are described in Supplemental Experimental Procedures.
Synaptic Quantification
Synaptic quantification is presented in Supplemental Experimental Procedures.
Electrophysiology
Recordings from intact spinal cord-hindlimb preparations were performed as described (Mentis et al., 2005), with connectivity to GL, TA, and IF peripheral muscles preserved. Recordings from V1 subsets were obtained in 300 μm thick slices as described (Gonzalez-Forero and Alvarez, 2005). Details are provided in Supplemental Experimental Procedures.
Statistics
Significance between means was assessed with an unpaired two-tailed Student's t-test, or a one-way ANOVA and Bonferroni post-hoc test. Fisher's exact test was used to compare the proportion of neurons receiving synaptic input from different muscles. All data are represented as mean ± SEM unless noted. Statistical methods for spatial distributions are presented in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to I. Schieren for help with FACS, B. Han and M. Mendelsohn for help generating transgenic mice, and K. MacArthur for manuscript preparation. We thank A. Joyner for advice on generating FlpoERT2, G. Fishell for RCE.dual.GFP mice, and K. Campbell for Sp8fl/fl mice. J. Nathans provided anti-Nr3b2, and S. Ross anti-Bhlhb5 and anti-Prdm8 antibodies. We thank R. Axel, E. Azim, N. Balaskas, R. Brownstone, and J. de Nooij for discussions and comments on the manuscript. F.J.A. was supported by NIH-NINDS (R01-NS047357.) G.Z.M. was supported by the NIH (R01-NS078375 and R21-NS079981) and the Department of Defense (GR.10235006). T.M.J was supported by NIH grant NS033245, the Brain Research Foundation, the Harold and Leila Y. Mathers Foundation and Project A.L.S., and is an HHMI investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.B.B. and T.M.J. devised the project. M.I.G., A.F.R., and E.D. contributed equally to this work. J.B.B. performed molecular, genetic, and anatomical experiments, with assistance from E.F. and C.D. M.I.G. contributed to the statistical analysis of V1 diversity. A.F.R. and F.J.A. characterized the firing features of V1 subsets. E.D. and G.Z.M. performed electrophysiological experiments in the spinal cord-hindlimb preparation. T.A.M. and A.M. helped analyze V1 spatial distributions, and S.B.M. developed antibodies. J.B.B. and T.M.J. wrote the manuscript, with input from all authors.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and one table, and can be found with this article online at:
REFERENCES
- Alvarez FJ, Fyffe REW. The continuing case for the Renshaw cell. J. Physiol. 2007;584:31–45. doi: 10.1113/jphysiol.2007.136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber S. Motor circuits in action: specification, connectivity, and function. Neuron. 2012;74:975–989. doi: 10.1016/j.neuron.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Baldissera F, Hultborn H, Illert M. Brooks VB, editor. Integration in spinal neuronal systems. Handbook of Physiology, The Nervous System. 1981:509–595. [Google Scholar]
- Benito-Gonzalez A, Alvarez FJ. Renshaw cells and Ia inhibitory interneurons are generated at different times from p1 progenitors and differentiate shortly after exiting the cell cycle. J. Neurosci. 2012;32:1156–1170. doi: 10.1523/JNEUROSCI.3630-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Wright CV, Kawaguchi Y, Erdelyi F, Szabo G, Jessell TM, Kaltschmidt JA. Stringent specificity in the construction of a GABAergic presynaptic inhibitory circuit. Cell. 2009;139:161–174. doi: 10.1016/j.cell.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Brown AG. Organization in the Spinal Cord: The Anatomy and Physiology of Identified Neurones. Springer-Verlag; New York: 1981. [Google Scholar]
- Dalla Torre di Sanguinetto SA, Dasen JS, Arber S. Transcriptional mechanisms controlling motor neuron diversity and connectivity. Curr. Opin. Neurobiol. 2008;18:36–43. doi: 10.1016/j.conb.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Demireva EY, Shapiro LS, Jessell TM, Zampieri N. Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell. 2011;147:641–652. doi: 10.1016/j.cell.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles JC, Eccles RM, Iggo A, Ito M. Distribution of recurrent inhibition among motoneurones. J. Physiol. 1961;159:479–499. doi: 10.1113/jphysiol.1961.sp006822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles JC, Eccles RM, Lundberg A. Synaptic actions on motoneurones caused by impulses in Golgi tendon organ afferents. J. Physiol. 1957;138:227–252. doi: 10.1113/jphysiol.1957.sp005849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles RM, Lundberg A. Integrative pattern of Ia synaptic actions on motoneurones of hip and knee muscles. J. Physiol. 1958;144:271–298. doi: 10.1113/jphysiol.1958.sp006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francius C, Harris A, Rucchin V, Hendricks TJ, Stam FJ, Barber M, Kurek D, Grosveld FG, Pierani A, Goulding M, et al. Identification of multiple subsets of ventral interneurons and differential distribution along the rostrocaudal axis of the developing spinal cord. PLoS ONE. 2013;8:e70325. doi: 10.1371/journal.pone.0070325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara K, Imai F, Ladle DR, Katayama K, Leslie JR, Arber S, Jessell TM, Yoshida Y. Specificity of monosynaptic sensory-motor connections imposed by repellent Sema3E-PlexinD1 signaling. Cell Rep. 2013;5:748–758. doi: 10.1016/j.celrep.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyffe RE. Evidence for separate morphological classes of Renshaw cells in the cat's spinal cord. Brain Res. 1990;536:301–304. doi: 10.1016/0006-8993(90)90038-d. [DOI] [PubMed] [Google Scholar]
- Gabitto MI, Pakman A, Bikoff JB, Abbott LF, Jessell TM, Paninski L. Bayesian sparse regression analysis documents the diversity of spinal inhibitory interneurons. Cell. 2016 doi: 10.1016/j.cell.2016.01.026. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Forero D, Alvarez FJ. Differential postnatal maturation of GABAA, glycine receptor, and mixed synaptic currents in Renshaw cells and ventral spinal interneurons. J. Neurosci. 2005;25:2010–2023. doi: 10.1523/JNEUROSCI.2383-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosgnach S, Lanuza GM, Butt SJ, Saueressig H, Zhang Y, Velasquez T, Riethmacher D, Callaway EM, Kiehn O, Goulding M. V1 spinal neurons regulate the speed of vertebrate locomotor outputs. Nature. 2006;440:215–219. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- Goulding M, Bourane S, Garcia-Campmany L, Dalet A, Koch S. Inhibition downunder: an update from the spinal cord. Curr. Opin. Neurobiol. 2014;26:161–166. doi: 10.1016/j.conb.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner S, Hongo T. Vestibulospinal effects on motoneurones and interneurones in the lumbosacral cord. Prog. Brain Res. 1972;37:243–262. doi: 10.1016/S0079-6123(08)63906-0. [DOI] [PubMed] [Google Scholar]
- Hultborn H, Jankowska E, Lindstrom S. Recurrent inhibition of interneurones monosynaptically activated from group Ia afferents. J. Physiol. 1971;215:613–636. doi: 10.1113/jphysiol.1971.sp009488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illert M, Wietelmann D. Distribution of recurrent inhibition in the cat forelimb. Prog. Brain Res. 1989;80:273–281. doi: 10.1016/s0079-6123(08)62221-9. [DOI] [PubMed] [Google Scholar]
- Jankowska E. Interneuronal relay in spinal pathways from proprioceptors. Prog. Neurobiol. 1992;38:335–378. doi: 10.1016/0301-0082(92)90024-9. [DOI] [PubMed] [Google Scholar]
- Knikou M, Rymer Z. Effects of changes in hip joint angle on H-reflex excitability in humans. Exp. Brain Res. 2002;143:149–159. doi: 10.1007/s00221-001-0978-4. [DOI] [PubMed] [Google Scholar]
- Lagerback PA, Kellerth JO. Light microscopic observations on cat Renshaw cells after intracellular staining with horseradish peroxidase. II. The cell bodies and dendrites. J. Comp. Neurol. 1985;240:368–376. doi: 10.1002/cne.902400405. [DOI] [PubMed] [Google Scholar]
- Lemon RN. Descending pathways in motor control. Annu. Rev. Neurosci. 2008;31:195–218. doi: 10.1146/annurev.neuro.31.060407.125547. [DOI] [PubMed] [Google Scholar]
- Matise MP, Joyner AL. Expression patterns of developmental control genes in normal and Engrailed-1 mutant mouse spinal cord reveal early diversity in developing interneurons. J. Neurosci. 1997;17:7805–7816. doi: 10.1523/JNEUROSCI.17-20-07805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy ML, Hansma DI, Houk JC, Gibson AR. Selective projections from the cat red nucleus to digit motor neurons. J. Comp. Neurol. 1987;265:367–379. doi: 10.1002/cne.902650306. [DOI] [PubMed] [Google Scholar]
- McCurdy ML, Hamm TM. Recurrent collaterals of motoneurons projecting to distal muscles in the cat hindlimb. J. Neurophysiol. 1992;67:1359–1366. doi: 10.1152/jn.1992.67.5.1359. [DOI] [PubMed] [Google Scholar]
- McHanwell S, Biscoe TJ. The localization of motoneurons supplying the hindlimb muscles of the mouse. Philos. Trans. R. Soc. of Lond. B, Biol. Sci. 1981;293:477–508. doi: 10.1098/rstb.1981.0082. [DOI] [PubMed] [Google Scholar]
- McVea DA, Donelan JM, Tachibana A, Pearson KG. A role for hip position in initiating the swing-to-stance transition in walking cats. J. Neurophysiol. 2005;94:3497–3508. doi: 10.1152/jn.00511.2005. [DOI] [PubMed] [Google Scholar]
- Mentis GZ, Alvarez FJ, Bonnot A, Richards DS, Gonzalez-Forero D, Zerda R, Donovan MJ. Noncholinergic excitatory actions of motoneurons in the neonatal mammalian spinal cord. Proc. Natl. Acad. Sci. USA. 2005;102:7344–7349. doi: 10.1073/pnas.0502788102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentis GZ, Siembab VC, Zerda R, O'Donovan MJ, Alvarez FJ. Primary afferent synapses on developing and adult Renshaw cells. J. Neurosci. 2006;26:13297–13310. doi: 10.1523/JNEUROSCI.2945-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecho-Vrieseling E, Sigrist M, Yoshida Y, Jessell TM, Arber S. Specificity of sensory-motor connections encoded by Sema3e-Plxnd1 recognition. Nature. 2009;459:842–846. doi: 10.1038/nature08000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille F, Marin-Burgin A, Adesnik H, Atallah BV, Scanziani M. Input normalization by global feedforward inhibition expands cortical dynamic range. Nat. Neurosci. 2009;12:1577–1585. doi: 10.1038/nn.2441. [DOI] [PubMed] [Google Scholar]
- Ryall RW, Piercey MF. Excitation and inhibition of Renshaw cells by impulses in peripheral afferent nerve fibers. J. Neurophysiol. 1971;34:242–251. doi: 10.1152/jn.1971.34.2.242. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The Types of Retinal Ganglion Cells: Current Status and Implications for Neuronal Classification. Annu. Rev. Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Sapir T, Geiman EJ, Wang Z, Velasquez T, Mitsui S, Yoshihara Y, Frank E, Alvarez FJ, Goulding M. Pax6 and Engrailed1 regulate two distinct aspects of Renshaw cell development. J. Neurosci. 2004;24:1255–1264. doi: 10.1523/JNEUROSCI.3187-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saueressig H, Burrill J, Goulding M. Engrailed-1 and Netrin-1 regulate axon pathfinding by association interneurons that project to motor neurons. Development. 1999;126:4201–4212. doi: 10.1242/dev.126.19.4201. [DOI] [PubMed] [Google Scholar]
- Stam FJ, Hendricks TJ, Zhang J, Geiman EJ, Francius C, Labosky PA, Clotman F, Goulding M. Renshaw cell interneuron specialization is controlled by a temporally restricted transcription factor program. Development. 2012;139:179–190. doi: 10.1242/dev.071134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sürmeli G, Akay T, Ippolito GC, Tucker PW, Jessell TM. Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template. Cell. 2011;147:653–665. doi: 10.1016/j.cell.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016 doi: 10.1038/nn.4216. doi:10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieseling E, Arber S. Target-induced transcriptional control of dendritic patterning and connectivity in motor neurons by the ETS gene Pea3. Cell. 2006;127:1439–1452. doi: 10.1016/j.cell.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Windhorst U. Muscle proprioceptive feedback and spinal networks. Brain Res. Bull. 2007;73:155–202. doi: 10.1016/j.brainresbull.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Zagoraiou L, Akay T, Martin JF, Brownstone RM, Jessell TM, Miles GB. A cluster of cholinergic premotor interneurons modulates mouse locomotor activity. Neuron. 2009;64:645–662. doi: 10.1016/j.neuron.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lupton R, Lanuza GM, Britz O, Wang Z, Siembab VC, Velasquez T, Alvarez FJ, Frank E, Goulding M. V1 and V2b interneurons secure the alternating flexor-extensor motor activity mice require for limbed locomotion. Neuron. 2014;82:138–150. doi: 10.1016/j.neuron.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.