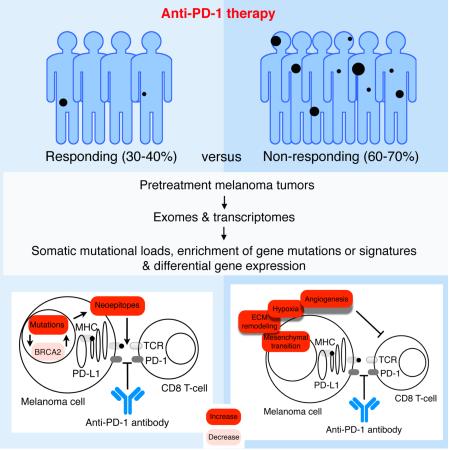
SUMMARY
PD-1 immune checkpoint blockade provides significant clinical benefits for melanoma patients. We analyzed the somatic mutanomes and transcriptomes of pretreatment melanoma biopsies to identify factors that may influence innate sensitivity or resistance to anti-PD-1 therapy. We find that, while overall high mutational loads associate with improved survival both in responding and non-responding patients, responding tumors are specifically enriched for mutations in the DNA repair gene BRCA2. Innately resistant tumors display a transcriptional signature (referred to as the IPRES or Innate anti-PD-1 Resistance) indicating concurrent upexpression of genes involved in the regulation of mesenchymal transition, cell adhesion, ECM remodeling, angiogenesis and wound-healing. Notably, MAPK-targeted therapy (MAPKi) induces similar signatures in melanoma, suggesting that a non-genomic form of MAPKi resistance mediates cross-resistance to anti-PD-1 therapy. Validation of the IPRES in other independent tumor cohorts defines a transcriptomic subset across distinct types of advanced cancer. These findings suggest that attenuating the biological processes that underlie IPRES may improve anti-PD1 response in melanoma and other cancer types.
Graphical Abstract
INTRODUCTION
PD-1 immune checkpoint blockade therapy induces a high rate of anti-melanoma response and provides unprecedented clinical benefits (Hamid et al., 2013; Topalian et al., 2012). This therapeutic approach has also been shown to be active against a growing list of human malignancies, and clinical testing of combinations of PD-1 (or PD-L1) with other treatment targets has already begun (Sharma and Allison, 2015). However, effective clinical use of anti-PD-1 agents is encumbered by a high rate of innate resistance (60-70%) in advanced metastatic melanoma. The mechanistic basis for the variation in response patterns or long-term clinical benefits (i.e., survival) remains poorly explained.
In melanoma, the extent of pretreatment and especially treatment-induced intra-tumoral T cell infiltration correlates with clinical responses (Tumeh et al., 2014), supporting unleashing of tumor-specific T cells as the primary mechanistic basis of anti-PD-1 therapy. Preliminary retrospective analyses of clinical data hinted at prior failure of MAPK-targeted therapy being a negative factor for subsequent response to immune checkpoint blockade in melanoma (Puzanov et al., 2015; Ramanujam et al., 2015; Simeone et al., 2015). In this context, acquired resistance to MAPK-targeted therapy has been correlated with depletion of intra-tumoral T cells, exhaustion of CD8 T cells and loss of antigen presentation (Hugo et al., 2015).
At the genomic level, the overall mutation load has been correlated with clinical responses to anti-PD-1 therapy and linked to smoking in non-small cell lung cancer or mismatch repair deficiency, in colon cancer. (Le et al., 2015; Rizvi et al., 2015). Whether it was non-small cell lung tumors on anti-PD-1 therapy or melanoma tumors on anti-CTLA-4 therapy (Snyder et al., 2014; Van Allen et al., 2015), the range of somatic mutation and neoepitope loads of the responding pretreatment tumors overlaps significantly with that of the non-responding tumors, despite statistically significant differences in their medians. As such, the lack of high mutational loads did not necessarily preclude clinical responses, and, conversely, the presence of high mutational loads not always correlates with responses. Thus, additional genomic or non-genomic features seem also to contribute to anti-PD-1 response patterns. Here, we sought to assess omic-scale features related to clinical response and survival patterns in order to gain insights into potential strategies for patient stratification and identification of anti-PD-1 combinatorial therapies.
RESULTS AND DISCUSSION
High Mutational Load Does Not Associate with Tumor Response But Correlates with Improved Patient Survival
We analyzed the whole exome sequences (WES) of 38 pretreatment (pembrolizumab, nivolumab) melanoma tumors (responding, n=21; non-responding, n=17; total 34 of 38 pretreatment; 4 of 38 early on-treatment; 14 of 38 patients with prior MAPKi treatment; Table S1A) and patient-matched normal tissues for germline references. Responding pretreatment tumors were derived from patients who went on to have complete or partial responses or stable disease control (with mixed responses excluded) in response to anti-PD-1 therapy. Non-responding tumors were derived from patients who had progressive disease. These response patterns were based on irRECIST (Hoos et al., 2015; Wolchok et al., 2009). We also analyzed the transcriptomes through RNASeq of responding (n=15) and non-responding (n=13) pretreatment tumors (total 27 of 28 pretreatment; 1 of 28 early on-treatment) with available high-quality RNA. WES achieved a median of 140X coverage in both tumor and normal tissues (Table S1B). We detected a median of 489 non-synonymous somatic mutations in the 38 tumors (range 73 to 3,985, which is similar to that in a different set of melanoma tissues (Van Allen et al., 2015)).
We found that responding pretreatment tumors on anti-PD-1 therapy harbored harbors more non-synonymous single nucleotide variants (nsSNVs) compared to the non-responding tumors, albeit the statistical significance cutoff was not met (median nsSNVs responding=495 and non-responding=281, P=0.30, Mann-Whitney; Table S1B). Increased predicted HLA class I and class II neoepitope loads were also detected in the responding pretreatment tumors, although these differences were not statistically significant either (median HLA class I neoepitopes responding=231 and non-responding=156, P=0.41; median HLA class II neoepitopes responding=130 and non-responding=95, P=0.36, Mann-Whitney; Table S1B). Even when we considered only expressed nsSNV and neoepitope loads, the statistical significance of the differences between the responding versus non-responding tumors was not augmented. The comparison of these two groups of tumors was not likely biased by small differences in mean tumor purities or depth of sequencing (Figure S1A and S1B). The numbers of predicted HLA class I and II neoepitopes were strongly correlated with the number of nsSNVs (Figure S1C). We did not identify any recurrent predicted neoepitope or experimentally validated neoantigens (Table S1C). Previous work analyzing melanoma tumors sampled prior to anti-CTLA-4 antibody therapy had associated responses with a tetrapeptide signature (Snyder et al., 2014). However, we did not observed enrichment of this peptide motif in the pretreatment tumors that responded to anti-PD-1 therapy (Figure S1D). Likewise, analysis of an independent cohort of 110 melanoma tumors pre-anti-CTLA-4 therapy also did not yield enrichment of this tetrapeptide motif among responding tumors (Van Allen et al., 2015).
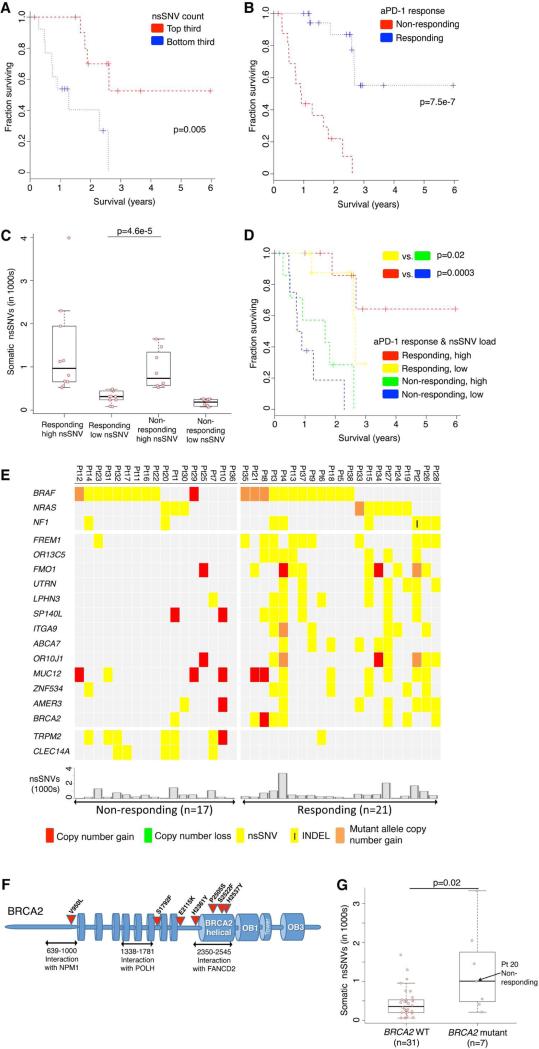
In addition to examining the relationship between non-synonymous somatic mutational loads in pretreatment tumors and anti-tumor responses (and lack thereof) elicited by anti-PD-1 antibodies, we also examined their relative potential influences on clinical benefits of anti-PD-1 immunotherapy as reflected by patient survival. Notably, a mutational load in the top third (compared to the bottom third) was significantly associated with improved survival (Figure 1A). We also observed a trend toward higher mutational load being associated with better survival among melanoma patients not treated with anti-PD-1 antibodies (TCGA, 2015), although this association did not reach statistical significance (Figure S1E), suggesting that the prognostic power of a high mutational load is augmented in the setting of anti-PD-1 therapy. As expected, a positive association between objective tumor responses and survival was highly statistically significant (Figure 1B). However, when we divided each non-responding and responding tumor group into sub-groups with low or high mutational loads (i.e., below or above the median total somatic nsSNVs of each response group) (Figure 1C), patients with responding tumors of low mutation loads significantly outlived patients with non-responding tumors of high mutation loads (Figure 1D). This is despite the fact that mutational loads of these two groups were significantly different, with no overlap across the two distributions (Figure 1C). Hence, factors beyond the mutational load also influence shorter-term tumor response patterns and longer-term patient survival.
Figure 1. Mutational Correlates of Innate Sensitivity to Anti-PD-1 Therapy.
(A) Overall survival of anti-PD-1-treated patients whose melanoma tumors harbored high (top third) versus low (bottom third) mutational (somatic nsSNVs) loads. P values, log-rank test.
(B) Overall survival of anti-PD-1-treated melanoma patients whose pretreatment tumors responded (n=20) or did not respond (n=17). P value, log-rank test.
(C) Total number of nsSNVs detected in anti-PD-1 responding and non-responding melanoma tumors harboring high (above the respective group's median) or low (below the group median) mutational loads. P value, log-rank test.
(D) Overall survival of anti-PD-1-treated melanoma patients whose pretreatment tumors responded or did not respond and harboring high (above the group median) or low (below the group median) mutational loads. P value, log-rank test.
(E) Recurrent exomic alterations (nsSNVs and small insertion/deletions or INDELs) in pretreatment tumors of responding versus non-responding patients on anti-PD-1 therapy. Copy number alterations were annotated for the same gene as a reference. Top, mutations of melanoma signature genes. Middle, mutations recurrent in responding versus non-responding tumors (recurrence in 25% in one group and at most one occurrence in the opposite group, Fisher exact test, FDR-corrected P≤0.05 on enrichment against the background mutation frequency). Bottom, the total nsSNV load of each melanoma tumor.
(F) Schematics of impact of non-synonymous missense and nonsense mutations in the BRCA2 protein and its domains.
(G) Total number of nsSNVs detected in melanomas with or without BRCA2 non-synonymous mutations. P value, Mann Whitney test.
Enrichment for BRCA2 Mutations in Anti-PD-1 Responsive Melanoma
We then sought to identify mutations (nsSNVs and small insertion-and-deletions or INDELs; Table S1D) that (i) were recurrently and selectively associated with either responding or non-responding tumors (recurrence ≥ 25% in one group and at most one hit in the other group) and (ii) occurred in genes at rates higher than background rates (Fisher exact test, FDR-corrected p≤0.05) (Figure 1E; Table S1E). The background mutation rate of each gene was calculated from the WES data of 469 melanoma tumors (Hodis et al., 2012; TCGA, 2015). Analysis of copy number variation (CNVs) did not identify any recurrent alterations exclusive to either group. BRCA2 harbored nsSNVs in six of 21 responding tumors (28%) but only one of 17 non-responding tumors (6%) (Figure 1E). With a background mutational rate estimated at 6% (28 of 469 melanoma tumors), BRCA2 was significantly more frequently mutated in the responding tumors than expected (Fisher P=0.002, odds ratio=6.2). The pattern of mutations in disparate BRCA2 protein domains suggested loss-of-function mutations (Figure 1F): one in the N-terminal NPM1-interacting region; one in the POLH-interacting domain; and four in the helical domain critical for FANCD2 interaction. Intriguingly, the somatic mutational load of the tumors with BRCA2 nsSNVs was significantly higher than those with wild type BRCA2 in this cohort of tumors (Figure 1G) as well as two additional cohorts of melanoma tumors (Figure S1F). Thus, BRCA2 LOF mutations, which are expected to produce defects in homologous recombination and double-stranded DNA break repair (Holloman, 2011), may produce specific mutational signatures or unknown effects (e.g., induction of cell death) which contribute to anti-PD-1 responsiveness.
Co-enriched Transcriptomic Signatures in a Major Subset of Anti-PD-1 Resistant Melanoma
We then addressed whether transcriptomic features would differentiate between responding (n=15) versus non-responding (n=13) tumors sampled prior to anti-PD-1 therapy (total 27 of 28 pretreatment tumors and 1 of 28 early on-treatment). We compared the transcriptomes of the two tumor groups using two approaches: (i) analysis of differentially expressed genes (DEGs) (Figure 2A top and Figure 2B) across the two aggregate groups (Table S2A) coupled with GO term enrichment analysis of DEGs (Figure 2C) and (ii) differential signature enrichment based on single-sample Gene Set Variance Analysis or GSVA scores using publicly available (C2 chemical and genetic perturbation C6 oncogenic, and C7 immunologic subsets of the Molecular Signature Database, Broad Institute) and self-curated (see below), perturbation-induced gene signatures (Table S2B; Figure 2D).
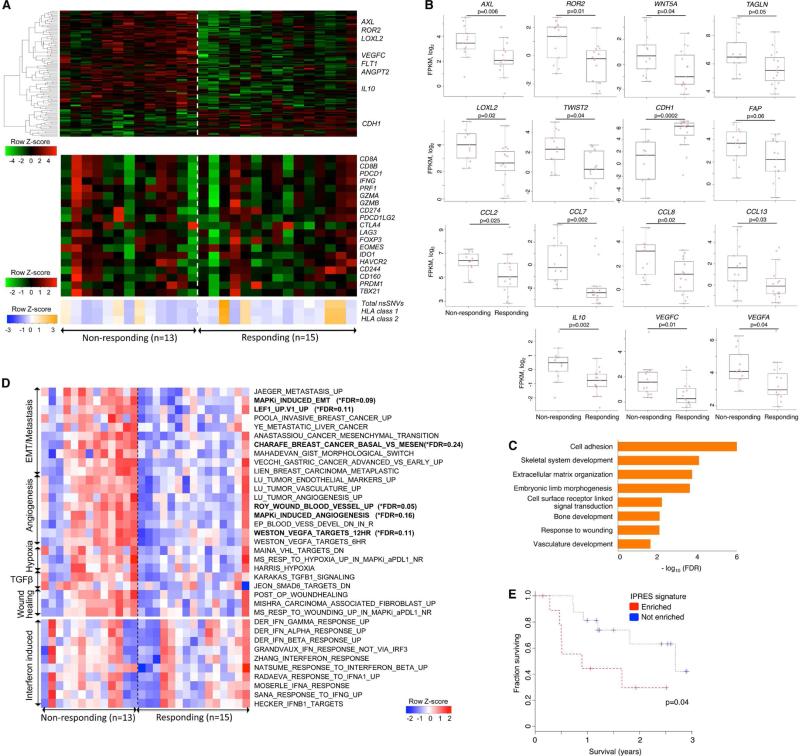
Figure 2. Transcriptomic Signatures of Innate Resistance to Anti-PD-1 Therapy.
(A) (Top) Heatmap showing differentially expressed genes in the pretreatment tumors derived from patients who responded versus who did not respond to anti-PD-1 treatment (gene expression with inter-quartile range (IQR) ≥ 2; median fold-change (FC) difference ≥ 2; Mann-Whitney P ≤ 0.05). (Middle) mRNA expression levels of genes with hypothetical roles in modulating response patterns to anti-PD-1 therapy. (Bottom) Overall number of nsSNVs, HLA class 1 and 2 neoepitopes (predicted).
(B) mRNA levels of genes (which control tumor cell mesenchymal transition, tumor angiogenesis and macrophage and monocyte chemotaxis) that were differentially expressed between the responding versus non-responding pretreatment tumors. P values, Mann Whitney test.
(C) GO enrichment of genes that were expressed higher in the responding tumors.
(D) Heatmap showing the Gene Set Variance Analysis (GSVA) scores of gene signatures differentially enriched in the responding versus non-responding pre-anti-PD-1 tumors (absolute median GSVA score difference ≥ 10%, FDR-corrected Welch t-test p≤0.25 or nominal Welch t-test p≤0.1). For comparison, enrichment scores of interferon signatures are also displayed.
(E) Overall survival of anti-PD-1-treated melanoma patients with presence (n=10) or absence (n=16) of co-enriched Innate Anti-PD-1 RESistance (IPRES) signatures. P value, log-rank test.
From analysis of DEGs (cutoff, two-fold difference between the absolute medians of normalized expressions in the two groups; nominal Mann-Whitney p≤0.1), we made observations suggesting that mesenchymal and inflammatory tumor phenotypes may be associated with innate anti-PD-1 resistance. First, 693 genes were differentially expressed between the responding versus non-responding pretreatment tumors, and the transcriptomes of non-responding tumors were dominated by relative gene up-expression events compared with the transcriptomes of responding tumors (Table S2A; Figure 2A top, showing only genes whose differential expression met nominal Mann-Whitney p≤0.05). Second, DEGs that were expressed higher in non-responding pretreatment tumors included mesenchymal transition genes (AXL, ROR2, WNT5A, LOXL2, TWIST2, TAGLN, FAP), immunosuppressive genes (IL10, VEGFA, VEGFC), and monocyte and macrophage chemotactic genes (CCL2, CCL7, CCL8 and CCL13) (Figure 2A and 2B). In addition to mesenchymal genes, genes associated with wound healing and angiogenesis, which are considered T cell-suppressive (Motz and Coukos, 2011; Schafer and Werner, 2008; Voron et al., 2014), were expressed higher among non-responding relative to responding pretreatment tumors. Interestingly, a recent study using a mouse melanoma model showed that VEGFA and CCL2 expression was associated with innate anti-PD-1 resistance (Peng et al., 2015). CDH1, which is typically down-expressed by mesenchymal cancer cells, was also down-expressed by non-responding (versus responding) pretreatment tumors. Third, genes with putative roles in modulating immune checkpoint sensitivity were not differentially expressed between responding versus non-responding tumor groups (Figure 2A bottom; Figure S2). GZMA, PRF1 (CD8 T cell cytolytic score), PDCD1LG2 (PD-L2) and CTLA4 were expressed higher in the pretreatment melanoma tumors of patients who derived benefit from CTLA-4 antibodies (Van Allen et al., 2015). However, these genes, along with other T cell-related genes such as CD8A/B, PD-L1, LAG3 (T cell checkpoint genes) and IFNG, did not present higher expression in anti-PD-1-responsive tumors (Figure 2A bottom; Figure S2A). Similarly, we did not observe higher enrichment of multiple interferon signatures in the anti-PD-1-responsive group (Figure 2C bottom). Previously, an interferon gamma signature was found to be differentially up-expressed in the pretreatment tumor biopsies from responding patients when a restricted set of immune genes were analyzed (Ribas et al., 2015). However, the technical approach may not be comparable to our whole tumor transcriptomic approach. We did note that the expression levels of HLA class I genes (HLA-A, -B, -C) trended higher among the responding tumors, although the differences were not statistically significant. Lastly, the complete loss of PTEN was reported to promote resistance to immune checkpoint blockade (Peng et al., 2015), but there was only one case of homozygous PTEN deletion (with nearly undetectable PTEN mRNA expression; Figure S2A) in our cohort (in the non-responsive sub-group), limiting our ability to draw meaningful associations in this dataset. Generally, we did not observe a statistically significant difference in PTEN expression between anti-PD-1 responding versus non-responding tumors. Thus, individual gene-based expression analysis suggested mesenchymal and T cell-suppressive inflammatory or angiogenic tumor phenotypes as being associated with innate anti-PD-1 resistance.
We then queried biological processes represented by DEGs. While gene ontology (GO) enrichment analysis of genes up-expressed among responding tumors produced no significantly enriched terms, genes up-expressed among non-responding tumors were enriched for cell adhesion, ECM organization, wound healing and angiogenesis (FDR-adjusted p-values of GO gene sets shown in Figure 2C). Using independently derived perturbation-based transcriptomic signatures (Molecular Signature Database; Table S2C), we tested for differentially enriched processes in the responding versus non-responding pretreatment tumors (cutoff, 10% difference between the absolute medians of GSVA scores in the two groups; FDR-corrected Welch t-test p≤0.25). Gene sets meeting these standard cutoffs formed the core sets (Figure 2D upper, in bold) from which we compiled additional concurrently enriched (nominal Welch t-test p≤0.1) and functionally related gene sets (Figure 2D upper, Table S2B). We considered these statistically weaker gene set enrichments biologically meaningful given the functional coherence of these gene signatures with the core signatures (Subramanian et al., 2005).
Importantly, a group of 26 transcriptomic signatures were co-enriched en bloc in 9 of 13 non-responding versus 1 of 15 responding pre-anti-PD-1 tumors (see Experimental Procedures). Co-enrichment of these signatures, collectively referred to as the Innate anti-PD-1 Resistance or IPRES signature, again indicated heightened mesenchymal transition, angiogenesis, hypoxia and wound healing. The concurrence of a tumor cell mesenchymal phenotype with an angiogenesis- and wound healing-related inflammatory microenvironment has been documented in the literature (Chen et al., 2015a; Chen et al., 2015b; Mak et al., 2015). Interestingly, this set of 26 IPRES signatures included signatures induced by MAPK inhibitor (MAPKi) treatment of melanoma tumors and cell lines (Table S2C). We have shown recently that MAPKi treatment of melanoma cells induces transcriptome-wide re-programming leading to concurrent phenotype switches (Song et al., 2015). Notably, MAPKi-induced signatures of mesenchymal-invasive transition, angiogenesis, and wound healing signatures were detected in the residual melanoma tumors from patients on MAPKi therapy, suggesting that induction of these signatures may negatively impact responsiveness to combinatorial anti-PD-1/L1 therapy.
IPRES (Innate anti-PD-1 Resistance) Signatures Define a Transcriptomic Subset Across Cancers
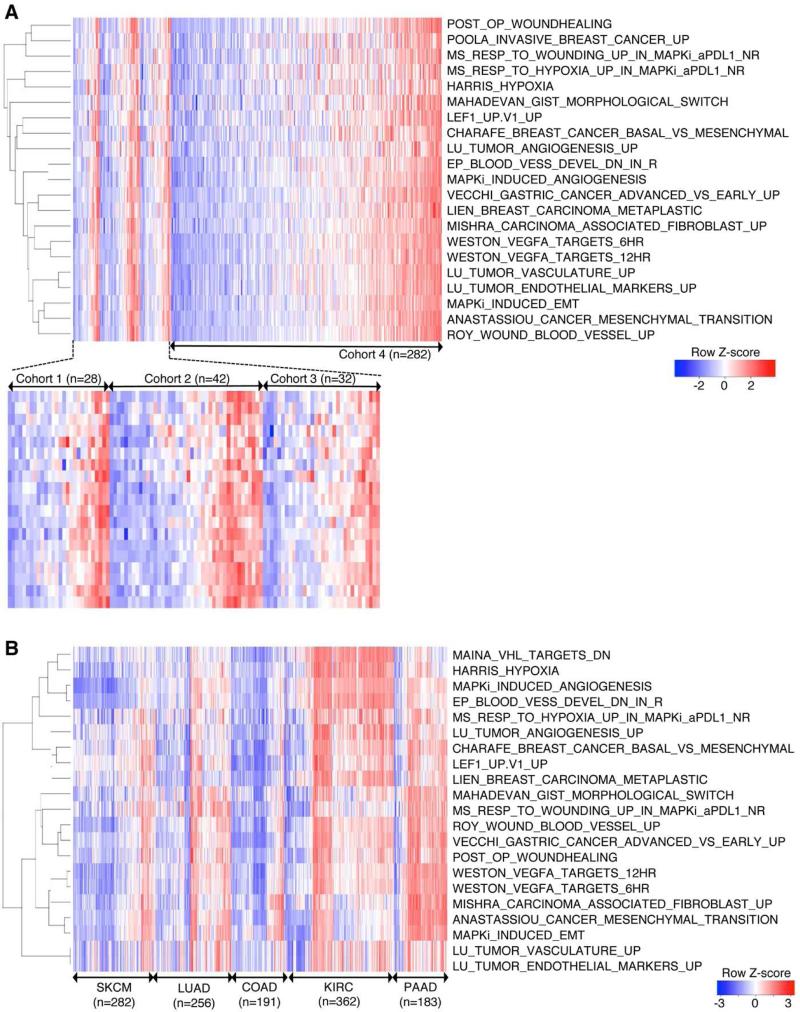
The observations that IPRES content signatures were co-enriched in the same tumors (Figure 2D) and that MAPKi induced these signatures concurrently (Table S2C) implied co-regulated tumor phenotypes that together define a transcriptomic subset. To evaluate whether co-enrichment of IPRES content signatures was an exclusive feature of our cohort, we queried three additional cohorts of metastatic melanoma-derived RNASeq (Hugo et al., 2015; TCGA, 2015; Van Allen et al., 2015), including a cohort consisting of only V600BRAF mutant melanomas (cohort 3) (Hugo et al., 2015). We found that IPRES content signatures co-enriched not only in the same tumors but also in about a third of total samples in each of the four independent transcriptomic data sets (cohort 1 from this study, 10 IPRES-enriched tumors of 28 total tumors; cohort 2, 15 of 42; cohort 3, 11 of 32; cohort 4, 90 of 282) (Figure 3A). Considering 126 among 384 total tumors as the background prevalence for co-enrichment of IPRES content signatures in metastatic melanoma, we determined that this IPRES-enriched transcriptomic subset was over-represented among the anti-PD-1 non-responding pretreatment tumors (Fisher P=0.013, odds ratio=4.6) and under-represented among the responding pretreatment tumors (Fisher P=0.04, odds ratio=0.15) within cohort 1. In contrast, co-enrichment of IPRES signatures was neither over- nor -under-represented among the responding or non-responding pre-anti-CTLA-4 melanoma tumors in cohort 2 (Figure S2B) (Van Allen et al., 2015), which suggests that mechanisms of innate resistance to anti-PD-1 and anti-CTLA-4 are not necessarily similar.
Figure 3. Co-enrichment of Innate Anti-PD-1 Resistance-associated Signatures Defines a Transcriptomic Subset in Melanoma and Multiple Cancers.
(A) Heatmap showing GSVA scores of IPRES signatures across four independent RNASeq data sets derived from metastatic melanoma. Cohort 1, pretreatment (anti-PD-1) tumors; cohort 2, pretreatment (anti-CTLA-4) tumors; cohort 3, pretreatment (MAPKi) tumors; cohort 4, TCGA cutaneous melanoma (metastatic only).
(B) Heatmap showing GSVA scores of IPRIM signatures across TCGA RNASeq data sets (metastatic melanoma or SKCM, lung adenocarcinoma or LUAD, colon adenocarcinoma or COAD, kidney clear cell carcinoma or KIRC, and pancreatic adenocarcinoma or PAAD).
See also Figure S3.
Furthermore, co-enrichment of the IPRES signatures defined a transcriptomic subset within not only melanoma but also all major common human malignancies analyzed (Figure 3B). The IPRES-enriched transcriptomic subset of certain cancers such as pancreatic adenocarcinoma made up the majority of tumors. Within a side-by-side comparison, only six of 69 primary cutaneous melanomas showed co-enrichment of IPRES signatures, in contrast to 90 of 282 metastatic (TCGA) melanomas (P=3.9e-5, odds ratio=0.2) (Figure S3), consistent with mesenchymal transition and metastasis gene sets among IPRES signatures. Thus, co-enrichment of IPRES signatures defines a distinct transcriptomic program that exists across cancers of distinct histology.
This study highlights the utility of both exome and transcriptome sequencing data generated from pretreatment tumor samples for the identification of potential determinants of response to anti-PD-1. Although the overall somatic mutational loads of anti-PD-1-responsive melanoma tumors were not significantly higher than those of non-responsive tumors, higher mutational loads associated significantly with better survival after anti-PD-1 therapy. This finding is still consistent with the notion that neoepitopes derived from somatic non-synonymous mutations are critical for deriving clinical benefits from anti-PD-1 therapy in melanoma. However, objective tumor responses, although strongly associated with survival benefits, did not appear to be driven overwhelmingly by the overall somatic mutational loads. That is to say, a relatively low mutational load did not preclude a tumor response. This is consistent with findings from gastrointestinal cancers where low mutational loads did not preclude tumor infiltration by mutation-reactive, class I and II-restricted T cells (Tran et al., 2015). Thus, overall somatic or predicted neoepitope loads of pretreatment melanoma tumors are not enough to predict response patterns to anti-PD-1 therapy.
In our cohort, responsive tumors were significantly enriched for (likely) loss-of-function mutations in BRCA2. As one would predict from the known function of BRCA2 in DNA repair, BRCA2-mutated melanomas harbored higher mutational loads than BRCA2-wildtype melanomas. Although it is conceivable that defective BRCA2-DNA repair results in specific mutational motifs (as opposed to the general increase in mutational load) that enhance responsiveness, it is also possible that cellular stress resulting from defective DNA repair could lead to increased cell death and anti-tumor immunity. Moreover, these data support the notion that tumor cell phenotypic plasticity (i.e., mesenchymal transition) and the resultant impacts on the microenvironment (e.g., ECM remodeling, cell adhesion, angiogenesis- features of immune suppressive wound healing) are critical barriers to anti-PD-1 responses. The limited number of patients in our melanoma cohort posed certain challenges to our analysis. For example, we relaxed the statistical stringency in single gene-based differential expression analysis (bypassing multiple hypothesis correction) to derive enough genes for gene ontology enrichment analysis. However, converging findings from alternative analysis (i.e., GSVA) of the transcriptome data helped to mitigate potential caveats. Finally, in separate work, we found that mutation-targeted therapy (i.e., MAPKi) induces tumor cell-autonomous changes (e.g., mesenchymal transition) (Song et al., 2015) and upregulates anti-PD-1 resistance-associated processes in residual tumors that have regressed in response to MAPKi treatment. Thus, while our findings in this study necessitate confirmation in independent tissue cohorts, the identification of transcriptomic features associated with anti-PD-1 resistance suggests that mitigation of IPRES-related biological processes may enhance response rates to anti-PD-1 (and anti-PD-1 plus MAPKi) therapy.
EXPERIMENTAL PROCEDURES
Tumor Specimens and Profiling
All tissues in this study were obtained with the approval of Institutional Review Boards and patients’ consents. All patients received either pembrolizumab or nivolumab as the anti-PD-1 therapy for their metastatic melanoma. Thirty-eight melanoma specimens (thirty-two pre-treatment tumors, two pretreatment tumor-derived cultures, three early on-treatment tumors without response, and one early on-treatment tumor with response) and their patient-matched normal tissues were analyzed by whole exome sequencing (WES). Among these thirty-eight samples with WES data, twenty-eight with sufficient RNA quality were also analyzed by RNASeq. This set include another RNASeq dataset derived from a second-site pre-treatment tumor biopsy from patient #27. However, this second-site, pre-treatment tumor-derived WES dataset was excluded in our aggregate mutation analysis to avoid double-counting two tumor exomes from the same patient (Table S1A).
Thirty eight tumor specimens and their respective normal tissues were subjected to whole exome sequencing (WES) (Table S1A). WES was performed using pair-end sequencing with read length of 2×100 bps based on the Illumina HiSeq2000 platform. RNA from a subset of twenty eight tumors were pair-end sequenced with read length of 2 × 100 bps (Illumina HiSeq2000). We included two tumors from Pt27 for transcriptomic analyses but not for mutation and neoepitope analyses since the tumors may not share the same transcriptomic profile but they essentially contain the same set of non-synonymous somatic mutations.
Whole Exome Sequencing
We called single nucleotide variant (SNV) and small insertion-deletion (INDEL) as reported (Shi et al., 2014) using a stand-alone version of Oncotator (Ramos et al., 2015). Copy numbers were called using the intersection of the copy number calls derived from Sequenza (Favero et al., 2015) and VarScan2 (Koboldt et al., 2012). Tumor purities and ploidies (Table S1B) were calculated based on the calls of Sequenza using WES data with default parameters. The impact of BRCA2 nsSNVs was visualized using the domain information in the INTERPRO protein domain database (Mitchell et al., 2015).
HLA Types and Neoepitopes
The 4-digit HLA class 1 and 2 types of each patient were called using ATHLATES (Liu et al., 2013) using the WES sequencing reads from the normal tissue. To ensure concordance, we manually compared ATHLATES’ calls of the normal versus tumor samples and ascertained there was at least no two-digit HLA typing discrepancy between any normal-tumor pair. For each non-synonymous coding mutation from a tumor, we predicted its impact on the patient's HLA class I and II binding using the stand-alone version of the programs NetMHCpan v2.8 (Hoof et al., 2009; Nielsen et al., 2007) and NetMHCIIpan v3.0 (Karosiene et al., 2013), respectively. Specifically, for HLA class I binding prediction using netMHCpan v2.8, we tested all 9-11-mer peptides containing the mutated amino acids for binding to the patient's HLA-A, -B and -C. A peptide was defined as a neoepitope based on two criteria : i) predicted binding affinity ≤ 500nM, and ii) rank percentage ≤ 2% (default cutoff). For HLA class II binding prediction using netMHCIIpan v3.0, we tested the 9-19-mers containing the mutated amino acids for binding to the patient-specific, ATHLATES-predicted DPA-DPB, DQA-DQB and DRA-DRB allele pairs. We also applied the same predicted binding affinity and rank percentage cutoff as we did for HLA class I to nominate the HLA class II-binding neoepitopes. Expressed non-synonymous mutations and neoepitopes were defined based on corresponding genes with normalized expression levels ≥ 1 (in FPKM). Statistical differences of nsSNV, HLA class I and II neoepitopes, WES coverages and tumor purities were computed using two-sided Mann-Whitney test.
Mutation Recurrence
To estimate the statistical significance of the recurrence of gene mutations in the responding or non-responding tumors, we used an independent batch of 469 melanomas’ whole exome sequence datasets (Hodis et al., 2012; TCGA, 2015) to estimate each gene's background mutation frequency. Significance was computed by Fisher exact test followed by FDR adjustment for multiple hypothesis testing. We listed genes that fulfilled the criteria: i) recurrence in at least 25% of the responder/non-responder, ii) occurrence of at most once in the opposite group and iii) Fisher exact test FDR adjusted p-value ≤ 0.05. These genes were illustrated in Figure 1A and all genes that fulfilled i) and ii) and tested for multiple hypotheses were listed in Table S1E. The association between BRCA2 nsSNVs and overall nsSNV counts were tested using two-sided Mann-Whitney test and validated in independent WES datasets (Hodis et al., 2012; TCGA, 2015).
RNASeq and Gene Set Enrichment
Paired-end transcriptome reads were mapped to the UCSC hg19 reference genome using Tophat2 (Kim et al., 2013). Normalized expression levels of genes were expressed in FPKM values as generated by cuffquant and cuffnorm. The program were run with the option “--frag-bias-correct” and “--multi-read-correct” to improve sensitivity (Roberts et al., 2011). A gene was defined as differentially expressed between the responding and non-responding tumor groups when its median expression differed by at least two-fold between the groups with a nominal two-sided Mann-Whitney p-value ≤ 0.1 (Table S2A). Applying multiple hypothesis correction of FDR p ≤ 0.25 only yielded 3 differentially expressed genes; ALDH1L2 and MFAP2 in the non-responding and CDH1 (E-cadherin) in the responding group. As such, the genes meeting the uncorrected, nominal Mann-Whitney p-value ≤ 0.1 that were expressed higher either in the responding or non-responding group were separately analyzed for GO term enrichments using the online functional annotation tools DAVID (Huang et al., 2008). Enriched GO terms were selected from the GO biological process terms in DAVID's Fat database (Huang et al., 2009). GO terms which were highly overlapping, as defined by functional clustering in DAVID's website, were represented by the terms with the best FDR-adjusted p-values.
To calculate single-sample gene set enrichment, we used the GSVA program (Hanzelmann et al., 2013) to derive the absolute enrichment scores of previously experimentally validated gene signatures as follow: i) the C2 CGP (chemical and genetic perturbation sets), ii) the C6 and C7 subset of the Molecular Signature Database (Subramanian et al., 2005), iii) self-curated MAPK inhibitor-induced gene signatures using cell lines and patient-derived tumors (Song et al., 2015), iv) post-operation wound signature (Inkeles et al., 2015), and v) melanoma invasive/proliferative signatures (Hoek et al., 2008). To derive the GSVA score of each signature in each tumor sample, we computed from raw RNASeq read counts by HTSEQ COUNT program and then normalized them to log2 CPM values using EdgeR (McCarthy et al., 2012). We removed batch effects using the edgeR function RemoveBatchEffect when we combined RNAseq data from multiple experiments (Figure 3A). The normalized log2 CPM values were then passed on as input for GSVA in the RNASeq mode. Differentially enriched core gene sets between the responding and non-responding tumor groups were defined by GSVA score differences of ≥ 10% and FDR-corrected, two-sided Welch T-test p-value ≤ 0.25 (we used T-test because the GSVA scores were normally distributed around 0). Two gene sets, INGRAM_SHH_TARGETS_DN and WONG_ENDMETRIUM_CANCER_DN, were not included in the core set because they did not specifically point to a cellular process and/or relate to the other six gene sets in the core set (Table S2B, top 8). We also collected gene sets that met the GSVA score differences of ≥ 10% and nominal Welch T-test p-value ≤ 0.1 (Table S2B) and included those which were concordantly enriched and functionally related to the core gene sets to make up the full list of IPRES signatures (Figure 2D).
To compare co-enrichment of IPRES signatures across multiple melanoma cohorts, we combined and batch-corrected the log2 CPM values of four melanoma transcriptome cohorts: i) our current pre-anti-PD-1 tumors (n=28), ii) pre-anti-CTLA-4 tumors (n=42), iii) pre-MAPKi tumors (n=32) and iv) the metastatic subset of TCGA melanoma (n=282). We row-normalized the GSVA scores of each gene set in the IPRES signature across the samples from the four cohorts. For this comparative study, we excluded the gene sets “JAEGER_METASTASIS_UP,” “YE_METASTATIC_LIVER_CANCER,” “KARAKAS_ TGFB1_SIGNALING,” and “JEON_SMAD6_ TARGETS_ DN” from the IPRES set because they showed weaker co-enrichment with rest of the gene sets (see Figure 2D upper panel). The IPRES (enrichment) score was defined as the average Z-score across all gene sets in the IPRES signature, and we applied an average Z-score of 0.35 as the cutoff for IPRES signature enrichment in a tumor sample. This resulted in IPRES co-enrichment in 9 non-responding tumors and 1 responding tumor in our anti-PD-1 cohort (this cutoff was chosen because it provided the largest average Z-score separation between the samples with and without IPRES co-enrichment). Since the average Z-score was not comparable between different cohorts, we used the 90th highest IPRES score in the TCGA metastatic melanoma cohort as the IPRES score cutoff (since there were 90 of 282 tumors showing IPRES co-enrichment in this TCGA metastatic cohort; Figure 3A) for analyses performed to yield Figures 3B and S3. This allowed for a non-parametric comparison across multiple TCGA datasets at the IPRES co-enrichment level established in our anti-PD-1 cohort.
Source Data
Analysis of differential non-synonymous mutational hits in responders versus non-responders to ipilimumab was based on the mutation calls as reported (Van Allen et al., 2015). We curated published CD8 T cell exhaustion genes (Wherry, 2011) to minimize those likely to be expressed by melanoma cells by excluding genes whose maximum log2 FPKM was 1 in an in-house melanoma cell line-derived RNASeq database (n = 26 cell lines). This resulted in the inclusion of genes for surface receptors PDCD1 (PD-1), LAG3, HAVCR2 (Tim-3), CD160, and CD244 as well as transcription factors EOMES, PRDM1 (Blimp-1) and TBX21 (T-bet). We assessed co-enrichment of IPRES content signatures in the i) anti-CTLA-4 pretreatment cohort (Van Allen et al., 2015), ii) MAPKi pretreatment cohort (Hugo et al., 2015; Song et al., 2015), iii) TCGA melanoma (metastatic and primary subsets separately analyzed) (TCGA, 2015), iv) TCGA pancreatic ductal adenocarcinoma (TCGA, provisional, see https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp), v) TCGA lung adenocarcinoma (TCGA, 2014), and vi) TCGA colorectal adenocarcinoma (TCGA, 2012), and vii) TCGA kidney clear cell carcinoma (TCGA, 2013).
Supplementary Material
Article Highlights.
High mutational loads may predict better survival, but not response to anti-PD-1
BRCA2 mutations are enriched melanomas responsive to anti-PD-1
A transcriptional signature (IPRES) is related to innate anti-PD-1 resistance
IPRES defines a transcriptomic subset across distinct types of advanced cancer
ACKNOWLEDGEMENTS
This work has been funded by the National Institutes of Health (R35 CA197633 to A.R., P01 CA168585 to A.R. and R.S.L., 1R01CA176111 to R.S.L; K12CA0906525 to D.B.J.), the Ressler Family Foundation (to R.S.L. and A.R.), Grimaldi Family Fund (to A.R. and R.S.L.), Burroughs Wellcome Fund (to R.S.L), the Ian Copeland Melanoma Fund (to R.S.L.), Wade F.B. Thompson/Cancer Research Institute CLIP Grant (to R.S.L.), Steven C. Gordon Family Foundation (to R.S.L.), American Skin Association (to H.W.), Dr. Robert Vigen Memorial Fund the Garcia-Corsini Family Fund (to A.R.), the ASCO Conquer Cancer Career Development Award (to K.B.D), and American Cancer Society Research Professorship (to J.A.S.). Patient-informed consents were obtained for the research performed in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTION
W.H., J.M.Z., L.S., C.S., A.R. and R.S.L. generated, analyzed and interpreted data. S. H-L., B.H.M., B.C., G.C., E.S., M.C.K., J.A.S., D.B.J., A.R. and R.S.L. evaluated patients and provided tissue reagents. B. H-M., J.P., S.L. and X.K. processed tissues for analysis. All authors contributed to manuscript preparation. W.H. and R.S.L developed the concepts, supervised the project, and wrote the paper.
ACCESSION NUMBER
The GEO accession number for the transcriptome data is GSE78220.
REFERENCES
- Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2015a;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Heymach JV, Qin FX, Gibbons DL. The mutually regulatory loop of epithelial-mesenchymal transition and immunosuppression in cancer progression. Oncoimmunology. 2015b;4:e1002731. doi: 10.1080/2162402X.2014.1002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ, Danilova L, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Morgan M, Gunaratne PH, Wheeler DA, Gibbs RA, Gordon Robertson A, Chu A, Beroukhim R, Cibulskis K, Signoretti S, et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, Szallasi Z, Eklund AC. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Holloman WK. Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol. 2011;18:748–754. doi: 10.1038/nsmb.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoos A, Wolchok JD, Humphrey RW, Hodi FS. CCR 20th Anniversary Commentary: Immune-Related Response Criteria--Capturing Clinical Activity in Immuno-Oncology. Clin Cancer Res. 2015;21:4989–4991. doi: 10.1158/1078-0432.CCR-14-3128. [DOI] [PubMed] [Google Scholar]
- Huang D,W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, Moriceau G, Hong A, Dahlman KB, Johnson DB, et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell. 2015;162:1271–1285. doi: 10.1016/j.cell.2015.07.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inkeles MS, Scumpia PO, Swindell WR, Lopez D, Teles RM, Graeber TG, Meller S, Homey B, Elder JT, Gilliet M, et al. Comparison of molecular signatures from multiple skin diseases identifies mechanisms of immunopathogenesis. J Invest Dermatol. 2015;135:151–159. doi: 10.1038/jid.2014.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 2013;65:711–724. doi: 10.1007/s00251-013-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, Pfeifer JD. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic Acids Res. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res. 2015 Sep;29 doi: 10.1158/1078-0432.CCR-15-0876. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motz GT, Coukos G. The parallel lives of angiogenesis and immunosuppression: cancer and other tales. Nat Rev Immunol. 2011;11:702–711. doi: 10.1038/nri3064. [DOI] [PubMed] [Google Scholar]
- Muzny DM, Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL, Lewis LR, Morgan MB, Newsham IF, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, Justesen S, Roder G, Peters B, Sette A, Lund O, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2:e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2015 Dec;8 doi: 10.1158/2159-8290.CD-15-0283. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzanov I, Ribas A, Daud A, Nyakas M, Margolin KA, ODay SJ, Lutzky J, Tarhini A, Li XN, Zhou H, et al. Pembrolizumab for advanced melanoma: effect of BRAFV600 mutation status and prior BRAF inhibitor therapy. Pigment Cell Melanoma Res. 2015;28:807. [Google Scholar]
- Ramanujam S, Lo S, Kong B, Liniker E, Carlino M, Guminski A, Menzies A, Long GV. Anti-PD1 responses in BRAF mutant advanced melanoma patients with prior BRAF inhibitor or combined BRAF and MEK inhibitor therapy. Pigment Cell Melanoma Res. 2015;28:808. [Google Scholar]
- Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, Getz G. Oncotator: cancer variant annotation tool. Hum Mutat. 2015;36:E2423–2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, Robert C, Hodi FS, Wolchok JD, Joshua AM, Hwu WJ, Weber JS, Zarour HM, Kefford R, Loboda A, et al. Association of response to programmed death receptor 1 (PD-1) blockade with pembrolizumab (MK-3475) with an interferon-inflammatory immune gene signature. J Clin Oncol. 2015;33 abstr 3001. [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer M, Werner S. Cancer as an overhealing wound: an old hypothesis revisited. Nat Rev Mol Cell Biol. 2008;9:628–638. doi: 10.1038/nrm2455. [DOI] [PubMed] [Google Scholar]
- Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone E, Grimaldi AM, Festino L, Giannarelli D, Palla M, Caraco C, Curvietto M, Esposito A, Grimaldi MC, Mozillo N, et al. Correlation between BRAF mutational status and response to pembrolizumab. Pigment Cell Melanoma Res. 2015;28:754. [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Sun L, Hugo W, Hong A, Moriceau G, Kong X, Yu CC, Damoiseaux R, Kelley MC, Dahlman KB, et al. Residual Melanoma Tumors on MAPKi Therapy Undergo Transcriptome-wide Phenotype Switching. 2015 Under revision. [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350:1387–1390. doi: 10.1126/science.aad1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MH, Goldinger SM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voron T, Marcheteau E, Pernot S, Colussi O, Tartour E, Taieb J, Terme M. Control of the immune response by pro-angiogenic factors. Front Oncol. 2014;4:70. doi: 10.3389/fonc.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Wolchok JD, Hoos A, O'Day S, Weber JS, Hamid O, Lebbe C, Maio M, Binder M, Bohnsack O, Nichol G, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.