Introduction
Head and neck squamous cell carcinoma (HNSCC) affects over 550,000 patients worldwide each year 1. Tobacco and alcohol consumption, together with human papillomavirus (HPV) infection are the main etiological factors 2-4. Many studies have found that patients with HPV-positive HNSCC have a better prognosis, with increased survival and less chance of recurrence 5-7. However, there have been contradictory reports showing either no or poor associations with outcome. Nevertheless, the growing proportion of HPV-positive cases poses an urgent need for prognostic biomarkers that guide patient management according to HPV status 3,6,8,9.
Small RNAs, particularly microRNAs (miRNAs), have proven to be promising prognostic tools in HNSCC patients 10,11. Piwi-interacting RNAs (piRNAs), an abundant class of small non-coding RNA that regulate genome stability 12-14, are emerging as a valuable addition to the category of small RNA biomarkers. Recently, piRNA expression patterns have been shown to be deregulated in a variety of cancer types, including head and neck tumors 15-22; however, the relationship between piRNA expression and clinicopathological features in HNSCCs has yet to be elucidated. Interestingly, we have reported the observation of an association between piRNA expression and nodal metastasis in HNSCC 16.
Here, we describe the expression pattern of piRNAs in HNSCC using a custom pipeline to generate piRNA transcriptomes for more than 20,000 human piRNAs from 498 non-malignant and tumor tissues from HNSCC patients, and investigate their association with HPV infection status and patient survival. Our results reveal that specific piRNA expression patterns are defined by HPV status and virus type. Importantly, a piRNA expression signature was able to stratify patients based on overall survival, suggesting the potential utility of piRNA in assessing HNSCC patient prognosis.
Materials and methods
Sample Acquisition
A total of 498 HNSCC small RNA sequencing libraries (455 tumors and 43 matched non-malignant tissue) generated by The Cancer Genome Atlas (TCGA) consortium were acquired from the Cancer Genomics Hub (cgHUB) Data Repository (dbgap Project ID: 6208). The demographics and clinical data of these cases were obtained from the TCGA data portal and the USCS Cancer browser (Supplementary Table S1) .
Assembly of piRNA Transcriptome
A custom sequence analysis pipeline was developed to deduce piRNA expression from raw sequencing data (FASTQ files) 16. Briefly, our pipeline includes sample accrual, quality control and data quantification. Raw sequence BAM files received from the cgHUB were converted to FASTQ files using the Picard analysis package (http://picard.sourceforge.net) and quality trimmed. Poor quality bases (Phred quality score ≥ 20) as well as potential miRNA-associated sequences (read length < 24 bp) were removed. The remaining reads were re-mapped to human genome hg19 using STAR aligner 23, with specific parameters for short reads mapping. piRNA were annotated using a custom piRNA reference transcriptome which was generated using the information from the functional RNA database (fRNAdb v 3.4) with corresponding NCBI accession IDs corresponding to piRNA sequences. Transcript abundance was determined using PartekFlow™ platform in conjunction with the Partek Genome Suite (Partek Inc., MO, USA). Expression values were represented using reads per kilobase of exon model per million mapped reads (RPKM) 24. piRNA with RPKM values ≥1 in at least 10% of samples were considered as robustly expressed and included in further analyses.
Global Analysis of piRNA Expression Patterns
Expression matrices were aligned to clinicopathological features in order to perform comparisons between piRNA expressions levels. Statistical analysis between groups were carried out using the non-parametric Mann-Whitney U test followed by false discovery rate (FDR) correction through the Benjamini-Hochberg method 25. Box plots were generated using Gene-E software (http://www.broadinstitute.org/cancer/software/GENE-E/). In all comparisons, a p-value≤0.05 was considered significant. Circular representation of genome-wide piRNA expression in non-malignant and malignant HNSCC tissue was generated using the CircosPlot software 26.
Survival Analysis
Overall and recurrence-free survival information for the 455 HNSCC cases was collected from the UCSC cancer genome browser (https://genome-cancer.ucsc.edu). The association of multiple piRNA with patient survival was first evaluated using a Cox proportional hazard model. Significantly associated piRNAs were used to calculate sample was multiplied by its corresponding Cox coefficient, to obtain an individual piRNA weight 27. The probability of death from disease was calculated by summing all the individual piRNA weights for a given patient, and then the patients were ranked based on these probabilities. Kaplan-Meier survival curves of patients belonging to the upper and lower tertiles were compared using the log rank test.
Results
piRNAs are Differentially Expressed in Head and Neck Non-Malignant and Tumor Tissue
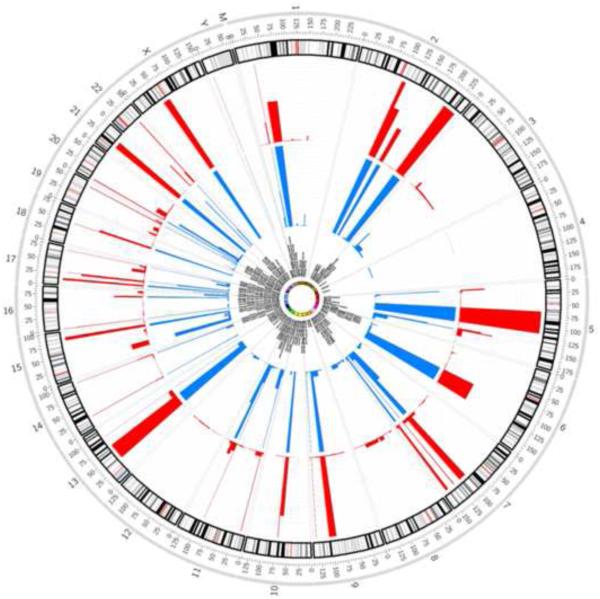
To identify piRNAs that might delineate tumor and non-malignant tissues, we analyzed piRNA expression in 43 non-malignant and 455 HNSCC samples. While the number of piRNAs expressed in either types of somatic tissue is small relative to the total number of piRNAs in the genome, we found intriguing patterns of differential expression between HNSCC tumors and their non-malignant counterparts. A total of 305 piRNAs were robustly expressed in both non-malignant and tumor tissue (Figure 1, Supplementary Table S2). Globally, we observed that piRNA expression was considerably more abundant in tumors relative to non-malignant tissue. A total of 247 were significantly differentially expressed between non-malignant and tumor tissues (Mann-Whitney U-test, FDR-BH p-value ≤ 0.05). Interestingly, 25 were exclusively expressed in non-malignant tissue, while 87 were only expressed in tumors.
Figure 1. Genome-wide expression of piRNAs in head and neck non-malignant and tumor tissue.
The expression of 305 piRNAs in non-malignant (blue) and tumors (red) are presented in a circos plot, which relates piRNA genes to the chromosomal location.
piRNAs Are Differentially Expressed in HPV Positive and Negative Cases
We next focused our attention on piRNA expression patterns in malignant tissues. Since HPV infection is an important driving event in HNSCC tumorigenesis 28-30, we investigated whether HPV status might be associated with a unique piRNA expression pattern. To this end, we compared the piRNA expression patterns between HPV-negative (HPV−, N=370) and HPV-positive (HPV+, N=83) tumors. Forty-one piRNAs were differentially expressed (Figure 2, Supplementary Table S3), with 30 being overexpressed in HPV+ cases and 11 overexpressed in HPV− samples.
Figure 2. HPV status and piRNA expression patterns.
Comparison of piRNA expression patterns between HPV negative (average of 370 HPV− HNSCC tumor piRNA transcriptomes, purple bars) and HPV positive cases (average of 83 HPV+ HNSCC tumor piRNA transcriptomes, yellow bars).
Since HPV16 and HPV18 have the highest oncogenic potential, we assessed whether expression of any of these piRNAs might specifically associate with these HPV types. We found that 11 out of 41 piRNAs (26.8%) were differentially expressed between HPV16 or 18 positive patients (N=67) compared to cases harboring other HPV types 33, 35, and 56 (N=22 combined) (Mann-Whitney U-test, FDR-BH p-value ≤ 0.05, Table 1). Remarkably, all these piRNAs were expressed at lower levels in HPV16/18 HNSCC samples (Supplementary Figure 1).
Table 1.
piRNAs differentially expressed in HPV positive cases with and without HPV16/18
| Tag | NCBI Accession |
Mean HPV16/ 18 |
Median HPV16/ 18 |
Mean other HPVs |
Median other HPVs |
p Value | Over- lapping gene |
|---|---|---|---|---|---|---|---|
| FR004819 | DQ596390 | 103.47 | 40.41 | 204.02 | 136.97 | 0.00904 | |
| FR018916 | DQ597887 | 3.48 | 2.00 | 2.89 | 1.00 | 0.00936 | RPL13A |
| FR136216 |
DQ598104, DQ598103 |
244.52 | 198.75 | 362.82 | 297.57 | 0.00417 | PRRC2B |
| FR140858 | DQ598918 | 2316.20 | 2366.00 | 3039.78 | 2636.00 | 0.04866 | MCM7 |
| FR197104 | DQ601526 | 1.78 | 1.00 | 3.61 | 2.50 | 0.00266 | RBM33 |
| FR213383 | DQ598648 | 99.87 | 53.59 | 176.97 | 143.06 | 0.00045 | RPL3 |
| FR222326 | DQ597945 | 37.69 | 3.00 | 96.94 | 10.50 | 0.03806 | |
| FR237180 | DQ598649 | 40.15 | 10.81 | 151.98 | 40.15 | 0.00507 | RPL3 |
| FR245310 | DQ596225 | 15.28 | 7.00 | 27.33 | 16.00 | 0.04054 | |
| FR279668 | DQ571955 | 8.69 | 6.00 | 10.78 | 8.00 | 0.01283 | |
| FR298757 | DQ570394 | 1.38 | 0.00 | 2.24 | 0.83 | 0.04258 |
A piRNA Signature is Associated with Patient Outcome in HPV-Positive HNSCC
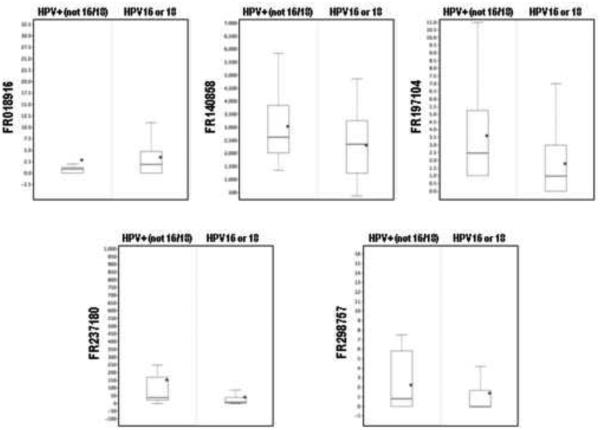
According to the literature, HPV+ patients have better overall survival compared to HPV− patients. Since we observed distinct piRNA expression patterns within the HPV+ cohort, we next evaluated the association of the 11 differentially expressed piRNAs with outcome. While we did not find any association with recurrence free survival, we found a five piRNA signature that was significantly associated with overall survival in these patients (p-value Chi2 test=0.002, Table 2 and Figure 3). Patients with high probability scores had significantly worse survival (log rank p-value=0.00092, Figure 4). We tested this signature in HPV− patients as well as all patients regardless of HPV status, and found no significant association with overall survival in these groups, indicating this signature is specific to HPV+ patients (Supplementary Table S4).
Table 2.
Five piRNAs significantly associated with patient overall survival
| Variable | Cox coefficient* |
Standard error |
Wald Chi- Square |
Pr > Chi2 |
Hazard ratio |
Hazard ratio Lower bound (95%) |
Hazard ratio Upper bound (95%) |
|---|---|---|---|---|---|---|---|
| FR018916 | −0.125 | 0.058 | 4.636 | 0.031 | 0.882 | 0.787 | 0.989 |
| FR140858 | −0.001 | 0.000 | 8.241 | 0.004 | 0.999 | 0.999 | 1.000 |
| FR197104 | 0.135 | 0.080 | 2.848 | 0.091 | 1.145 | 0.978 | 1.340 |
| FR237180 | 0.003 | 0.001 | 5.580 | 0.018 | 1.003 | 1.001 | 1.006 |
| FR298757 | 0.103 | 0.075 | 1.878 | 0.171 | 1.109 | 0.957 | 1.285 |
Regression coefficient for piRNA association with patient overall survival. 11 piRNAs from Table 1 were initially tested for correlation with survival using a Cox regression model. A combination of the five piRNAs shown was significantly associated with patient overall survival.
Figure 3. Expression boxplots of the five survival-associated piRNAs (RPKM) in HPV+ tumors.
On each boxplot, the average is denoted by a black diamond, the line within each boxplot indicates the median value, the border of the box indicates the 25th and 75th percentiles, and the dashed line represents the range.
Figure 4. A five piRNA signature delineates the outcome of patients with HPV+ HNSCC.
The expression values of the piRNAs in the model were transformed into a score representing the probability of death by disease by multiplying the expression values of each piRNA by their respective Cox proportional hazard coefficient (Table 2), and then summing their values. The Kaplan-Meier curve shows patients with a high score (top 30th percentile) had worse survival than those with a low score (bottom 30th percentile).
Discussion
Considerable advances have been made in the detection and chemoprevention of head and neck cancer 4,31; however, major challenges still remain regarding the improvement of patient management once the disease is diagnosed. Additionally, the interaction between biological risk agents, such as HPV, and cellular mechanisms are still not completely understood. In this proof of principle study, we show that another important component of the small RNA transcriptome, piRNA, is likely to play a role in patient outcome.
We assessed piRNA expression of 498 non-malignant and tumor tissues from TCGA consortium using a custom pipeline to assemble a piRNA transcriptome. Our findings revealed that, 1) piRNAs are differentially expressed between malignant HNSCC and non-malignant tissues, 2) piRNA expression patterns are specific to the HPV status of the tumor, and 3) a five piRNA expression signature can be used to predict HPV+ HNSCC overall survival.
It has been widely established that HPV+ HNSCC is a distinct disease and develops through different mechanisms than carcinomas with other etiologies 3,32,33. miRNA dysregulation has been previously implicated in the development and progression of HPV+ HNSCC tumors. For example, members of the mir106b~25 cluster were considerably up-regulated in HPV associated HNSCC; while mir-199-1 was highly downregulated in HPV+ patient samples 34. These HPV-associated miRNAs may play a mechanistic role in the distinct clinical behavior of HPV-infected tumors, and suggest potential utility of miRNAs as biomarkers 34,35. While initial studies have shown that HPV infection affects the expression of some small non-coding RNAs, such as miRNAs, its effect on piRNA expression is unknown. The association of piRNA with patient outcome has been previously demonstrated in other cancer types, such as breast and gastric, among others 16,17,19. Thus, it is of great relevance to characterize changes in piRNA expression patterns in the context of HPV status, as these piRNAs may represent novel predictors of clinical outcome in HPV+ HNSCC.
A deeper investigation of piRNA expression within malignant tissues revealed a group of piRNAs whose expression was significantly associated with the presence of oncogenic HPV types HPV16/18. One of the most highly differentially expressed piRNAs is FR140858 (DQ598918) (Table 1). The 31 nucleotides of the piRNA sequence displays 100% complementarity with positions 99,691,656 to 99,691,686 in chromosome 7, which encompasses the sequences of different transcripts of the minichromosome maintenance complex component 7 (MCM7) gene. Interestingly, the expression of MCM7 is induced through the activity of the HPV viral oncogene E7 36 and significantly correlates with HPV infection in cervical carcinoma 37. Although the mechanisms of piRNA-mediated silencing of mRNA targets are still emerging 38, the loss of the piRNA FR140858 in HPV16/18-associated HNSCC could potentially promote MCM7 expression, a possibility that may be assessed in future mechanistic studies. Although more validation studies are needed, piRNAs hold great promise as potential prognostic biomarkers, owing to their small non-coding RNA features such as small size, stability in biofluids and archival materials, and variety of detection methods 39. Also, considering there are 10–25 times more piRNA species (20,000–50,000) than microRNAs, the impact of their deregulation is likely at least as relevant.
In summary, we have identified loci that actively express piRNA in non-malignant and malignant head and neck tissues. Although it is generally accepted that HPV+ HNSCC patients have a better prognosis compared to HPV− cases 5-7, our results indicate this cohort can be further stratified based on piRNA expression, suggesting a potential clinical utility of piRNAs in head and neck cancer.
Supplementary Material
Highlights of results from “HPV status alters PIWI-interacting RNA expression pattern in head and neck cancer”.
piRNAs are differentially expressed between HNSC and non-malignant tissues
piRNA expression patterns are specific to the HPV status of the tumor
A piRNA expression signature can predict overall survival in HPV+ HNSC patients
Acknowledgements
This work was supported by grants from the National Institutes of Health (NIH-NIDCR R01 DE015965), Canadian Institutes for Health Research (CIHR, FDN143345) and the Canadian Cancer Society (CCSRI), and CIHR Frederick Banting and Charles Best Canada Graduate Scholarships to K.S.S.E. and D.A.R. K.L.B. is a Michael Smith Foundation for Health Research Biomedical Research Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011 Mar-Apr;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- D'Souza G, Dempsey A. The role of HPV in head and neck cancer and review of the HPV vaccine. Preventive medicine. 2011 Oct;53(Suppl 1):S5–S11. doi: 10.1016/j.ypmed.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marur S, D'Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. The Lancet. Oncology. 2010 Aug;11(8):781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez VD, MacAulay CE, Guillaud M, et al. Targeting of chemoprevention to high-risk potentially malignant oral lesions: challenges and opportunities. Oral oncology. 2014 Dec;50(12):1123–1130. doi: 10.1016/j.oraloncology.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009 Apr 20;27(12):1992–1998. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. The New England journal of medicine. 2010 Jul 1;363(1):24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. International journal of cancer. Journal international du cancer. 2007 Oct 15;121(8):1813–1820. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- Stein AP, Saha S, Kraninger JL, et al. Prevalence of Human Papillomavirus in Oropharyngeal Cancer: A Systematic Review. Cancer J. 2015 May-Jun;21(3):138–146. doi: 10.1097/PPO.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008 Feb 1;26(4):612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. The American journal of pathology. 2009 Mar;174(3):736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervigne NK, Reis PP, Machado J, et al. Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Human molecular genetics. 2009 Dec 15;18(24):4818–4829. doi: 10.1093/hmg/ddp446. [DOI] [PubMed] [Google Scholar]
- Mei Y, Wang Y, Kumari P, et al. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nature communications. 2015;6:7316. doi: 10.1038/ncomms8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annual review of biochemistry. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annual review of cell and developmental biology. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer letters. 2013 Aug 9;336(1):46–52. doi: 10.1016/j.canlet.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Scientific reports. 2015;5:10423. doi: 10.1038/srep10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez VD, Enfield KS, Rowbotham DA, Lam WL. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric cancer : official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2015 Mar 17; doi: 10.1007/s10120-015-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clinical & translational oncology : official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2013 Jul;15(7):563–568. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- Hashim A, Rizzo F, Marchese G, et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014 Oct 30;5(20):9901–9910. doi: 10.18632/oncotarget.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SL, Ricciardelli C, Oehler MK, Tan IM, Russell D, Grutzner F. Overexpression of piRNA pathway genes in epithelial ovarian cancer. PloS one. 2014;9(6):e99687. doi: 10.1371/journal.pone.0099687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer letters. 2015 Jan 28;356(2 Pt B):561–567. doi: 10.1016/j.canlet.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015 Jan 29;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013 Jan 1;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008 Jul;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome research. 2009 Sep;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. The New England journal of medicine. 2007 Jan 4;356(1):11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nature communications. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olthof NC, Huebbers CU, Kolligs J, et al. Viral load, gene expression and mapping of viral integration sites in HPV16-associated HNSCCC cell lines. International journal of cancer. Journal international du cancer. 2015 Mar 1;136(5):E207–218. doi: 10.1002/ijc.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebbers CU, Preuss SF, Kolligs J, et al. Integration of HPV6 and downregulation of AKR1C3 expression mark malignant transformation in a patient with juvenile-onset laryngeal papillomatosis. PloS one. 2013;8(2):e57207. doi: 10.1371/journal.pone.0057207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny P, Zhang L, Fan YH, et al. Gene expression profiling predicts the development of oral cancer. Cancer Prev Res (Phila) 2011 Feb;4(2):218–229. doi: 10.1158/1940-6207.CAPR-10-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partlova S, Boucek J, Kloudova K, et al. Distinct patterns of intratumoral immune cell infiltrates in patients with HPV-associated compared to non-virally induced head and neck squamous cell carcinoma. Oncoimmunology. 2015 Jan;4(1):e965570. doi: 10.4161/21624011.2014.965570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrjanen S. The role of human papillomavirus infection in head and neck cancers. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2010 Oct;21(Suppl 7):vii243–245. doi: 10.1093/annonc/mdq454. [DOI] [PubMed] [Google Scholar]
- Miller DL, Davis JW, Taylor KH, et al. Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the Cancer Genome Atlas. The American journal of pathology. 2015 Mar;185(3):679–692. doi: 10.1016/j.ajpath.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajer CB, Nielsen FC, Friis-Hansen L, et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: a prospective translational study. British journal of cancer. 2011 Mar 1;104(5):830–840. doi: 10.1038/bjc.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton K, Peh W, Southern S, et al. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. Journal of virology. 2003 Oct;77(19):10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake T, Connor JP, Petereit DG, Lambert PF. Comparative analysis of cervical cancer in women and in a human papillomavirus-transgenic mouse model: identification of minichromosome maintenance protein 7 as an informative biomarker for human cervical cancer. Cancer research. 2003 Dec 1;63(23):8173–8180. [PubMed] [Google Scholar]
- Zhang P, Kang JY, Gou LT, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell research. 2015 Feb;25(2):193–207. doi: 10.1038/cr.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ren C, Han J, et al. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. British journal of cancer. 2014 Apr 29;110(9):2291–2299. doi: 10.1038/bjc.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.