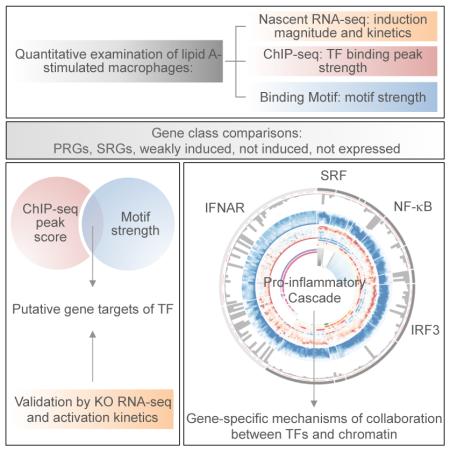
Abstract
Much has been learned about transcriptional cascades and networks from large-scale systems analyses of high-throughput data sets. However, analysis methods that optimize statistical power through simultaneous evaluation of thousands of ChIP-seq peaks or differentially expressed genes possess substantial limitations in their ability to uncover mechanistic principles of transcriptional control. By examining nascent transcript RNA-seq, ChIP-seq, and binding motif data sets from lipid A-stimulated macrophages with increased attention to the quantitative distribution of signals, we identified unexpected relationships between the in vivo binding properties of inducible transcription factors, motif strength, and transcription. Furthermore, rather than emphasizing common features of large clusters of co-regulated genes, our results highlight the extent to which unique mechanisms regulate individual genes with key biological functions. Our findings demonstrate the mechanistic value of stringent interrogation of well-defined sets of genes as a complement to broader systems analyses of transcriptional cascades and networks.
INTRODUCTION
The molecular biology revolution of the 1970s was followed by a 20-year period during which gene regulation was studied at the level of individual model genes. Near the turn of the century, the emergence of DNA microarrays and whole-genome sequences opened avenues toward the study of gene regulation at a global scale, making it possible to identify genes and networks that characterize a cell type, environmental response, or disease state. More recently, RNA sequencing (RNA-seq) has emerged as a method that allows global transcript levels to be evaluated with greater accuracy (Marioni et al., 2008). RNA-seq also provides an opportunity to monitor nascent transcripts in addition to mRNA (e.g. Bhatt et al., 2012; Core et al., 2008; Rabani et al., 2011). For studies of stimulus-induced transcription, nascent transcript levels provide more accurate information about the kinetics with which gene transcription is activated, and they allow transcription to be studied independently of mRNA stability.
Transcriptional cascades induced by inflammatory stimuli in cells of the mouse innate immune system have been especially well-studied at a global scale, with most studies focusing on cells stimulated with lipopolysaccharide (LPS) or lipid A. LPS and lipid A engage Toll-like receptor 4 (TLR4), which then activates common signaling pathways via the MyD88 and TRIF adaptors. The TLR4-induced cascade has been monitored by DNA microarray, RNA-seq, and nascent transcript RNA-seq (e.g. Amit et al., 2009; Bhatt et al., 2012; Ramsey et al., 2008). Binding sites for several transcription factors are enriched within the promoters of defined clusters of co-regulated genes, and distinct subsets of promoters contain features of either active or inactive chromatin prior to cell stimulation (Hargreaves et al., 2009; Ramirez-Carrozzi et al., 2009). Moreover, thousands of inducible enhancers have been defined, with some enhancers poised for activation and others lacking chromatin marks prior to stimulation (Ghisletti et al., 2010; Heinz et al., 2010; Ostuni et al., 2013). Gene expression profiles have been further integrated with ChIP-seq data sets and siRNA knockdown experiments for transcription factors and chromatin regulators (e.g. Amit et al., 2009; Garber et al., 2012).
Although conventional systems analyses have provided considerable insight into the logic underlying the transcriptional response to a stimulus, the results are often limited to statistical trends and lack the precision needed to fully uncover molecular mechanisms. Moreover, for most systems analyses, all genes that are induced or differentially expressed by a magnitude exceeding a low threshold – often 2-fold – are considered equally. This approach enhances statistical power and provides an opportunity to simultaneously examine an entire “system.” However, the results tend to be strongly biased toward genes that are differentially expressed by small magnitudes; these genes are far more prevalent - and may be regulated by different mechanisms – than genes differentially expressed by large magnitudes.
Here, we describe an analysis of lipid A-induced transcription using gene-centric approaches that place greater emphasis on quantitative aspects of nascent transcript RNA-seq, ChIP-seq, and binding motif data sets. In addition to providing insight into a number of unanswered mechanistic questions, these approaches allowed us to move beyond the identification of common features of large clusters of co-regulated genes and toward an appreciation of the unique molecular mechanisms used to regulate individual genes within the inflammatory cascade.
RESULTS
Basic Properties of the Transcriptional Cascade
We first performed RNA-seq with mouse bone marrow-derived macrophages (BMDMs) treated with lipid A for 0, 15, 30, 60, and 120 min. To separate transcription from mRNA stability, we analyzed nascent, chromatin-associated transcripts. 3,863 (14.1%) of the 27,384 annotated Refseq genes (prior to removal of duplicate isoforms) reached an expression level of at least 3 RPKM in at least one time point. We used a high expression threshold because our subsequent analysis emphasized induction magnitudes, which can be quantified most accurately when both basal and induced transcript levels can be measured with confidence.
Of the 3,863 expressed genes, 1,340 (34.7%) were induced by at least 2-fold (p<0.01) (Figure 1A). Importantly, however, 79.5% of these genes were induced less than 10-fold (Figure 1A). If all genes induced by 2-fold or greater were evaluated together, the analysis would be dominated by weakly induced genes. Notably, most induced genes encoding key cytokines, chemokines, and transcription factors were induced by >10-fold (data not shown). We therefore focused on the potently induced genes, with the resulting insights then examined in the context of the weakly induced genes (see below). Notably, the basal transcript levels of the weakly induced genes were generally higher than those of the strongly induced genes (Figure 1B).
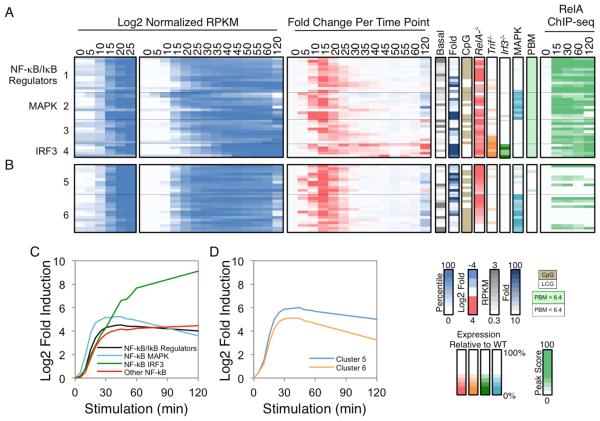
Figure 5. Kinetic and Functional Analysis of Putative NF−κB Target Genes.
(A) The 37 PRGs containing strong NF−κB promoter motifs and RelA ChIP-seq promoter peaks were grouped into four categories: those that encode NF-κB/IκB family members and regulators (group 1), those that exhibit either MAPK or IRF3 dependence (groups 2 and 4), and the remaining genes (group 3). Normalized expression values from 0-25 min (left panel) and 0-120 min (second panel), and the fold change relative to the previous time point (third panel) are shown. To the right of the heatmaps, the basal expression values, fold induction magnitudes, promoter CpG contents, and expression values in Rela−/−, Trif−/−, Irf3−/−, and MAPK-inhibited BMDMs are shown. The presence of a p50:RelA motif based on PBM datasets and the RelA ChIP-seq binding peak scores are indicated in the far right panels. See also Figure S4.
(B) Examples of PRGs that exhibited similar activation kinetics and/or RelA dependence to the 37 genes with strong NF−κB motifs and ChIP-seq peaks are shown. See also Figure S4.
(C) The average activation kinetics of the NF-κB subgroups is shown as log2 fold inductions relative to basal during the 120-min lipid A treatment period.
(D) The average activation kinetics of the two additional clusters from Figure 5B (Cluster 5 and 6) are shown.
With the above considerations in mind, we focused on 226 genes, 215 of which were induced (p<0.01) >10-fold during the 2-hr induction period. The remaining 11 genes were transiently induced by 5-10-fold at the 15-min time point; these genes were added to capture a larger number of genes that are rapidly downregulated after their early induction. Although the analysis focuses on only 226 genes, their basal and peak transcript levels were distributed over more than two orders of magnitude (Figure 1C).
Separation of Primary and Secondary Response Genes
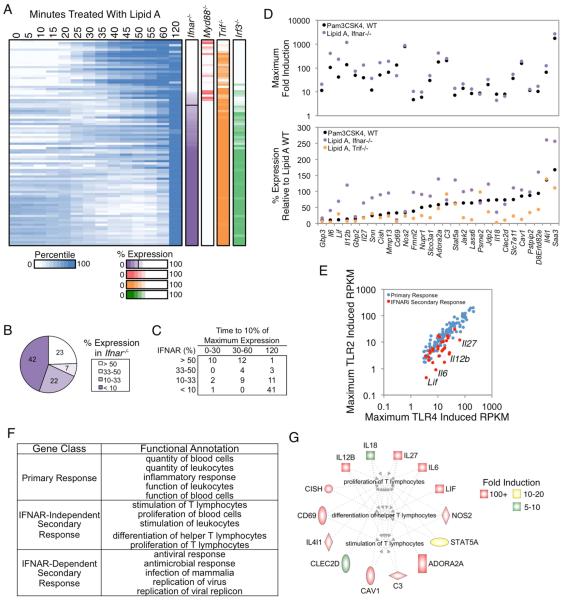
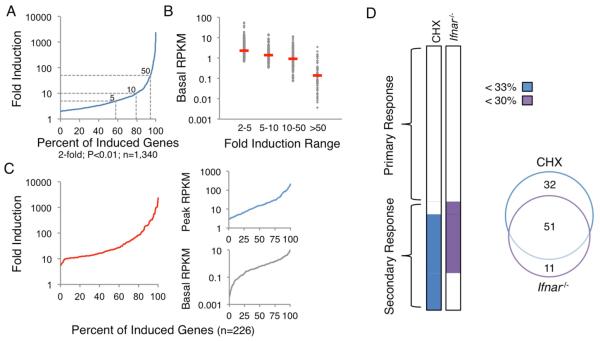
We next separated primary response genes (PRGs) and secondary response genes (SRGs) by performing RNA-seq with nascent transcripts from BMDMs stimulated with lipid A in the presence of cycloheximide (CHX). This analysis revealed 83 genes that were expressed at a level in CHX-treated cells that was <33% of the expression level in untreated cells (Figure 1D). These 83 genes were included in the SRG group (Figure 1D).
IFN-β expression is induced by lipid A and activates a Type I IFN gene program. RNA-seq analysis of nascent transcripts from Type I IFN receptor (IFNAR)-deficient (Ifnar−/−) BMDMs stimulated with lipid A revealed 62 genes that were expressed <30% of wild-type (WT) (Figure 1D). Interestingly, 11 of these IFNAR-dependent genes were classified as PRGs in the CHX analysis because they exhibited expression levels in the presence of CHX that placed them just above the threshold used for SRG classification. Nevertheless, an analysis of their induction kinetics revealed greater similarity to the other IFNAR-dependent SRGs than to the PRGs (data not shown; see Figure S1). Because of their strong IFNAR-dependence and kinetic profiles, these 11 genes were added to the SRG category (Figure 1D). Thus, 132 and 94 genes, respectively, were defined as PRGs and SRGs for the current analysis. Because some genes possess both primary and secondary response components (data not shown), the classification assignments will need to be re-evaluated as our knowledge increases.
Separation of IFNAR-Dependent and -Independent SRGs
As described above, a central feature of the secondary response to lipid A is the activation of Type I IFN. Therefore, we separated SRGs into IFNAR-dependent and -independent groups. Forty-two of the 94 SRGs were expressed <10% of WT in Ifnar−/− BMDMs, with an additional 22 expressed between 10 and 33% (Figure 2A,B). Kinetic analyses revealed that 41 of the 42 genes expressed <10% of WT failed to reach an expression level in WT cells corresponding to 10% of the maximum level until the 120-min time point (Figure 2C), indicating that a robust transcriptional response to IFNAR signaling begins between 60 and 120 min post-stimulation. In contrast, 22 of the 23 SRGs that were largely unaltered in the Ifnar−/− cells (expression level >50% of WT) reached an expression level in WT cells corresponding to 10% of their maximum within 60 min (Figure 2C). Thus, the CHX-sensitive events needed for activation of IFNAR-independent SRGs generally occur more rapidly than the autocrine/paracrine loop that activates IFNAR-dependent genes.
Figure 2. Analysis of IFNAR-Independent and -Dependent SRGs.
(A) Activation kinetics are shown for SRGs from BMDMs stimulated at 5-min intervals from 0-60 min, with an additional 120-min time point. Shades of blue indicate percentile values. Genes were sorted on their maximum percent expression in Ifnar−/− BMDMs relative to WT BMDMs (purple column). The maximum percent expressions in Myd88−/−, Trif−/−, and Irf3−/− are shown to the right. See also Figure S1.
(B) The distribution of genes in IFNAR-dependence bins based on their expression in Ifnar−/− BMDMs is shown.
(C) The time point at which each SRG in the IFN-dependence bins reached 10% of its maximum expression is indicated.
(D) The maximum fold induction of the 29 IFNAR-independent genes in PAM-stimulated (black) and lipid A-stimulated Ifnar−/− (purple) BMDMs is shown (top), along with the percent expression of these genes in PAM-stimulated (black), lipid A-stimulated Ifnar−/− (purple), and lipid A-stimulated Trif−/− (orange) BMDMs relative to WT BMDMs stimulated with lipid A (bottom). IFNAR-independent genes were defined as those induced >10-fold and expressed >3 RPKM in the absence of IFNAR signaling, or expressed at greater than 50% of WT in Ifnar−/− BMDMs stimulated with lipid A or WT BMDMs stimulated with PAM.
(E) A scatterplot comparing the maximum RPKMs in PAM-stimulated BMDMs (y-axis) and lipid A-stimulated BMDMs (x-axis) for PRGs (blue) and the IFNAR-independent SRGs (red) is shown.
(F) Ingenuity Pathway Analysis was used to identify the top functional annotations for PRGs and the IFNAR-dependent and -independent SRGs.
(G) The IFNAR-independent genes involved in the proliferation, differentiation, and activation of T lymphocytes (Ingenuity Pathway Analysis) are colored based on their fold induction in Ifnar−/− BMDMs.
To separate IFNAR-dependent and -independent genes more carefully, we further examined the RNA-seq data sets from lipid A-stimulated Ifnar−/− BMDMs, as well as RNA-seq data sets from WT BMDMs stimulated with Pam3CSK4 (PAM), a TLR2 ligand that does not induce IFNAR signaling (Toshchakov et al., 2002). Twenty-nine SRGs remained strongly induced in these data sets (Figure 2D, top).
Interestingly, although these 29 SRGs were strongly induced in the absence of IFNAR signaling, a subset, including the critical T cell polarizing cytokines Il12b, Il6, Lif, and Il27 (Metcalfe, 2011; Shih et al., 2014), were induced much less potently by PAM than by lipid A (Figure 2D, bottom). In fact, Il12b, Il6, Lif, and Il27 exhibited greater differential induction by TLR4 versus TLR2 ligands than any other PRG or SRG (Figure 2E). This finding suggests that the TRIF pathway activated by lipid A but not by PAM may be important for the activation of these genes, but not due to its role in activating IFNAR signaling. Consistent with this possibility, a direct comparison of WT to Trif−/− BMDMs revealed strong TRIF-dependence of these genes (Figure 2D, bottom). Together, the data suggest that lipid A induces the expression of key T-cell polarizing cytokines (Il12b, Il6, Lif, and Il27) much more potently than does PAM because the TRIF pathway strongly promotes the expression of these genes in an IFNAR-independent manner.
To better understand the significance of the above regulatory strategies, we performed gene ontology analysis with the 132 PRGs, 65 IFNAR-dependent SRGs, and 29 IFNAR-independent SRGs (Figure 2F). The PRG analysis suggested broad roles in regulating inflammation and the functions of blood cells. As expected, the IFNAR-dependent SRGs were implicated in anti-viral responses. Most interestingly, the small group of IFNAR-independent SRGs exhibited highly significant enrichment for genes that regulate T cell proliferation, differentiation, and activation. Specifically, 14 of the 29 IFNAR-independent SRGs are involved in the regulation of T cell responses (Figure 2G). Eleven of these 14 genes are among the 13 IFNAR-independent SRGs that are most potently induced by lipid A. Thus, these results reveal common regulatory features of a prominent group of genes that helps bridge the innate and adaptive immune systems. Nevertheless, a careful examination reveals that the induction kinetics for each of these genes is unique (Figure S1), suggesting that gene-specific regulatory events are superimposed on top of their common characteristics of potent and rapid CHX-sensitive yet IFNAR-independent induction.
Initial Analysis of PRGs
Shifting our attention to the 132 PRGs, we first examined their expression kinetics in greater detail by nascent transcript RNA-seq from lipid A-stimulated BMDMs collected every five min during the first hr of activation, with an additional 120-min time point. We also performed nascent transcript RNA-seq with BMDMs from Myd88−/−, Trif−/−, Myd88−/−Trif−/−, and Irf3−/− mice, and with WT BMDMs stimulated with lipid A in the presence of ERK and p38 MAPK inhibitors; the two inhibitors were analyzed together because little effect was observed in pilot experiments with each inhibitor alone (data not shown). The results consider the maximum induced RPKM in WT cells for each gene to be 100% and the basal RPKM in unstimulated WT cells to be 0%; the maximum induced RPKM observed in each mutant strain for each gene is then displayed as a percentage of the maximum WT RPKM.
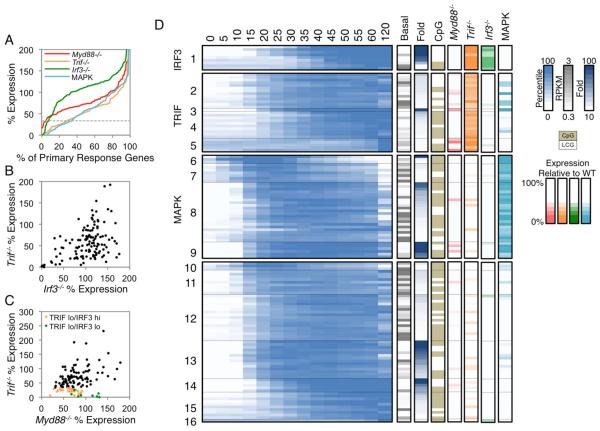
Figure 3A (see also, Figure S2) shows that each perturbation resulted in a continuum of effects. For this study, genes expressed <33% of WT were considered to be dependent on the factor that was absent. By combining these data sets with k-means cluster analysis of expression kinetics, an initial classification of the 132 PRGs emerged (Figure 3D; see Figure S2 for gene names). Cluster 1 includes nine genes that exhibited reduced expression (<33% of WT) in both Trif−/− and Irf3−/− BMDMs. Clusters 2-5 include 28 genes that exhibited reduced expression in Trif−/− but not in Irf3−/− BMDMs (Figure 3B,D); these genes were then subdivided by k-means clustering on the basis of their expression kinetics. Clusters 6-9 include 38 genes that exhibited reduced expression in WT BMDMs treated with MAPK inhibitors, but without strongly reduced expression in Trif−/− BMDMs; as above, the genes were subdivided by k-means clustering (Figure 3D). Finally, Clusters 10-16 include the remaining 57 genes that did not exhibit reduced expression in the presence of MAPK inhibitors or in Trif−/− or Irf3−/− cells; these genes were divided into seven kinetic clusters. It is noteworthy that only five of the 132 PRGs exhibited reduced expression in Myd88−/− cells (Figure 3C,D). No genes were induced in Myd88−/−Trif−/− mutant cells (data not shown).
Figure 3. Properties of PRGs.
(A) The distribution of the maximum percent expressions in Myd88−/− (red), Trif−/− (orange), Irf3−/− (green), and MAP kinase inhibitor-treated (light blue) BMDMs stimulated with lipid A are shown for the 132 PRGs. The horizontal dashed grey line indicates the 33% expression threshold.
(B-C) The percent expression of each PRG is shown in Trif−/− versus Irf3−/− cells (B), or in Trif−/− versus Myd88−/− cells (C). TRIF lo (<33% relative to WT) IRF3 hi (>33% relative to WT) genes are in orange, and TRIF lo (<33% relative to WT) IRF3 lo (<33% relative to WT) genes are in green.
(D) Activation kinetics are shown (log2-normalized and mean-centered RPKMs) for the PRGs in BMDMs stimulated for 5-min intervals between 0-60 min, and for 120 min. The PRGs were broadly classified based on their expression in Myd88−/− (red), Trif−/− (orange), Irf3−/− (green), and MAP kinase inhibitor-treated (light blue) BMDMs with the following order: IRF3-dependent (cluster 1; <33% in both Trif−/− and Irf3−/−), TRIF-dependent (cluster 2-5; <33% in Trif−/− only), and MAPK-dependent (cluster 6-9; <33% in MAPK inhibitor-treated samples). The remaining PRGs were not dependent on any perturbation examined (cluster 10-16; >33% in all perturbed datasets). The genes in each class were subclustered (k-means) on their expression kinetics. The properties of each gene are shown to the right of the heat map: basal expression value (grey), fold induction magnitude (blue), promoter CpG-island (beige), and the maximum percent expression in Myd88−/− (red), Trif−/− (orange), Irf3−/− (green), and MAPK inhibitor-treated (light blue) BMDMs. See also Figure S2 and S3.
In addition to the degree of dependence of each PRG on MyD88, TRIF, IRF3, and MAPKs, Figure 3D indicates basal transcript and fold-induction values. Furthermore, Figure 3D indicates which genes contain CpG-island or low CpG (LCG) promoters. As shown previously (Bhatt et al., 2012), all early transiently induced genes (e.g. Clusters 6 and 10) contain CpG-island promoters and a high percentage of the most potently induced genes contain LCG promoters (e.g. Clusters 1 and 14), whereas the two promoter types are distributed fairly randomly among the other clusters.
Initial Transcription Factor Binding Motif and ChIP-Seq Analyses
To extend the above foundation, we evaluated the over-representation of transcription factor binding motifs within the promoters of the PRGs within each of the 16 clusters in Figure 3D. This analysis (Figure S3) provided insight into transcription factors that may regulate each cluster. However, toward the goal of elucidating molecular mechanisms, these statistical enrichments were unsatisfying. For example, although NF-κB motifs are enriched in the promoters of genes in several clusters, a closer analysis revealed considerable heterogeneity within each cluster, with only a subset of promoters in a cluster generally containing a strong NF-κB motif (data not shown). Imprecise correlations were also apparent when examining ChIP-seq data sets for NF-κB and other transcription factors (data not shown). Therefore, additional strategies are needed to move beyond statistical enrichments toward more meaningful mechanistic insights.
Quantitative Analysis of NF-κB’s Contribution to the Transcriptional Cascade
We next focused on NF-κB. Prior studies showed that a large percentage of ChIP-chip and ChIP-seq peaks for NF-κB family members do not coincide with strong binding motifs (Lim et al., 2007; Martone et al., 2003; Zhao et al., 2014), raising questions about NF-κB’s DNA recruitment and transcriptional activation mechanisms. However, when focusing attention on the promoters of our well-defined set of strongly induced PRGs, a different relationship between NF-κB binding and motifs emerged.
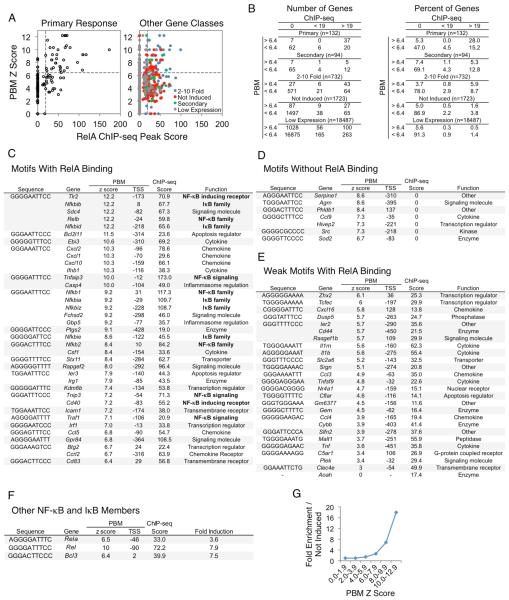
Specifically, Figure 4A examines NF-κB ChIP-seq peak scores versus motif scores for the promoters (−500 to +150 relative to the TSS) of each of the 132 PRGs. The NF-κB motif scores were derived from protein binding microarray (PBM) results obtained with a recombinant RelA:p50 heterodimer, the most abundant NF-κB dimer involved in TLR4-induced transcription (Siggers et al., 2012). RelA ChIP-seq experiments were performed with BMDMs stimulated with lipid A for 0, 15, 30, 60, and 120 min (followed by stringent peak-calling and a focus on peaks observed in multiple biological replicates). This analysis revealed 8,458 total peaks, with 942 promoter peaks.
Figure 4. NF−κB Interactions at the Promoters of Defined Gene Classes.
(A) PBM Z scores of p50:RelA (y-axis) and RelA ChIP-seq peak scores (x-axis) in the promoters of the PRGs (left) and all remaining genes in the genome (right) were plotted. The remaining genes were assigned to 2-10-fold induced (blue), not induced (red), SRG (green), or low expression (grey) categories. The horizontal dashed line indicates the PBM Z score threshold (6.4), and the vertical dashed line indicates the ChIP-seq peak score threshold (19).
(B) Tables are shown indicating the distribution of genes from panel (A) for both numbers (left) and percentages (right) of genes.
(C-F) Tables are shown indicating the best matching κB motif in each promoter (column 1), the gene name (column 2), the PBM p50:RelA Z score (column 3), the position of the motif relative to the TSS (column 4), the RelA ChIP-seq peak score (column 5), and either the function or fold induction (column 6). This information is included for the PRGs with: (C) strong κB motifs and strong RelA binding, (D) strong κB motifs that do not support RelA binding, (E) weak κB motifs and strong RelA binding, and (F) other NF-κB and IκB family members.
(G) A line graph is shown indicating the p50:RelA motif Z score enrichment in the promoters of the PRGs relative to the promoters of uninduced genes. See also Figure S5.
When focusing on the promoters of the 132 strongly induced PRGs, a motif Z score threshold readily emerged that resulted in a high probability of a strong ChIP-seq peak; 37 of 44 promoters (84%) containing an NF−κB motif exceeding a Z score of 6.4 supported strong RelA binding (ChIP-seq peak >19), whereas only 20 of 88 promoters (23%) whose strongest NF-κB motif was below this motif threshold supported strong binding (Figure 4A, left, 4B, left). These results suggest that, although a high percentage of NF-κB genomic interactions do not coincide with strong binding motifs (see Figure 4A, right), most interactions observed at the promoters of a well-defined set of PRGs are associated with strong motifs. Thus, NF-κB function may often require binding to a near-consensus motif. The results also suggest that, at the promoters of this well-defined set of genes, a surprisingly strict motif strength threshold exists, in which promoter motifs exceeding this threshold almost always support strong in vivo binding (see below). This in vivo threshold contrasts with the continuum of binding affinities observed in vitro (Siggers et al., 2012).
To evaluate the significance of these findings, we examined promoters for all other annotated genes separated into five groups: the 132 strongly induced PRGs, the 94 strongly induced SRGs, 732 genes induced between 2 and 10 fold, 1732 genes that were expressed at a nascent transcript level >3 RPKM but without induction, and the remaining 18,487 annotated genes. Promoters within each group were separated into six classes on the basis of their ChIP-seq peak scores and motif scores, including three ChIP-seq categories (no binding, peak strength <19, and peak strength >19) combined with two motif categories (Z score <6.4 and >6.4) (Figure 4B).
An examination of the ChIP-seq/motif categories for the five groups of annotated genes revealed extensive enrichment of genes whose promoters combined strong ChIP-seq peaks and NF-κB motifs among the strongly induced PRG class. Specifically, whereas 28% (37/132) of the strongly induced PRGs combined strong ChIP-seq peaks and motifs, only 1.6% (27/1723) of expressed but uninduced genes combined strong peaks and motifs. Importantly, little or no enrichment of strongly induced PRGs was observed in four of the other ChIP-seq/motif categories (weak peak/strong motif, weak peak/weak motif, no peak/strong motif, no peak/weak motif). Substantial but lesser enrichment in the PRG class was observed for only one other ChIP-seq/motif category: those that combined a strong ChIP-seq peak with a weak motif (15.2% of strongly induced PRGs versus 3.8% of expressed uninduced genes).
The strong enrichment of promoters that combine strong ChIP-seq peaks and motifs in the group of 132 PRGs suggests that most or all of the 37 PRGs possessing these properties may be directly activated by RelA-containing dimers via direct promoter binding. Furthermore, the ability to define a motif Z-score threshold above which 84% of promoters supported strong NF-κB binding suggests that a single strong NF-κB motif is usually sufficient to support strong binding. Notably, although several of the 37 promoters contain two or more near-consensus NF-κB binding motifs, a strong correlation was not found between the number of NF-κB motifs and either the strength of the RelA ChIP-seq peak or the magnitude of transcriptional induction (data not shown). It is also noteworthy that ChIP-seq experiments examining the NF-κB p50 subunit revealed strong peaks at all 37 promoters that contain RelA peaks and motifs (data not shown), suggesting that the promoters are typically bound by RelA:p50 heterodimers.
The substantial but lesser enrichment of promoters with strong NF-κB peaks (score >19) and weak binding motifs (Z score <6.4) among the strongly induced PRGs is also of interest. In these promoters, NF-κB may bind directly to weak motifs. Alternatively, NF-κB may be recruited by other transcription factors, or the NF-κB ChIP-seq signal could be due to looping of an NF-κB-bound enhancer to the promoter. Although the significance of these interactions remains unknown, our ability to classify these promoters and distinguish them from the more prevalent promoters that combined strong ChIP-seq peaks and motifs will facilitate future studies of their regulation.
An examination of the 732 genes induced by 2-10-fold provides additional insights. A higher percentage of genes in this weakly induced class (5.9%) contain strong NF-κB peaks and motifs than in the class of genes that is expressed but not induced (1.6%). This enrichment suggests that a subset of weakly induced genes is regulated by NF-κB binding to strong motifs. However, a much smaller percentage of genes in this 2-10-fold induced class (5.9%) combine strong NF-κB peaks and motifs than in the strongly induced PRG class (28%), suggesting that a much smaller fraction of the weakly induced genes is regulated by NF-κB promoter binding.
Examination of NF-κB Regulated Genes
A major goal of this study was to elucidate the logic through which the lipid A-induced transcriptional cascade is regulated. The identities of the 37 strongly induced PRGs that combine strong ChIP-seq peaks and strong motifs provide compelling evidence of an underlying logic; specifically, more than a third (13 of 37, Figure 4C) encode NF-κB or IκB family members or key regulators of NF-κB activation, including three NF-κB family members (Nfkb1, Nfkb2, and RelB), five IκB family members (Nfkbia, Nfkbib, Nfkbid, Nfkbie, and Nfikbiz), two NF-κB-inducing receptors (Tlr2 and Cd40), and three regulators of NF-κB signaling (Tnfaip3, Tnip3, and Traf1). Strikingly, these 13 genes include all of the NF-κB/IκB family members and direct regulators of NF-κB signaling found among the 132 PRGs. Notably, the promoters of genes encoding the two NF-κB family members and one IκB family member missing from this list also combine a strong RelA ChIP-seq peak with a strong NF-κB motif (Figure 4F); these genes were not among the 132 PRGs because they were only weakly induced.
The 37 PRGs in Figure 4C contain only 21 distinct motifs, which adhere to one of two motif definitions: (G/T)GG(G/A)(N)(A/T)(T/G)(T/C)CC (17 motifs) or (G/A)GGGG(G/A)(T/A)TT(T/C) (4 motifs). The finding that a high level of similarity to the optimal NF-κB consensus is usually associated with NF-κB binding in the RelA ChIP-seq experiments was initially surprising. However, support for the significance of this finding emerged from an examination of binding motif enrichment at the 132 PRGs in comparison to the 1,723 expressed but uninduced genes, without any consideration of ChIP-seq data. Specifically, motifs with Z scores above 8.0 were strongly enriched among the promoters of the 132 PRGs. Motifs with Z scores between 6.0 and 7.9 were weakly enriched, but no enrichment was observed with motifs with Z scores below 6.0 (Figure 4G).
One remaining question is the reason seven promoters with motifs exceeding the threshold of 6.4 did not support RelA binding (Figure 4A,B). The motifs in three of these promoters possess very high Z scores (8.4-8.6, Figure 4D). However, two of these motifs are at a distance upstream of their TSS (−310 and −395) that exceeds the distance observed in all but five of the 37 promoters that support NF-κB binding (Figure 4D). We hypothesize that these two motifs do not support binding in vivo because they are occluded by nucleosomes. The third strong motif is located farther downstream from the TSS (+137) than the motifs found in any of the promoters that support strong NF-κB binding, suggesting that this motif may also be masked by a nucleosome.
The three remaining motifs possess Z scores between 6.7 and 7.4 (Figure 4D). We speculate that their Z scores may be defined imperfectly due to limitations of the PBM method. One of these motifs is found in two different promoters, neither of which supports binding, and the other two do not conform to the motif definitions derived from the 21 motifs that support binding (see above). Detailed affinity measurements will be needed to better understand why a few motifs fail to support NF-κB binding, but this quantitative analysis reveals a remarkably strong ability to predict NF-κB promoter binding in vivo on the basis of in vitro motif strength, as well as a motif strength threshold below which the probability of in vivo binding is greatly diminished.
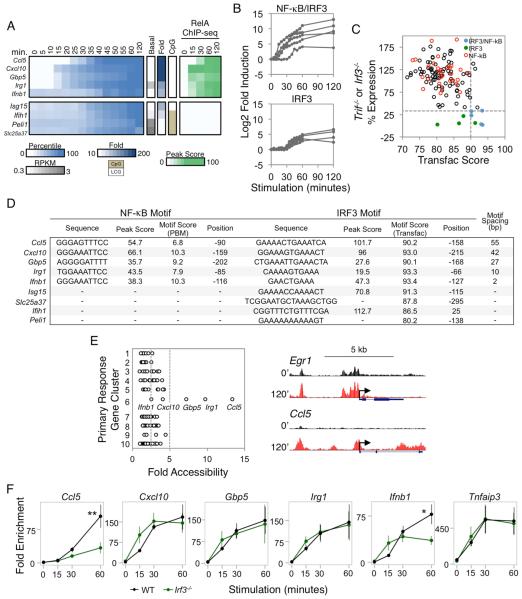
Kinetic and Functional Analysis of Putative NF-κB Targets
To test the prediction that the 37 PRGs described above are regulated by NF-κB, we examined their activation kinetics and RelA dependence. The initial upregulation of most of the genes occurred 10-20 min post-stimulation, as is evident from the third panel in Figure 5A, in which the fold-increase in RPKM relative to the preceding time point is highlighted. Although most of these genes are initially upregulated at the same time, their overall expression kinetics are diverse (Figure 5A, second panel; see also Figure S4), implicating other factors in their regulation. Consistent with this suggestion, the NF-κB target genes that depend on MAPK signaling were, on average, induced slightly earlier than the other putative target genes (Figure 5A, Cluster 2; Figure 5C).
To examine RelA dependence, we compared WT and Rela−/− fetal liver-derived macrophages by RNA-seq. Most of the putative NF-κB targets exhibited RelA dependence (Figure 5A, RelA−/− column), although the degree of dependence varied considerably, possibly due to redundancy between RelA and other NF-κB family members.
We next asked whether the activation kinetics and RelA-dependence observed in Figure 5A are unique to genes whose promoters contain strong ChIP-seq peaks and motifs. Interestingly, several other PRGs exhibited similar activation kinetics and/or degrees of RelA dependence (Figure 5B,D). A subset of these genes contains RelA ChIP-seq peaks in their promoters without strong motifs, but most lack RelA peaks (Figure 5B, right). We speculate that NF-κB directly regulates these genes by binding to distant enhancers. Consistent with this possibility, strong RelA ChIP-seq non-promoter peaks (peak score >19) were found in the vicinity of many of the PRGs (Figure S5). Thus, NF-κB may regulate strongly induced PRGs through either promoter or enhancer binding, with an underlying logic suggested by the fact that promoter binding characterizes genes encoding NF-κB/IκB family members and other NF-κB regulators.
Gene-Specific Regulation of IRF3-Dependent Genes
Although most studies emphasize large clusters of co-regulated genes, the above data suggest that, when induction magnitudes are considered, the unique features of individual genes and small clusters of genes begin to emerge. This concept is further exemplified by an examination of PRGs dependent on the transcription factor, IRF3. As shown in Figure 3, only 9 strongly induced PRGs exhibited expression levels in both Irf3−/− and Trif−/− BMDMs that fail to reach 33% of the level observed in WT. Five of these genes are within the group of 37 PRGs containing strong NF-κB ChIP-seq peaks and motifs in their promoters (Figure 5A). One notable difference between the five genes containing NF-κB motifs and the four lacking NF-κB motifs is that the induction magnitude of the former group is much higher than that of the latter, with average induction magnitudes of 643- and 40-fold, respectively (Figure 6A,B).
Figure 6. Analysis of IRF3 Target Genes.
(A) PRGs exhibiting IRF3 dependence (<33% expression in both Irf3−/− and Trif−/− macrophages) were separated based on the presence or absence of strong NF-κB promoter motifs and RelA ChIP-seq peaks. Colors indicate the percentile of the relative expression. Also shown are the basal RPKM, fold induction magnitude, and promoter CpG content. The rightmost heatmap indicates the RelA ChIP-seq binding peak scores.
(B) The fold induction for each IRF3-dependent gene is shown over the 2-hr time period, grouped based on their additional requirement for NF-κB.
(C) For each PRG, the higher maximum percent expression from either Trif−/− or Irf3−/− BMDMs (y-axis) was assessed against the best scoring IRF3 motif (x-axis) within the promoter based on the IRF Transfac PWM. The five IRF3/NF-κB genes are shown in blue, and the four IRF3 genes are shown in green. The PRGs containing strong NF-κB promoter motifs and RelA ChIP-seq peaks are shown in red. The horizontal dashed line indicates the expression threshold (33%), and the vertical dashed line indicates the Transfac threshold (90).
(D) For each IRF3-dependent gene, the IRF3 and RelA:p50 binding sites (for the IRF3/NF-κB groups of genes) were identified. The spacing between the NF-κB and IRF3 motifs is indicated at the right. The strengths of the κB motifs are represented by PBM Z scores, and the strengths of the IRF motifs are represented by PWM Transfac scores. For the four genes lacking NF-κB motifs, the best IRF promoter motif is shown.
(E) Left: The fold increase in ATAC-seq RPM at gene promoters (x-axis) is shown according to the PRG clusters 1-10 (y-axis) where the cluster designations denote 1:SRF, 2:MAPK, 3:MAPK/NF-κB, 4:NF-κB/Iκβ regulator, 5:NF-κB/Other, 6:NF-κB/IRF3, 7:NF-κB/Enhancer, 8:TRIF, 9:IRF3, 10:Unknown (see also Figure S6). The vertical dashed lines indicate the 2.5- and 5-fold cutoffs. Right: UCSC Genome Browser tracks of chromatin accessibility in resting and 120-min stimulated BMDMs at the promoters of two genes from different gene clusters are shown.
(F) RelA ChIP-qPCR was performed using WT and Irf3−/− BMDMs stimulated with lipid A. The relative enrichment of RelA binding was normalized to a negative control region. The RelA binding kinetics at the promoters of the 5 NF-κB/IRF3 genes were compared to the Tnfaip3 promoter as a control (far right). The data shown represent an average of 3 biological replicates. Error bars indicate the standard error. ** P <0.01; * P <0.05.
An examination of the five genes exhibiting both NF-κB promoter binding and IRF3 dependence reveals the extent to which genes have evolved unique regulatory strategies. Within this group, the expression kinetics of Ccl5 and Ifnb1 are each unique, whereas Cxcl10, Gbp5, and Irg1 are similar (Figure 6A). These latter three genes were initially induced 10-15 min post-stimulation along with most NF-κB-dependent genes. Consistent with the hypothesis that NF-κB contributes to this early induction, RelA ChIP-seq peaks were observed at these genes by 15 min post-stimulation (Figure 6A, right), and their induction at early time points was unaltered in Irf3−/− macrophages (data not shown). IRF3-dependence was observed only at later times, consistent with prior knowledge that IRF3 activation is relatively slow (Kagan et al., 2008).
Interestingly, Ccl5 is unique in that RelA binding was not observed until the 30-min time point; at all other PRGs bound by RelA, RelA binding was readily detected at the 15-min time point (Figure 5A,B, right; Figure 6A, right). The delay in RelA binding correlates with the delayed Ccl5 activation. Thus, RelA binding to this promoter requires an additional event that is unique among PRGs.
Ifnb1 regulation also appears unique. Ifnb1 induction was not observed until the 35-min time point, but RelA binding was observed by 15 min (Figure 6A, right). This early binding is consistent with evidence that the promoter lacks a nucleosome in unstimulated cells (Agalioti et al., 2000). Nevertheless, the delayed induction is consistent with evidence that activation is strongly dependent on IRF3 (Panne et al., 2007).
Figure 6C shows the distribution of promoter IRF motif scores relative to functional dependence on IRF3. IRF motifs with Transfac Position Weight Matrix (PWM) scores of 90 or greater accompany the strong NF-κB motifs in all five promoters (Figure 6C,D). The distances between the IRF3 and NF-κB motifs range from 2 to 55 bp (Figure 6D). Notably, of the four IRF3-dependent genes that do not contain NF-κB promoter motifs, only one (Isg15) contains an IRF3 motif of similar strength to those found in the genes with strong NF-κB motifs (Figure 6C,D).
The above results support a hypothesis in which multiple distinct mechanisms regulate the nine IRF3-dependent genes. To examine this hypothesis further, three additional experiments were performed. First, IRF3 ChIP-seq experiments revealed that strong IRF3 peaks (>19) coincide with strong IRF motifs (>90) at the promoters of only six of the 132 primary response genes, including the five NF-κB/IRF3 genes described above and the IRF3-dependent Isg15 gene that lacks NF-κB binding (see peak scores in Figure 6D; a detailed analysis of the IRF3 ChIP-seq data will be presented elsewhere). An IRF3 ChIP-seq peak was also observed in the promoter of one of the IRF3-dependent genes that lacks a strong IRF motif (Ifih1; Figure 6D).
Second, ATAC-seq experiments revealed weak increases in chromatin accessibility upon lipid A stimulation at PRGs in many different classes (Figure 6E). However, the largest increase was observed at the Ccl5 promoter, with large increases also observed at the Irg1 and Gbp5 promoters (Figure 6E). The large increase at the Ccl5 promoter is consistent with the hypothesis that a nucleosome remodeling requirement is responsible for the delayed binding of RelA. Furthermore, the absence of an inducible ATAC-seq signal at the Ifnb1 promoter is consistent with prior evidence that the promoter is nucleosome-free prior to stimulation. However, the strong increases in ATAC-seq signal at the Irg1 and Gbp5 promoters were surprising, given the rapid RelA binding and induction of these genes.
The third experiment performed was ChIP-qPCR examining RelA binding in Irf3−/− macrophages. This experiment revealed strong IRF3-dependence of RelA binding to the Ccl5 promoter (Figure 6F), consistent with our evidence from nuclease accessibility experiments that IRF3 is important for nucleosome remodeling at this promoter (Ramirez-Carrozzi et al., 2009). At the Ifnb1 promoter, the initial binding of RelA was not dependent on IRF3 (Figure 6F); however, the increase in RelA binding at later time points exhibited IRF3-dependence, consistent with the notion that IRF3 stabilizes RelA binding while promoting synergistic transcriptional activation (Agalioti et al., 2000; Panne et al., 2007). Finally, although potently induced ATAC-seq signals were observed at the Irg1 and Gbp5 promoters, RelA binding to these promoters was not IRF3-dependent (Figure 6F). Thus, the nucleosome remodeling observed at these promoters by ATAC-seq is likely to be dictated by NF-κB itself or by other rapidly induced factors.
Together, these results support a model in which the mechanisms by which NF-κB and IRF3 regulate the Ccl5 and Ifnb1 genes are unique, with these two transcription factors contributing to Irg1 and Gbp5 activation (and possibly Cxcl10 activation) by a third distinct mechanism. To determine whether these mechanisms appear to be unique only because we focused on a stringently defined group of PRGs, we asked whether any additional annotated promoters throughout the genome could be identified that possess the basic DNA properties of the five NF-κB/IRF3 genes (i.e. a strong RelA ChIP-seq peak [>19], a strong NF-κB motif [Z score >6.4], a strong IRF3 motif [Transfac score ≥90], and a distance between the NF-κB and IRF3 motifs of less than 100 bp [see Figure 6D]). Strikingly, only six additional promoters from among the 21,168 annotated promoters share these properties (data not shown).
Together, these results reveal the extent to which a quantitative, gene-centric analysis can begin to move toward an understanding of the unique molecular mechanisms used to regulate key genes in the transcriptional cascade. Although previous ChIP-seq studies led to the hypothesis that IRF3 and NF-κB cooperatively activate hundreds of genes (Freaney et al., 2013), the results presented here demonstrate that only five PRGs induced greater than 10-fold by lipid A combine strong NF-κB promoter binding, strong IRF3-dependence, a strong IRF3 promoter motif, and strong IRF3 binding, yet with at least three distinct modes of collaboration between NF-κB and IRF3 among these five genes. Although IRF3 can also bind many enhancers (Freaney et al., 2013), these interactions may have more subtle modulatory functions in lipid A-stimulated macrophages or may represent opportunistic binding events that lack functional consequences.
Regulation of Transiently Transcribed Genes by Serum Response Factor (SRF)
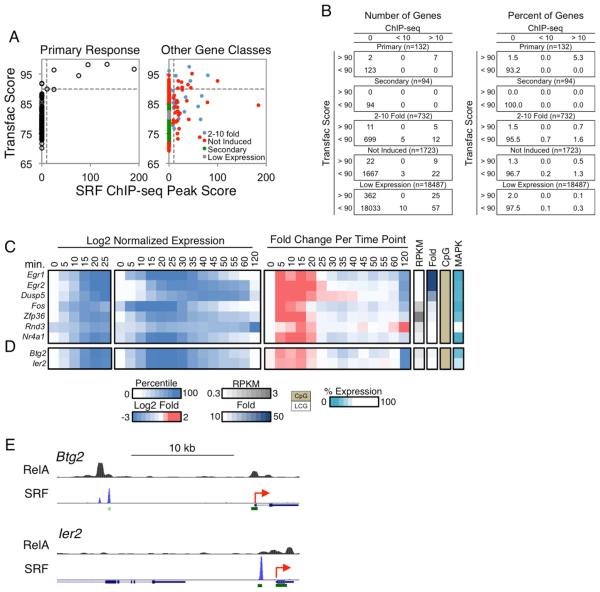
The most distinctive cluster of genes in Figure 3 is arguably the MAPK-dependent Cluster 6, which contains genes that exhibit rapid yet transient upregulation within 5 min of lipid A stimulation. This cluster contains only three genes, Egr1, Fos, and Nr4a1, yet the initial motif analysis (Figure S3) suggests enrichment of promoter binding sites for SRF. We therefore examined SRF binding by ChIP-seq in BMDMs stimulated with lipid A for 0, 15, 30, 60, and 120 minutes. SRF peaks remained unchanged through the time course, consistent with knowledge that SRF binds its targets constitutively, with inducible activity due to the induction of co-regulatory ternary complex factors (TCFs, Treisman, 1994).
The SRF ChIP-seq data sets yielded the strongest peaks we have detected and the greatest specificity of binding, with only a small number of strong peaks and very little background. A simultaneous examination of ChIP-seq peaks and Transfac PWM-defined motifs revealed that only seven of the 132 PRGs contain promoters with strong ChIP-seq peaks (peak score >10); all seven promoters contain strong motifs (Transfac score >90) (Figure 7A). No strong ChIP-seq peaks were observed at these promoters in the absence of a strong motif and only two promoters contained a strong motif without a strong ChIP-seq peak; both of these motifs are far from their TSS (−306 and −331), suggesting that they may be occluded by nucleosomes. Thus, to even a greater extent than observed with NF−κB, strong binding of SRF correlated closely with motif strength, leading to a motif threshold that may be both necessary and sufficient for SRF binding in the context of a well-defined set of promoters.
Figure 7. Analysis of SRF Target Genes.
(A) Scatterplots comparing the Transfac PWM scores of SRF binding motifs (y-axis) versus the SRF ChIP-seq peak scores (x-axis) in the promoters (−500 to +150) of the PRGs (left) and all remaining genes in the genome (right) is shown. The genes in the latter graph were divided into categories as in Figure 4A. The horizontal and vertical dashed lines indicate the SRF motif (90) and ChIP-seq peak (10) thresholds.
(B) Tables are shown indicating the distribution of genes from panel (A), for both numbers (left) and percentages (right) of genes.
(C) Log2 normalized expression values from 0-25 min (first panel), 0-120 min (second panel), and the fold induction relative to the expression level at the previous time point (third panel) are shown for the seven putative SRF target genes. To the right are columns indicating the basal expression level, fold induction magnitude, promoter CpG content, and MAPK dependence for each gene.
(D) Two genes that exhibited similar activation kinetics as the putative SRF target genes are shown, with the same layout as in Figure 7C.
(E) The two genes from panel (D) were examined on UCSC Genome Browser to identify distal SRF binding peaks. RelA binding peaks were also examined for these genes. The TSSs of the genes are indicated as red arrows, and the green rectangles indicate CpG islands. See also Figures S6 and S7.
Surprisingly, only 39 additional promoters within the remaining 21,036 annotated genes reached the same peak and motif thresholds achieved by the seven binding events at the primary response genes (Figure 7A,B). Instead, the vast majority of binding events at other gene classes coincided with weak motifs (Figure 7A,B).
A closer examination of the seven genes that combine strong SRF ChIP-seq peaks and motifs supports the hypothesis that at least six are functional targets of SRF. This group of seven genes includes the three found in cluster 6 of Figure 3A (Egr1, Fos, and Nr4a1) and four additional genes (Egr2, Dusp5, Zfp36, and Rnd3). All but Rnd3 were initially upregulated during the first 5 min of lipid A stimulation (Figure 7C, third panel), and all but Rnd3 exhibited MAPK dependence. MAPKs are responsible for activation of the TCFs (Treisman, 1994). The fact that Rnd3 exhibited different properties suggests that this gene may instead require a second class of SRF co-activator proteins that are not activated by MAPKs (Posern and Treisman, 2006).
An examination of the expression kinetics of the seven genes explains why only three were placed in the same kinetic cluster in Figure 3A: these three genes exhibited relatively uniform induction and repression kinetics, whereas Egr2, Dusp5, and Zfp36, although initially induced by 5-min, were either further upregulated at later time points or were upregulated less potently and downregulated more slowly, presumably due to the contributions of other factors.
Lastly, an analysis of the 132 PRGs led to the identification of only two additional genes that exhibit similarly rapid induction kinetics as the six genes discussed above: Btg2 and Ier2 (Figure 7D). These two genes lack SRF ChIP-seq peaks and motifs in their promoters, but instead contained strong promoter NF-κB ChIP-seq peaks. This finding raises the question of how these two genes achieve induction kinetics similar to those of the genes whose promoters are directly bound by SRF. Interestingly, both of these genes contain strong SRF ChIP-seq peaks at upstream regions (Figure 7E); in both instances, the SRF peaks coincide with CpG islands and are conserved through evolution (data not shown). The SRF peaks at the Btg2 and Ier2 loci are 10 kb and 1 kb upstream of their TSSs, respectively. Remarkably, only three other PRGs contained SRF ChIP-seq peaks within the region 10 kb upstream of their TSS, indicating that this property is rare. These results suggest that SRF contributes to the early transient induction of these genes by cooperating with NF-κB bound to the promoters.
DISCUSSION
Broad systems analyses of gene expression cascades and networks continue to provide important biological and mechanistic insights. However, the focus of most conventional studies on large numbers of genes or ChIP-seq peaks meeting low-stringency criteria, for the purpose of optimizing statistical power, possesses significant limitations. The results described here demonstrate that, toward the goal of a mechanistic understanding of transcriptional control at a genome-wide level, it is not only possible, but often preferable, to use more stringent and quantitative approaches to examine RNA-seq, ChIP-seq, and binding motif data sets.
This approach allowed us to obtain evidence that a single NF-κB or SRF motif that reaches a defined threshold consistently supports factor binding and function in vivo. Moreover, we obtained evidence of an underlying logic through which NF-κB may regulate distinct sets of genes by binding to promoters versus enhancers. We speculate that promoter binding may be compatible with transcriptional induction in response to any stimulus that induces NF-κB activity, whereas enhancer binding may often be preferred at genes that require cell type-specific induction. Of greatest interest, our results reveal that, even when two inducible factors (e.g. NF-κB and IRF3) act in concert to regulate a small cluster of only five genes, individual genes within the cluster are regulated by unique mechanisms. Overall, the results of this analysis provide a wealth of mechanistic insights that are accessible to future exploration.
Figures S6 and S7 display summaries of the results obtained in this study. Notably, although the study derived great benefit from detailed kinetic analyses of chromatin-associated transcripts, diverse overall expression kinetics are observed within each cluster. This observation supports the long-standing view that multiple transcription factors act in concert to shape the expression pattern of each gene.
The findings described here contrast with ChIP-seq studies that implicate key transcription factors in the regulation of hundreds or thousands of genes. It is important to emphasize that our study focused on the properties of the limited number of potently induced genes for which NF-κB, IRF3, and SRF appear to be major regulators, but they do not rule out the possibility that these same factors play more subtle roles in the regulation of hundreds of additional genes. For example, although only a small fraction of genes induced by 2-10-fold contain promoter binding sites for NF-κB, this factor may contribute to the induction of a large fraction of weakly induced genes by binding to distant enhancers. Nevertheless, the current results document a clear distinction between strongly induced and weakly induced genes with respect to the prevalence of NF-κB promoter binding and promoter motifs, providing a framework for studies to elucidate the diverse mechanisms by which NF-κB contributes to the lipid A response. Similarly, the ability to identify consistent properties of genes that appear to be regulated by SRF, IRF3, and IRF3/NF-κB provides a step toward a precise understanding of the broad mechanisms regulating the transcriptional cascade. In addition to exploring the insights obtained in greater depth, an important goal for the future will be to continue building on this framework by using stringent approaches to examine additional signaling pathways, transcription factors, and chromatin regulators, while extending the analysis to enhancers, weakly induced genes, and gene expression cascades induced by diverse stimuli in diverse cell types and physiological settings.
EXPERIMENTAL PROCEDURES
Cell Culture
BMDMs were prepared from 6-week-old C57BL/6, Myd88−/−, Trif−/−, Irf3−/−, or Ifnar−/− male mice. Fetal liver macrophages were from D14.5 C57BL/6 or RelA−/− embryos. Macrophages were activated on day 6 with 100 ng/ml lipid A (Sigma) or Pam3CSK4 (InvivoGen). When indicated, cells were preincubated for 15 min with 10 mg/ml CHX or 1 hr with 10 μM PD0325901 (Sigma) and 1 μM BIRB0796 (AXON Medchem).
RNA-seq
Total RNA and chromatin-associated RNA were prepared as described (Bhatt et al., 2012). Strand-specific libraries were generated from 60 ng chromatin RNA or 400 ng total RNA using the TruSeq RNA Sample Preparation Kit v2 (Illumina) and the dUTP second strand cDNA method (Levin et al., 2010). cDNA libraries were single-end sequenced (50bp) on an Illumina HiSeq 2000.
Reads were aligned to the mouse genome (NCBI37/mm9) with TopHat v1.3.3 and allowed one alignment with up to two mismatches per read. Chromatin RNA RPKM values were calculated by dividing all mapped reads within the transcription unit by the length of the entire locus. mRNA RPKM values were calculated by dividing mapped exonic reads by the length of the spliced product.
All RPKMs represent an average from two or three biological replicates. A gene was included in the analysis if it met all of the following criteria: The maximum RPKM reached 3 at any time point, the gene was induced at least 10-fold, and the induced expression was significantly different from the basal (P<0.01) as determined by the edgeR package in R Bioconductor (Robinson et al., 2010). Additionally, a gene was included if its induction reached 5-fold at the 15-min time point. P-values were adjusted using the Benjamini-Hochberg procedure of multiple hypothesis testing (Benjamini and Hochberg, 1995).
To determine the impact of a perturbation, the basal RPKM in WT samples was set at 0% and the maximum RPKM at 100% for each gene. The maximum RPKMs in the mutant samples were converted to percent expression using this scale. For the Rela−/− analysis, the RelA-dependence of a gene was determined by the percent expression in Rela−/− samples at the earliest time point in which the WT samples were induced by at least 3-fold.
ChIP-seq
ChIP-seq libraries were prepared using the Kapa LTP Library Preparation Kit (Kapa Biosystems). ChIP-seq was performed as described (Barish et al., 2010; Lee et al., 2006) with minor modifications, using anti-RelA (Santa Cruz, sc-372), anti-IRF3 (Santa Cruz, sc-9082), or anti-SRF (Santa Cruz, sc-335) antibodies.
Reads were aligned to the mouse genome (NCBI37/mm9) with Bowtie2. Uniquely mapped reads were used for peak calling and annotation using HOMER (Heinz et al., 2010). Peaks were called if they passed a false discovery rate of 0.01 and were enriched over input. Called peaks were considered for downstream analysis if peaks from at least 4 of 7 replicates were overlapping within 200 bp for RelA and 5 of 5 replicates were overlapping within 300 bp for SRF. Peaks were annotated to the nearest TSS.
ATAC-seq
ATAC-seq libraries were prepared using the Nextera Tn5 Transposase kit (Illumina) as described (Buenrostro et al., 2015) with slight modifications. Libraries were single-end sequenced (50bp) on an Illumina HiSeq 2000. Reads were mapped to the mouse genome (NCBI37/mm9) using Bowtie2. Reads were removed from the subsequent analysis if they were duplicated, mapped to mitochondrial genome, or aligned to unmapped contiguous sequences. Promoter accessibility was calculated by totaling all reads within −500 to +150 bp relative to the TSS. The reads were converted to reads per million (RPM) by dividing by the total number of reads per sample. The average RPM from four replicates was used to quantify the fold increase in promoter accessibility.
Motif Analysis
The promoters of genes (−500 to +150 bp) were used for motif analysis unless otherwise indicated. The strongest p50:RelA binding site within each promoter was identified using a PBM dataset (Siggers et al., 2012). Transfac PWMs were used to identify the best matching SRF and IRF3 binding sites in promoters using Pscan (Zambelli et al., 2009).
Supplementary Material
Figure 1. Properties of the Lipid A-Induced Transcriptional Cascade.
Chromatin-associated transcripts from BMDMs stimulated with lipid A were analyzed by RNA-seq.
(A) The distribution of maximum fold induction values over the 2-hr stimulation period is shown for the 1,340 significantly induced (2-fold, p<0.01) and expressed (3 RPKM) genes. With multiple hypothesis testing, two weakly induced genes exhibited q values >0.01. The dashed gray lines represent 5-, 10-, and 50-fold induction thresholds.
(B) The 1,340 induced genes were grouped into bins, with basal RPKMs shown for each bin and red dashes indicating median RPKMs.
(C) The distributions of maximum fold inductions (left), peak RPKMs (top right), and basal RPKMs (bottom right) are shown for the 226 genes selected for analysis.
(D) The 226 genes were separated into PRG and SRG groups on the basis of their expression in CHX-treated and Ifnar−/− BMDMs. Genes were classified as SRGs if they were expressed <33% in CHX or <30% in Ifnar−/− samples. The Venn diagram indicates the number of genes affected by CHX treatment, the absence of IFNAR, or both.
Highlights.
An underlying logic distinguishes NF-κB promoter binding from enhancer binding NF-κB and IRF3 collaborate via diverse mechanisms to activate key inflammatory genes Genes induced most rapidly and transiently by lipid A invariably support SRF binding
ACKNOWLEDGMENTS
We thank Christopher Glass, Alexander Hoffmann, Steven Ley, Ranjan Sen, and Trevor Siggers for helpful discussions, and the UCLA Broad Stem Cell Research Center Core for sequencing. This work was supported by NIH grants R01GM086372 (S.T.S.), P50AR063030 (S.T.S.), T32CA009120 (A.J.T.), and T32GM008042 (B.J.T.), and by the China Scholarship Council and Whitcome pre-doctoral training program (X.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All data are accessible through GEO Series accession number GSE67357.
AUTHOR CONTRIBUTIONS
A.J.T., X.L., B.J.T., and M.M.L. designed and performed most experiments and wrote the manuscript. M.R.B., M.D.S., A.L.A., and G.D.B performed the RelA ChIP-seq experiments, and S.T.S. designed experiments and wrote the manuscript.
REFERENCES
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered Recruitment of Chromatin Modifying and General Transcription Factors to the IFN-β Promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-κB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. Current Protocols in Molecular Biology. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2015. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide; pp. 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freaney JE, Kim R, Mandhana R, Horvath CM. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 2013;4:959–973. doi: 10.1016/j.celrep.2013.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. A High-Throughput Chromatin Immunoprecipitation Approach Reveals Principles of Dynamic Gene Regulation in Mammals. Mol. Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei C-L, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of Inducible Gene Expression by Signal-Dependent Transcriptional Elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C-A, Yao F, Wong JJ-Y, George J, Xu H, Chiu KP, Sung W-K, Lipovich L, Vega VB, Chen J, et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-kappaB upon TLR4 activation. Mol. Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone R, Euskirchen G, Bertone P, Hartman S, Royce TE, Luscombe NM, Rinn JL, Nelson FK, Miller P, Gerstein M, et al. Distribution of NF-kappaB-binding sites across human chromosome 22. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12247–12252. doi: 10.1073/pnas.2135255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe SM. LIF in the regulation of T-cell fate and as a potential therapeutic. Genes Immun. 2011;12:157–168. doi: 10.1038/gene.2011.9. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Sciumè G, Poholek AC, Vahedi G, Hirahara K, Villarino AV, Bonelli M, Bosselut R, Kanno Y, Muljo SA, et al. Transcriptional and epigenetic networks of helper T and innate lymphoid cells. Immunol. Rev. 2014;261:23–49. doi: 10.1111/imr.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 2012;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov V, Jones BW, Perera P-Y, Thomas K, Cody MJ, Zhang S, Williams BRG, Major J, Hamilton TA, Fenton MJ, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat. Immunol. 2002;3:392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Barrera LA, Ersing I, Willox B, Schmidt SCS, Greenfeld H, Zhou H, Mollo SB, Shi TT, Takasaki K, et al. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014;8:1595–1606. doi: 10.1016/j.celrep.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.