Summary
Estimation of the skeleton of a directed acyclic graph (DAG) is of great importance for understanding the underlying DAG and causal e ects can be assessed from the skeleton when the DAG is not identifiable. We propose a novel method named PenPC to estimate the skeleton of a high-dimensional DAG by a two-step approach. We first estimate the non-zero entries of a concentration matrix using penalized regression, and then fix the difference between the concentration matrix and the skeleton by evaluating a set of conditional independence hypotheses. For high dimensional problems where the number of vertices p is in polynomial or exponential scale of sample size n, we study the asymptotic property of PenPC on two types of graphs: traditional random graphs where all the vertices have the same expected number of neighbors, and scale-free graphs where a few vertices may have a large number of neighbors. As illustrated by extensive simulations and applications on gene expression data of cancer patients, PenPC has higher sensitivity and specificity than the state-of-the-art method, the PC-stable algorithm.
Keywords: DAG, Penalized regression, log penalty, PC-algorithm, skeleton, high dimensional
1. Introduction
Many statistical methods have been developed to identify the associations between genomic features and disease outcomes or cancer subtypes. However, such association results are descriptive in their nature, and they cannot deliver “actionable” conclusions for disease treatment. Many recently developed cancer drugs are so-called “targeted drugs” that target particular (mutated) proteins in cancer cells, and the mechanism of such drugs can be understood as direct interventions on tumor cells. To characterize or predict the consequences such drug interventions, statical methods that allow causal inference based on high dimensional genomic data are urgently needed.
One of the most commonly used tools for causal inference among a large number of random variables is the probabilistic directed acyclic graph (DAG) (also known as Bayesian Network) (Lauritzen, 1996; Pearl, 2009). In a DAG, all the edges are directed, and the direction of an edge implies a direct causal relation. There is no loop in a DAG. Such “acyclic” property is necessary to study causal relations (Spirtes et al., 2000). When we remove the directions of all the edges in a DAG, the resulting undirected graph is the skeleton of the DAG.
Estimation of the skeleton of a DAG is of great importance because it is a crucial step towards estimating the underlying DAG and skeleton itself may provide a limited amount of information for causal inference (Maathuis et al., 2009, 2010). Several methods have been developed to estimate DAGs or their skeletons from observational data (Heckerman et al., 1995; Spirtes et al., 2000; Chickering, 2003; Kalisch and Bühlmann, 2007), however most of them are not suitable for the high dimensional genomic problems that motivate our study. In this paper, we proposed a new method named PenPC to address this challenging problem. We proved the estimation consistency of PenPC for high dimensional settings of p = O (exp{na}) for 0 ≤ a < 1, and we also derived the conditions for estimation consistency for two types of graphs: random graphs where all the vertices have the same expected number of neighbors, and scale-free graphs where a few vertices have much larger number of neighbors than other vertices. As verified by both simulation and real data analyses, PenPC provides more accurate estimates of DAG skeletons than existing methods. In addition to skeleton, PenPC can further estimate the complete partially directed acyclic graph (CPDAG), which can used to estimate causal effects (Maathuis et al., 2009).
The remaining parts of this paper are organized as follows. In Section 2, we give a brief review of DAG estimation methods and the conceptual advantages of our PenPC algorithm. We present the details of the PenPC algorithm and its theoretical properties in Sections 3 and 4, followed by simulations and real data analyses in Section 5 and Section 6, respectively. Finally, we conclude with some discussions in Section 7.
2. Review of DAG Estimation
2.1 Directed Acyclic Graph (DAG)
A DAG of random variables X1, ..., Xp can be denoted by G = (V, E), where V contains p vertices 1, 2, ...., p that correspond to X1, ..., Xp, and E contains all the directed edges. In a DAG, a chain of length n from i to j is a sequence i = i0 – i1 – · · · – in–1 – in = j of distinct vertices such that il–1 → il ∈ E or il il–1 ∈ E for l = 1, . . . , n; and a path of length n from i to j is a sequence i = i0 → i1 → · · · → in = j of distinct vertices such that il–1 → il ∈ E for l = 1, ..., n. Given this path, il–1 is a parent of il, il is a child of il–1, i0, i1, ..., il–1 are ancestors of il, and il+1, ..., in are descendants of il.
Given a DAG G for random variables X1, . . . , Xp and assume that with density fX. Let Xpai be the parents of Xi. We say that the distribution PX is Markov to G if the joint density fX satisfies the recursive factorization: The factorization naturally implies acyclic restriction of the graph structure. Equivalently PX is Markov to G if every variable is conditionally independent of its non-descendants given its parents. A related concept is the so-called faithfulness:
Definition 1
Let PX be Markov to G. < G, PX > satisfies the faithfulness condtiontion if and only if every conditional independence relation true in PX is entailed by the Markov property applied to G (Spirtes et al., 2000).
This means that if a distribution PX is faithful to a DAG G, all conditional independences can be read o from the DAG G using d-separation defined in the following definition 2, and thus the faithfulness assumption requires stronger relationship between the distribution PX and the DAG G than the Markov property.
Definition 2
(d-separation). A vertex set S block a chain p if either (i) p contains at least one arrow-emitting vertex belonging to S, or (ii) p contains at least one collision vertex (j is a collision vertex if the chain includes i → j ← k) that is outside S and no descendant of the collision vertex belongs to S. If S blocks all the chains between two sets of random variables X and Y , we say “S d-separates X and Y” (Pearl, 2009).
Not all the distributions can be faithfully represented by a DAG. In this paper, we assume the random variables follow multivariate Gaussian distribution, then the faithfulness assumption can be justified by the fact that among all the multivariate Gaussian distributions associated with G, the non-faithful ones form a Lebesgue null set (Meek, 1995).
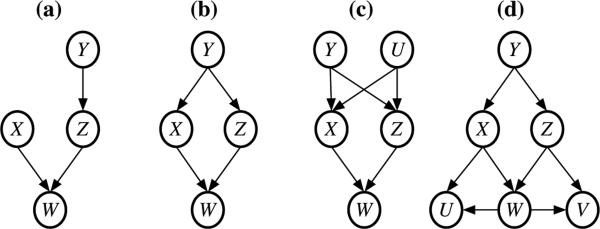
Given multivariate Gaussian distribution assumption, a commonly used graphical model is Gaussian Graphic Model (GGM), where two vertices are connected if the corresponding two variables are dependent, given all the other variables. A GGM can be constructed by a concentration matrix (i.e., precision matrix or inverse of covariance matrix) in that two vertices are connected if the corresponding elements in the concentration matrix is non-zero. The skeleton of a DAG is di erent from its GGM because of v-structures. In a v-structure X → W ← Z, co-parent X and Z are marginally independent or conditionally independent given their parents, but given every vertex set that contains W (a collision vertex) or any descendant of W , X and Z are dependent with each other. Note that by the definition of v-structure, the co-parents X and Z are not connected. A few examples are shown in Figure 1, and instances of the covariance and concentration matrices of the GGM in Figure 1(a) are shown in the Supplementary Materials, Section 1.
Figure 1.
Four DAGs where X and Z are not connected in the skeleton, but are connected in the corresponding GGMs. The true relation between X and Z can be revealed by appropriate conditional independence testing. For example, X ⊥ Z in Figure 1(a), X ⊥ Z|Y in Figure 1(b), X ⊥ Z|(Y, U) in Figure and 1(c), and X ⊥ Z|Y in Figure 1(d).
2.2 DAG estimation using observational data
In this paper we focus on DAG skeleton estimation using observational data instead of interventional data. When the p variables have a nature ordering (i.e., all the parents or ancestors of Xi are among the vertices X1, ..., Xi–1, and all the children or descendants of Xi are among vertices Xi+1, ..., Xp), the problem of skeleton estimation is greatly simplified because a regression of Xi versus X1, ..., Xi–1 can be used to identify the true skeleton (Shojaie and Michailidis, 2010). However, in many high-dimensional problems, such a nature ordering is not available. Throughout this paper, we assume no knowledge of nature ordering. Then the underlying DAG is not identifiable from observational data, because conditional dependencies implied by the Markov property on the observational distribution PX only determine the skeleton and v-structures of the graph (Pearl, 2009). All the DAGs with the same skeleton and v-structures correspond to the same probability distribution and they form a Markov equivalence class. After estimating skeleton, the v-structures can be identified by a set of deterministic rules, and thus we do not distinguish the estimation of a DAG skeleton and a Markov equivalence class.
In general, there are two approaches for DAG or DAG skeleton estimation. The first one is the search-and-score approach that searches for the DAG that maximizes or minimizes a pre-defined score, such as BIC (Bayesian Information Criterion) or L0-penalized maximum likelihood estimates (van de Geer and Bühlmann, 2013). Instead of searching across all the DAGs, which is often computationally infeasible, elegant methods have been developed to search across Markov equivalence classes (Chickering, 2003) or the nature orderings of the variables (Teyssier and Koller, 2005). However, these methods are still computationally very challenging for genomic applications with thousands of variables.
The second approach for DAG (skeleton) estimation is constraint-based approach that constructs DAGs by assessing conditional independence of random variables. One representative method is the PC algorithm (named after its authors, Peter Sprites and Clark Glymour) (Spirtes et al., 2000). Starting with a complete undirected graph where any two vertices are connected with each other, the PC algorithm first thins the complete graph by removing edges between vertices that are marginally independent. Then it removes edges by assessing conditional independence given one vertex, two vertices, and so on. Kalisch and Bühlmann (2007) proved the consistency of the PC-algorithm in high-dimensional settings where p = O(na) for a > 0. The results of the PC algorithm depend on the order of the edges to be assessed. Colombo and Maathuis (2012) proposed PC-stable algorithm, which modified the PC algorithm to remove such order dependency and substantially improve the performance of the PC algorithm. We consider the PC-stable algorithm as the state-of-theart method and compare our method with the PC-stable algorithm.
The Independence Graph (IG) algorithm (Chapter 5.4.3 of Spirtes et al. (2000)) modifies the PC algorithm by using a different initial graph: an undirected independence graph where two vertices are connected if the corresponding two variables are conditionally dependent given all the other variables, i.e., a GGM under multivariate Gaussian distribution assumption. In such an independence graph, the neighbors of a vertex Yj include its parents, children, and co-parents of v-structures in the underlying DAG, which constitute the so-called Markov blanket of Yj such that Yj is independent of all the other vertices given its Markov blanket.
The Max-Min Hill-Climbing (MMHC) algorithm is a popular hybrid method that first estimates DAG skeleton using a constraint-based method (the Max-Min part of the algorithm), and then orient the edges using a search-and-score technique (the Hill-Climbing part of the algorithm) (Tsamardinos et al., 2006). Schmidt et al. (2007) proposed to replace the Max-Min part of the MMHC algorithm by a penalized regression with Lasso (l1) penalty, i.e., neighborhood selection (Meinshausen and Bühlmann, 2006). Variable selection consistency of Lasso requires the irrepresentable condition (Zhao and Yu, 2006): there is weak correlation between the variables within and outside a Markov blanket. This is a strong condition and it generally does not hold for the genomic problems that motivate this study.
We propose a PenPC algorithm for DAG skeleton estimation in two steps. It first adapts neighborhood selection method to select Markov blanket of each vertex, and then it applies a modified PC-stable algorithm to remove false positive edges between co-parents of v-structures. Although the two-step approach of the PenPC algorithm shares similar spirit to the IG algorithm (Spirtes et al., 2000) and the modified MMHC algorithm (Schmidt et al., 2007), we have made the following novel contributions. First, we employ the log penalty p(|b|; λ, τ)= λlog(|b|+τ) (Mazumder et al., 2011) for neighborhood selection, which significantly improves the accuracy of Markov blanket search for higher dimensional problems, e.g., n = 30 and p = 100, or n = 300 and p = 1000. In contrast, Schmidt et al. (2007) explicitly assume n p in their paper. The resulting PenPC algorithm outperforms the state-of-the-art PC-stable algorithm and also enjoys some advantage in terms of computational efficiency in high dimensional settings. Second, we provide theoretical justifications of the estimation consistency of the PenPC algorithm in high dimensional settings where p = O (exp{na}) for 0 ≤ a < 1. We also discuss the implications for estimation consistency for two types of graphs: traditional random graph where all the vertexes have the same expected number of connections, and scale-free graph where a few vertices can have much larger number of neighbors than the other vertices. Whereas non-scale-free graph is often assumed in previous studies (Kalisch and Bühlmann, 2007), scale-free graph is more frequently observed in gene networks as well as many other applications (Barabási and Albert, 1999).
3. Methods
We adopt a multivariate Gaussian distribution assumption: X = (X1, . . . , Xp)T ~ N(0,Σ). Let X = (x1, ..., xp) be the n × p observed data matrix. Our PenPC algorithm proceeds in two steps: (1) neighborhood selection, and (2) application of a modified PC-stable algorithm to remove false connections. Theoretical justification of our algorithm is presented in Section 4.
Step 1. (Neighborhood Selection)
We first select the neighborhood of vertex i by a penalized regression with Xi as response variable and all the other variables corresponding to vertices V \ {i} as covariates:
| (1) |
where X–i is an n × (p – 1) matrix for n measurements of the remaining p – 1 covariates, bi = (bi,1, ..., bi,i–1, bi,i+1, ..., bi,p)T, and denotes a penalty function with one or more tuning parameters, denoted by . We consider a class of folded concave penalty functions satisfying the following condition:
Condition 1
The penalty function is concave in β ∈ [0, ∞), with continuous derivative , and .
This is a generalization of the Condition 1 in Fan and Lv (2011). In this study, we employ the log penalty p(|b|; λ τ) = λlog(|b| + τ), which has been demonstrated to have good performance in high-dimensional genetic studies (Sun et al., 2010). We solve penalized regression with log penalty using a coordinate descent algorithm (Sun et al., 2010), and the two tuning parameters λ and τ are selected by two-grid search to minimize extended BIC (Chen and Chen, 2008). After p penalized regressions for each of the p variables, we construct the GGM by adding an edge between vertices i and j if or .
Step 2. (Modified PC-stable algorithm)
We apply a modified PC-stable algorithm to remove the false edges between parents of v-structures. For each edge i – j, we first assess marginal association between vertices i and j. If they remain dependent, we test whether they are conditionally dependent. The conditional set should be selected from the Markov blanket of i and j, after excluding i and j's common children or descendants. Specifically. we use the following strategy to search for candidate separation sets. Let Ai,j be the Markov blanket of i and j, and let Ci,j be the set of vertices that could be common children or descendants of i and j. Then the candidate conditional sets are
| (2) |
Each element of Πi,j is a set Ai,j \ Di,j, where Di,j is exhaustively searched across all subsets of Ci,j. More details are described in the Supplementary Materials, Section 2.
We test the conditional independence of Xi and Xj given K ∈ Πi,j using Fisher transformation of partial correlation. Specifically, denote the partial correlation between Xi and Xj given K ∈ Πi,j by . With the significance level α, we reject the null hypothesis H0 : against the alternative hypothesis Ha : if , where and ϕ(·) is the cdf of N(0, 1).
The final output of PenPC algorithm is the estimated skeleton and separation sets S(i, j) for all (i, j). If vertices i and j are connected in the skeleton, the separate set is an empty set, otherwise Xi and Xj are independent given S(i, j), hence the name separation set. Given the skeleton and the separation sets, one can estimate CPDAG (Complete Partially Directed Acyclic Graphs) (Supplementary Materials Section 3) and then apply the idaFast or ida functions of R package pcalg (Kalisch et al., 2012) to estimate multi-set of possible causal effects.
4. Theoretical Properties
4.1 Fixed Graphs
We first introduce the following notations. For an m × n matrix A, denote the matrix Lb norm of A by , where x is a vector of length n, and . In particular, is the spectral norm of A, is the maximum absolute column summation, and is the maximum absolute row summation. Denote the vector Lb norm of A by |A|b. In particular, , and . Denote by Ai,–i the submatrix of A that includes the i-th row and excludes the i-th column of A. Ai,i, A–i,i and A–i,–i are defined similarly. For any compatible subsets S1 and S2, AS1S2 is the submatrix that contains all the rows with indices in S1 and all the columns with indices in S2.
We denote p as pn to emphasize it is a function of sample size n. Let a DAG and the corresponding GGM be Gn = (Vn, En) and CGn = (Vn, Fn), respectively. We further denote the skeleton of Gn by where or b → a ∈ En. For any vertex i, denote the observed centralized data of the variables within and outside of the neighbors of i in CGn (denoted by adj(i, CGn)), but not including Xi, by and , respectively, i.e., and where S0 = {1, 2, ..., n} and Si = {j : j ∈ adj (i,CGn)}.
The following Lemma 1 is a well-known conclusion that gives the relation between concentrate matrix of multivariate Gaussian distribution and the regression coe cients when we regress one variable versus all the other variables (Anderson, 2003).
Lemma 1
Suppose and . Then where , and independent of X–i.
With the aforementioned notations and definitions, we can state the following conditions that are needed for the consistency of the PenPC algorithm.
-
(A1)
Dimensionality of the problem. pn = O (exp{na}) with a ∈ [0, 1).
-
(A2)
Sparseness assumption. Let , i.e., the maximum degree of the Gaussian graphical model CGn. qn = O(nb) for some 0 ≤ b < (1 – a)/2. Let . By the following Lemma 2, M ≤ qn = O(nb).
-
(A3)Minimum e ect size for neighborhood selection.
-
(A4)
Conditions for the population covariance matrix . Let λmin(ΣS,S) be the minimum eigen-value of a sub matrix ΣS,S. For any S with . We also assume maxi σii < C2. Here C1 and C2 are two positive constants. Consequently C2 is also an upper bound of all the off-diagonal elements of Σ because
-
(A5)
Conditions for penalty function. Let for . Thus if the penalty function has continuous second derivative. Let and Ni is a hypercube around the vector such that . We assume , where C1 is defined in (A4) , , and .
-
(A6)Restriction on the size of conditional partial correlation. Denote the partial correlations between Xi and Xj given a set of variables for by . For K ∈ Πij (Πi,j was defined in equation (2)), the absolute values of 's are bounded:
where cn = O(n−d2) for some 0 < d2 < min{(1 – a)/2, (1 – b)/2}.
The sparseness assumption (A2) will be replaced by tighter assumptions for two specific random graph models later. Assumptions (A3)-(A5) ensure that the step 1 of PenPC can recover the partial correlation graph. There are fairly reasonable conditions to ensure the identifiability of the problem. Assumption (A3) requires the minimum e ect size is larger than noise level (e.g., larger than O(n−1/2) when p = O(1)). Assumption (A4) requires the covariance matrix for those important covariates in a neighborhood selection problem is not singular. Assumption (A5) are conditions for the penalty function, which can be easily satisfied by adjusting the two tuning parameters of the Log penalty (Chen et al., 2014). Assumption (A6) ensures the summation of the mistaken probabilities of the step 2 of the PenPC algorithm goes to 0 asymptotically. The condition of assumption (A5) deserves more discussions because it corresponds to the irrepresentable condition that limits the performance of Lasso regression. Specifically, in Supplementary Materials, we show that with probability approaching to 1. The assumption is needed so that . For the Lasso, = 1, and thus this is a very strong assumption for the size of . In contrast, for the log penalty, , and thus , which can goes to infinity if τi = o(δn). We can show that the log penalty satisfies other assumptions and refer the readers to Chen et al. (2014) for details.
Consider the neighborhood selection problem for the i-th variable versus all the other variables. Recall that is the support of the true regression coe cient bi with size . Let bi1 and be respectively the sub-vectors of bi and corresponding to Si.
Theorem 1
Given Assumptions (A1) - (A5), with probability at least 1 – C exp{–na} for a constant 0 < C < ∞, there exists a local minimizer that satisfies the following conditions for any i = 1, . . . , pn,
-
(a)
Sparsity:
-
(b)
L∞ loss: , where d1 is defined in (A3).
Therefore, if we denote the estimate of by the neighborhood selection as , where are tuning parameters of the penalty function, .
The proof is in the Supplementary Materials. The following Lemma 2 and 3 provide the theoretical justifications for using GGM as a starting point of our modified PC-algorithm.
Lemma 2
If the distribution PX is Markov to G, i.e., if the joint density fX satisfies the recursive factorization, the set of edges Fn of CGn includes all edges of plus the edges between co-parents of v-structures in Gn.
Lemma 3
Assume (A1). If (i, j) ∈ Fn of CGn but (i, j) of , the conditioning set Πi,j in (2) includes at least one set which d-separates vertices i and j in G.
Lemma 2 has been proved in Lemma 3.21 of Lauritzen (1996). The proof of Lemma 3 is presented in the Supplementary Materials. Lemma 2 shows that the concentration matrix recovers all the edges in the skeleton with no false negatives, but some false positives between the co-parents of v-structures. Lemma 3 shows that we can remove such false positives by examining partial correlation conditioning on some set in Πi,j.
Next we discuss the theoretical property of the modified PC-stable algorithm given a perfect estimation of GGM.
Theorem 2
Let αn be the p-value threshold for testing whether a partial correlation is 0. Let be the estimates of from the second step of the PenPC algorithm given a perfect estimation of GGM from the first step of the PenPC algorithm. Assume (A1), (A2) and (A6), then there exists αn → 0, such that where 0 < C < ∞ is a constant.
The proof is in the Supplementary Materials. Similar theorem has been proved in Kalisch and Bühlmann (2007) with pn at polynomial order of n. By starting with GGM, we extend the theorem to pn = O (exp{na}) case. Combining the results of Theorem 1 and Theorem 2, corollary 1 show that the summation of mistaken probabilities of GGM estimation and skeleton estimation given GGM goes to 0 as n → ∞.
Corollary 1
Let be the estimates of from the two-step approach PenPC algorithm. Assume (A1)-(A6), then there exists an αn → 0, such that , where 0 < C < ∞ is a constant.
4.2 Random Graphs
Next we extend our theoretical results to two commonly used models for random graphs: Erdös and Rényi (ER) Model (Erdös and Rényi, 1960) and Barabási and Albert (BA) Model (Barabási and Albert, 1999). Let and In general, assumption (A2) no longer holds for random graphs. It is easy to see that assumption (A2) can be relaxed to (A2’) and we introduced an additional assumption (A7)
-
(A2')
for some .
-
(A7)
, where 0 < C < 1 is a constant.
Assumption (A7) says that the maximal degree in the GGM is not dominated by the edges induced by co-parents of v-structures as n goes to infinity, which is a reasonable assumption. Given this assumption, Mn and qn are on the same scale.
4.2.1 Erdös and Rényi (ER) Model
The ER model constructs a graph G(pn, pE) of pn vertices by connecting vertices randomly. Each edge is included in the graph with probability pE independent from all other edges. By law of large numbers, such vertex is almost surely connected to (pn – 1)pE edges. Erdös and Rényi (1960) proved the following results about Mn, the maximal degree of the graph.
Lemma 4
In the graph G(pn, pE) following the ER model, the maximal degree Mn almost surely converges to mn, where if pnpE < 1, if pnpE = 1, and mn = O(pn) if .
When pn = O{exp(na)}, by Lemma 4 and assumption (A7), assumption (A2’) holds if pnpE < 1 and b ≥ a. When pnpE ≥ 1, our proof cannot handle the general case pn = O{exp(na)}. However, when the number of vertices is of the polynomial order of n, assumption (A2’) may still hold. In particular, suppose pn = O(nr). When pnpE < 1, assumption (A2’) holds for any b ∈ [0, ∞). When pnpE = 1, assumption (A2’) holds if b ≥ 2r/3. When pnpE → c > 1, assumption (A2’) holds if r < 1 and b ≥ r.
4.2.2 Barabási and Albert (BA) Model
The BA model is used to generate scale free graphs whose degree distribution follows a power law: , with a normalizing constant γ0 and a exponent γ1. Specifically, BA model generates a graph by adding vertices into the graph over time and when each new vertex is introduced into the graph, it is connected with larger probability to the existing vertices with larger number of connections. Since the distribution does not depend on the size of the network (or time), the graph organizes itself into a scale free state (Barabási and Albert, 1999). Móri (2005) showed that Mn (the maximal degree of the graph) almost surely converges to O(p1/2). Thus, assumption (A2’) holds for the case pn = O(nr) with b ≤ r/2.
5. Simulation Studies
We evaluated the performance of the PenPC algorithm and the PC-stable algorithm in terms of sensitivity and specificity of skeleton estimation using DAGs simulated by the ER model or the BA model. In both simulations and real data analysis, we used the implementation of the PC-stable algorithm by function skeleton in R package pcalg (version 1.1-6), and we have implemented PenPC algorithm in R package PenPC.
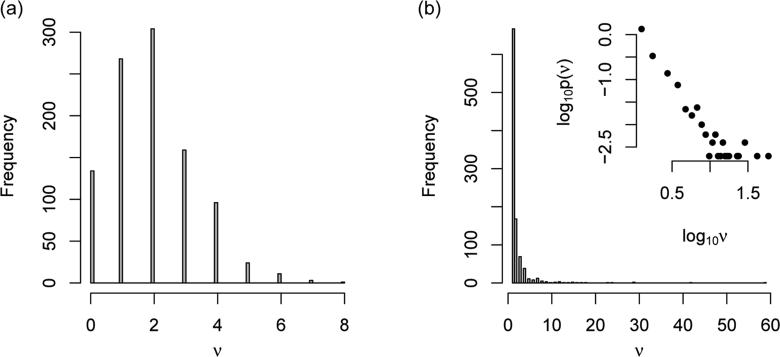
Following Kalisch and Bühlmann (2007), we simulated DAGs of p vertices by the ER model as follows. For any vertex pair (i, j) where i < j, we added an edge i → j with probability pE. For the BA model, the DAGs were simulated following Barabási and Albert (1999). The initial graph had one vertex and no edge. In the (t + 1)-th step, e edges were proposed. For each edge, the new vertex was connected to the i-th (1 ≤ i ≤ t) existing vertex with probability , where , and was the DAG at the t-th step. The distribution of the degrees ν from simulated DAGs under ER model (p = 1000 and pE = 2/p) and BA model (p = 1000 and e = 1) are shown in Figure 2 and similar graph for BA model (p = 1000 and e = 2) is shown in Figure S2 of Supplementary Materials.
Figure 2.
Histograms of the degree ν. (a) ER model with p = 1000 and pE = 2/p. (b) BA model with p = 1000 and e = 1 and the log10 scale density of log10ν in its subplot.
The probability of finding a highly connected vertex decreases exponentially with ν for the graphs generated by the ER model (Figure 2(a)). However, for the graphs generated by the BA model, there is a linear relation between degree and degree probability in log-log scale, confirming its scale-free property (Figure 2(b)).
After constructing the DAGs, the observed data were simulated by structure equations under multivariate Gaussian assumption. For example, denote the parents of Xj by paj, then where In our simulations, all bjk's and σ2 were set to be 1. For either ER or BA model, we considered low dimension setting where p = 11, n = 100 and high-dimension settings where p = 100, n = 30 and p = 1000, n = 300 with various sparsity levels determined by pE for ER model and e for BA model (Table 1).
Table 1.
Simulation Setting
| p | n | pE (ER) | e (BA) |
|---|---|---|---|
| 11 | 100 | 0.2 | 1,2 |
| 100 | 30 | 0.02, 0.03, 0.04, 0.05 | 1,2 |
| 1000 | 300 | 0.002, 0.005, 0.01 | 1,2 |
Due to limited space, here we only show the results for the simulation setups using ER model with p=1000, n=300, and pE=0.005; and BA model with p=1000, n=300, and e=1. The remaining results are presented in Figure S3 - S14 of the Supplementary Materials.
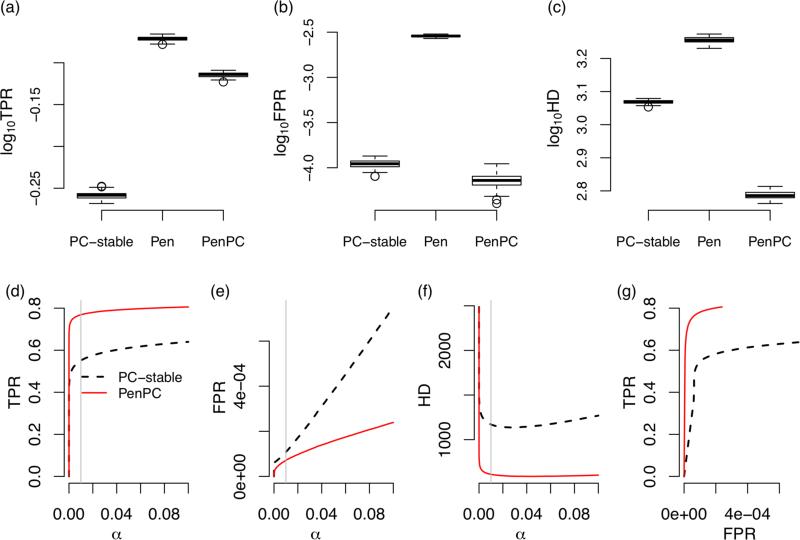
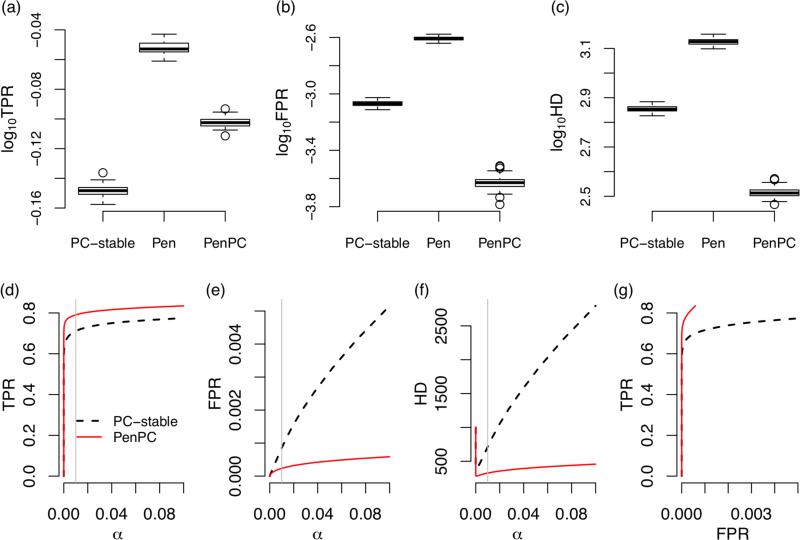
First consider the results from the ER model when α = 0.01. Recall that α is the p-value threshold for conditional independence testing. The penalized regression (step 1 of the PenPC algorithm) identifies more true positives than the PC-stable algorithm, but also introduce more false positives (Figure 3 (a-b)), while PenPC algorithm significantly reduces the number of false positives, though some true positives are also removed. At the end, the PenPC has the lowest number of false positives plus false negatives, as measured by Hamming distance (HD) (Figure 3 (c)). Figures 3(d-f) show that across various values of α, PenPC consistently has better performance than the PC-stable algorithm. Finally, Figure 3(g) shows the ROC curves for the PenPC and the PC-stable algorithms, which illustrate that PenPC has better sensitivity and specificity than the PC-stable algorithm regardless of the cuto α. Similar conclusions can be drawn for the simulation results shown in Figure 4, where the DAGs are simulated by the BA model. We note that although PenPC performs well for the BA model, further improvement is possible by incorporating special consideration for the scale-free structure of BA graphs (Liu and Ihler, 2011).
Figure 3.
Performance of ER model (p = 1000, n = 300, pE = 0.005). The upper panels are box plots (in log10 scale) of true positive rate (TPR) (a), false positive rate (FPR) (b) and hamming distance (HD) (c) from 100 replications at α = 0.01. The lower panels are average true positive rate (d), false positive rate (e), and Hamming distance (f) from 100 replications when the tuning parameter α is changed from 0 to 0.1 (the grey vertical line are at α = 0.01). ROC curves are shown in panel (g). This figure appears in color in the electronic version of this article.
Figure 4.
Performance of BA model (p=1000,n=300,e=1). The upper panels are box plots (in log10 scale) of true positive rate (TPR) (a), false positive rate (FPR) (b) and hamming distance (HD) (c) from 100 replications at α = 0.01. The lower panels are average true positive rate (d), false positive rate (e), and Hamming distance (f) from 100 replications when the tuning parameter α is changed from 0 to 0.1 (the grey vertical line are at α = 0.01). ROC curves are shown in panel (g). This figure appears in color in the electronic version of this article.
6. Application
We applied the PC-stable algorithm and the PenPC algorithm to study gene-gene network using gene expression data from tumor tissue of 550 TCGA (The Cancer Genome Atlas) breast cancer patients (Cancer Genome Atlas Network, 2012). Gene expression were measured by RNA-seq. We quantified the expression of each gene within each sample by log(total read count) (logTReC). After removing genes with low expression across most samples, we ended up with 18,827 genes. We first removed the e ects of several covariates by taking residuals of logTReC for each gene using a linear regression with the following covariates: 75 percentile of logTReC per sample, which captures read depth, plate, institution, age, and six PCs from the corresponding germline genotype data.
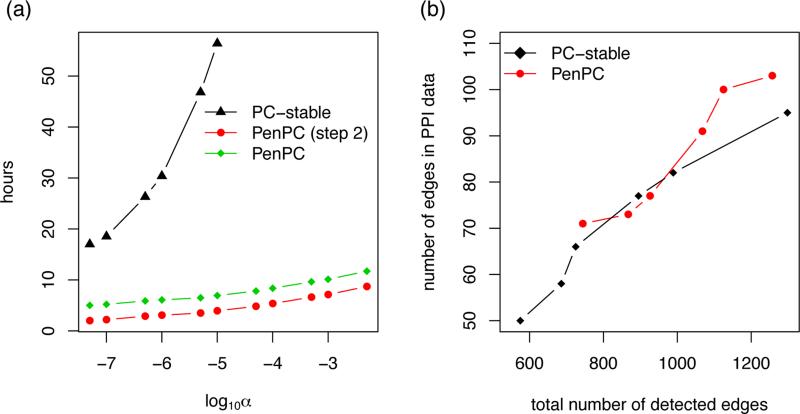
The computational cost of the PC-stable algorithm increases quickly as the number of vertices or the p-value cuto increases. When we study 410 genes, the computational time of the PC-stable and PenPC algorithms are both within an hour. When we expand the number of genes to 8,261. The step 1 in PenPC algorithm took 3 hours in total while searching for 1000 combinations of tuning parameters for each gene. Given the GGM, the 2nd step of the PenPC is computationally much more e cient than the PC-stable algorithm (Figure 5 (a)). For example, with p-value threshold varies from 10−7 to 10−5, the computational time of the PC-stable algorithm increases from 20 to 50 hours. In contrast, the computation time of the PenPC remains below 10 hours even for p-value cuto 5 × 10−3. All the computation are done in Linux server with an 2.93 GHz Intel processor and 48GB RAM.
Figure 5.
Comparing PenPC algorithm with PC-stable algorithm in terms of skeleton estimation by changing the significance levels for partial correlation testings: α=0.0001, 0.0005, 0.001, 0.005, 0.01 and 0.05. (a) The computational time at different α values when we consider 8,261 genes. (b) The number of detected edges vs. the number of edges in the PPI (protein-protein interaction) data when we consider 410 genes. This figure appears in color in the electronic version of this article.
Since PC-stable algorithm is not computationally feasible for larger gene set, we first discussed the results on 410 genes from the cancer Gene Census in http://cancer.sanger.ac.uk/cancergenome/projects/census/. For α = 0.0001 to 0.05, we estimated the skeleton by the PC-stable and PenPC algorithms. The estimated skeletons were evaluated by comparing the estimated edge sets with protein-protein interaction (PPI) database at http://www.pathwaycommons.org/pc2/downloads.html. PPI is a reasonable resource to evaluate graphical model estimates because genes with PPI tend to co-express (Rhodes et al., 2005). There were 3315 PPIs where both proteins belong to the 410 genes. Figure 5(a) shows the total number of detected edges versus the number of edges in PPI data. For both methods, the total number of detected edges increase monotonically as increases. The PenPC results have higher sensitivity to detect PPI given the number of edges discovered.
Next we applied PenPC to 8,261 genes with PPI annotation. Using α = 0.001, we detected 12,150 edges, that is 0.03% of the total number of edges. We arbitrarily define the genes with more than 7 neighbors as hub genes and there are 46 hub genes (Supplementary Table 1). Interestingly, many of the hub genes are cancer-related. For example, all the hub genes with more than 9 neighbors, MYC, ELF3, and RAB15, are cancer-related. MYC encodes Myc proto-oncogene protein, which are associated with multiple types of human cancers including breast cancer. ELF3 is one of the ETS transcription factor and it modulates breast cancer-associated gene expression. RAB15 is a member RAS oncogene family.
7. Discussions
The seminal works of Kalisch and Bühlmann (2007) have greatly advanced our understating of the PC-algorithm and provided well-designed and user-friendly software packages (Kalisch et al., 2012). Our PenPC algorithm provides some helpful improvements, especially in high dimensional settings. The PenPC algorithm has three tuning parameters, two for step 1 (tuning parameters of the Log penalty for neighborhood selection) and one for step 2 (p-value cutoff) of the PenPC algorithm. The selection of tuning parameters for step 1 and step 2 of PenPC are two independent procedures. For step 1, it is a classical problem of tuning parameter selection for penalized regression. We chose to use extended BIC as it delivers the best performance and it has sound theoretical justifications (Chen and Chen, 2008). Choosing the best combination of the two tuning parameters for log penalty using extended BIC does not induce heavy computational cost. For example, we use 1,000 combinations of λ and τ for each of the penalized regressions with Log penalty, and for our real data analysis with n = 550 and p = 8, 261, it takes about 3 hours for all the p = 8,261 penalized regressions. In the second step of PenPC, we need to choose a p-value threshold for conditional independence tests, similar to the tuning parameter for the PC algorithm. How to choose this p-value cuto is an open problem that warrants further research.
We have compared PenPC with the approach of replacing the Log penalty with the Lasso penalty. As expected, the Lasso penalty leads to much worse performance in high dimensional settings (Figures S15-S16 in Supplementary Materials). Following Kalisch and Bühlmann (2007), we assume a multivariate Gaussian assumption so that we may test conditional independence by assessing conditional correlation. The first step of the PenPC algorithm (neighborhood selection) does not require this assumption. Our method is robust to this multivariate Gaussian assumption. For example, Figures S15-S16 show that the performance of PenPC algorithm is comparable when the data are simulated from multivariate Gaussian and multivariate t-distribution (df=5). Assuming the observed data are generated by linear, potentially non-Gaussian structural equation model (SEM), Loh and Bühlmann (2014) proved that the moral graph of a DAG (i.e., the DAG skeleton plus the edges that connect the co-parents of v-structures) can be estimated by the support of the inverse covariance matrix and they estimated inverse covariance matrix using graphical Lasso. Borrowing their theoretical justifications, we can extend PenPC to non-Gaussian cases. The first step of PenPC is similar to graphical Lasso, but with log penalty instead of Lasso penalty. The second step of PenPC can be modified by using a conditional independent test that does not rely on Gaussian assumption. Finally, our work assumes no hidden confounders or latent variables, which may be justified by the fact that we examine the expression of all the genes and the e ects of confounders or latent variables may be manifested by the expression of certain genes.
Supplementary Material
Acknowledgements
This research is supported in part by NIH R01 GM105785-01 and HG006292-03. We are grateful for the constructive comments from two anonymous reviewers and the editors.
Footnotes
8. Supplementary Materials
The Supplementary Figures and Results referenced in Sections 2-7 are available with this paper at the Biometrics website on Wiley Online Library http://www.biometrics.tibs.org, along with our method in an R package named PenPC.
Contributor Information
Min Jin Ha, Department of Biostatistics, MD Anderson Cancer Center, Houston, Texas, MJHa@mdanderson.org.
Wei Sun, Department of Biostatistics, Department of Genetics, UNC Chapel Hill, North Carolina, weisun@email.unc.edu.
Jichun Xie, Department of Biostatistics & Bioinformatics, Duke University, Durham, North Carolina, jichun.xie@duke.edu.
References
- Anderson T. An Introduction to Multivariate Statistical Analysis. 2003. Wiley; New York: 2003. [Google Scholar]
- Barabási A, Albert R. Emergence of scaling in random networks. science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chen Z. Extended bayesian information criteria for model selection with large model spaces. Biometrika. 2008;95:759–771. [Google Scholar]
- Chen T, Sun W, Fine J. Technical report. University of North Carolina; Chapel Hill: 2014. Designing penalty functions in high dimensional problems: The role of tuning parameters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chickering DM. Optimal structure identification with greedy search. The Journal of Machine Learning Research. 2003;3:507–554. [Google Scholar]
- Colombo D, Maathuis M. A modification of the pc algorithm yielding order-independent skeletons. arXiv preprint arXiv. 2012:1211.3295. [Google Scholar]
- Erdőos P, Rényi A. On the evolution of random graphs. Publications of the Mathematical Institute of the Hungarian Academy of Sciences. 1960;5:17–61. [Google Scholar]
- Fan J, Lv J. Nonconcave penalized likelihood with np-dimensionality. Information Theory, IEEE Transactions on. 2011;57:5467–5484. doi: 10.1109/TIT.2011.2158486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckerman D, Geiger D, Chickering D. Learning bayesian networks: The combination of knowledge and statistical data. Machine learning. 1995;20:197–243. [Google Scholar]
- Kalisch M, Bühlmann P. Estimating high-dimensional directed acyclic graphs with the pc-algorithm. The Journal of Machine Learning Research. 2007;8:613–636. [Google Scholar]
- Kalisch M, Mächler M, Colombo D, Maathuis M, Bühlmann P. Causal inference using graphical models with the R package pcalg. Journal of Statistical Software. 2012;47:1–26. [Google Scholar]
- Lauritzen S. Graphical models, volume 17. Oxford University Press; USA: 1996. [Google Scholar]
- Liu Q, Ihler AT. Learning scale free networks by reweighted l1 regularization. International Conference on Artificial Intelligence and Statistics. 2011:40–48. [Google Scholar]
- Loh P-L, Bühlmann P. High-dimensional learning of linear causal networks via inverse covariance estimation. Journal of Machine Learning Research. 2014;15:3065–3105. [Google Scholar]
- Maathuis M, Colombo D, Kalisch M, Bühlmann P. Predicting causal effects in large-scale systems from observational data. Nature Methods. 2010;7:247–248. doi: 10.1038/nmeth0410-247. [DOI] [PubMed] [Google Scholar]
- Maathuis M, Kalisch M, Bühlmann P. Estimating high-dimensional intervention effects from observational data. The Annals of Statistics. 2009;37:3133–3164. [Google Scholar]
- Mazumder R, Friedman JH, Hastie T. Sparsenet: Coordinate descent with nonconvex penalties. Journal of the American Statistical Association. 2011;106 doi: 10.1198/jasa.2011.tm09738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek C. Proceedings of the Eleventh conference on Uncertainty in artificial intelligence. Morgan Kaufmann Publishers Inc.; 1995. Strong completeness and faithfulness in bayesian networks. pp. 411–418. [Google Scholar]
- Meinshausen N, Bühlmann P. High-dimensional graphs and variable selection with the lasso. The Annals of Statistics. 2006;34:1436–1462. [Google Scholar]
- Móri T. The maximum degree of the barabási-albert random tree. Combinatorics Probability and Computing. 2005;14:339–348. [Google Scholar]
- Pearl J. Causality: models, reasoning and inference. Cambridge Univ Press; 2009. [Google Scholar]
- Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM. Probabilistic model of the human protein-protein interaction network. Nature biotechnology. 2005;23:951–959. doi: 10.1038/nbt1103. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Niculescu-Mizil A, Murphy K. Learning graphical model structure using l1-regularization paths. AAAI. 2007;7:1278–1283. [Google Scholar]
- Shojaie A, Michailidis G. Penalized likelihood methods for estimation of sparse high-dimensional directed acyclic graphs. Biometrika. 2010;97:519–538. doi: 10.1093/biomet/asq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirtes P, Glymour C, Scheines R. Causation, prediction and search. Vol. 81. The MIT Press; 2000. [Google Scholar]
- Sun W, Ibrahim JG, Zou F. Genomewide multiple-loci mapping in experimental crosses by iterative adaptive penalized regression. Genetics. 2010;185:349–359. doi: 10.1534/genetics.110.114280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier M, Koller D. Ordering-based search: A simple and effective algorithm for learning bayesian networks. In UAI. 2005:584–590. [Google Scholar]
- Tsamardinos I, Brown LE, Aliferis CF. The max-min hill-climbing bayesian network structure learning algorithm. Machine learning. 2006;65:31–78. [Google Scholar]
- van de Geer S, Bühlmann P. l0-penalized maximum likelihood for sparse directed acyclic graphs. The Annals of Statistics. 2013;41:536–567. [Google Scholar]
- Zhao P, Yu B. On model selection consistency of lasso. The Journal of Machine Learning Research. 2006;7:2541–2563. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.