Abstract
Metastasis frequently develops years after the removal of a primary tumor, from a minority of disseminated cancer cells that survived as latent entities through unknown mechanisms. We isolated latency competent cancer (LCC) cells from early-stage human lung and breast carcinoma cell lines and defined the mechanisms that suppress outgrowth, support long-term survival, and maintain tumor-initiating potential in these cells during the latent metastasis stage. LCC cells show stem cell-like characteristics and express Sox2 and Sox9 transcription factors, which are essential for their survival in host organs under immune surveillance and for metastatic outgrowth under permissive conditions. Through expression of the WNT inhibitor DKK1, LCC cells self-impose a slow-cycling state with broad downregulation of ULBP ligands for NK cells and evasion of NK cell-mediated clearance. By expressing a Sox-dependent stem-like state and actively silencing WNT signaling, LCC cells can enter quiescence and evade innate immunity to remain latent for extended periods.
INTRODUCTION
Cancer patients with no clinical evidence of disease after the initial treatment frequently relapse with distant metastasis years later. Prior to diagnosis and treatment, primary tumors may release large numbers of cancer cells into the circulation. Although a majority of the dispersed cells perish in the bloodstream or soon after infiltrating distant organs, a minority may survive as latent seeds in host tissues. As a result, people who are clinically considered disease-free after cancer treatment may carry thousands of disseminated tumor cells (DTCs) in the bone marrow and other organs (Braun et al., 2005). Latent metastasis is a major concern in the clinic, yet little is known about the nature of dormant DTCs and the mechanisms that allow these cells to remain quiescent, evade immunity, retain tumor-initiating capacity, and evolve into aggressive metastasis (Massague and Obenauf, 2016).
A leading hypothesis posits that dormant DTCs are tumor-initiating cells that enter quiescence by the action of growth inhibitory signals from the host tissue stroma (Sosa et al., 2014). Recent studies have identified stromal TGF-β and BMP as inhibitors of DTC growth (Bragado et al., 2013; Gao et al., 2012; Kobayashi et al., 2011). However, organs that host DTCs, such as the bone marrow, liver, and lungs, support cell proliferation as part of their normal tissue homeostasis and regenerative processes, raising questions as to whether stromal growth inhibitory signals are persistent enough to enforce long-term metastatic latency.
Another important consideration is the role of immunity in latent metastasis. The interplay between cancer cells and the various components of the immune system plays a crucial role in tumor progression (Dunn et al., 2004; Eyles et al., 2010; Kitamura et al., 2015). Notably, organ transplants from donors who had been cured of melanoma, or who suffered glioblastoma, considered a non-metastatic tumor, developed donor-derived metastasis in immunosuppressed recipients, suggesting that immune surveillance prevents the outgrowth of dormant DTCs (MacKie et al., 2003; Xiao et al., 2013). Metastatic latency may therefore require DTCs to be in equilibrium with the immune system.
Our understanding of the molecular basis for latent metastasis has been limited by a scarcity of preclinical models that recapitulate key features of this metastatic stage (Massague and Obenauf, 2016). To address this problem, we isolated latency competent cancer (LCC) cells by in vivo selection of human tumor cell populations in mice. Using these models, we show that LCC cells are a distinct class of stem-like cells primed to enter quiescence and evade innate immunity. LCC cells express Sox transcription factors that impart tumor-initiating stem/progenitor cell identity. These cells can actively self-impose a slow-cycling state by producing DKK1, an inhibitor of the WNT signaling pathway. We propose a quiescence-linked mechanism for evasion of NK cell-mediated immunity, long-term survival and evolution of latent metastasis-initiating cells.
RESULTS
Latency competent cells isolated from early-stage breast and lung cancers
We isolated cancer cells that are competent to seed relevant organs with latent metastasis (latency competent cancer cells, LCC cells). As sources, we used H2087, a cell line derived from stage I lung adenocarcinoma (Gazdar and Minna, 1996), and HCC1954 from a stage IIA HER2+ breast tumor (Gazdar et al., 1998) (Figure S1A). Nearly half of early-stage lung adenocarcinoma cases develop distant relapse despite surgical resection of the primary tumor, implying latent disseminated disease (Maeda et al., 2010). The HER2+ breast cancer patient population is experiencing a marked increase in the incidence of brain metastasis after anti-HER2 therapies that suppress extracranial relapse and extend survival (Duchnowska et al., 2009). Thus, both cancer types are important sources of latent metastasis in the clinic.
H2087 and HCC1954 cells transduced with a GFP-luciferase reporter and antibiotic resistance vectors were intracardially injected into the arterial circulation of Foxn1nu mice (Pelleitier and Montplaisir, 1975). As monitored by bioluminescence imaging (BLI), most mice remained signal-free and healthy for 3 months. Organs from these mice were dissociated into single-cell suspensions in culture to recover antibiotic-resistant cancer cells from lungs (H2087-LCC1 cell line) and kidneys (H2087-LCC2) of mice injected with H2087, and from brain of mice injected with HCC1954 (HCC1954-LCC1) (Figure S1A-B). The tumorigenic activity of LCC cells after orthotopic implantation was similar to that of the parental populations (Figure S1C-D). Thus, LCC cells retained tumor initiating potential after months of latency in mouse tissues.
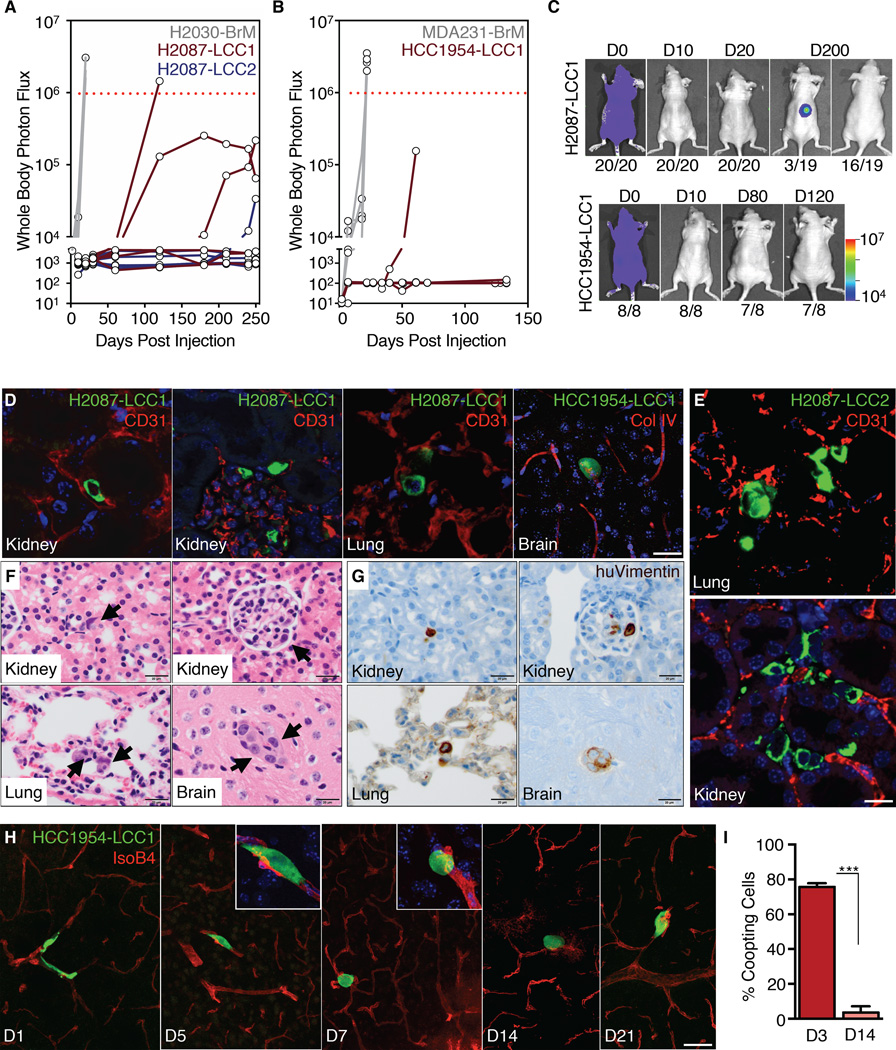
Of 20 athymic mice injected with H2087-LCC cells, one developed overt metastasis, one a spinal metastasis after 4 months that did not progress, two developed incipient lesions after 7 months, and 16 remained metastasis-free for over 8 months (Figure 1A,C). Only one of 8 mice injected with HCC1954-LCC1 developed overt metastasis over 4 months (Figure 1B-C). In comparison, aggressive breast (MDA-MB-231) and lung adenocarcinoma (H2030) metastatic lines formed extensive metastases within 3 weeks (Figure 1A-B, S1E).
Figure 1. Localization and dormancy of disseminated LCC cells.
(A-B) Bioluminescence Imaging (BLI) tracking of mice injected with the indicated cell lines. Each line represents an individual mouse. Mice with BLI signal above the dotted red line were euthanized.
(C) Representative images of BLI signal in mice injected with the indicated LCC cell lines. The number of mice represented by each image is indicated at the bottom.
(D) Immunofluorescence staining of disseminated LCC cells (green) next to capillaries (red) in the parenchyma of organ sites.
(E) Infrequent H2087-LCC2 cell (green) clusters in the lung (top panel) or kidney (bottom panel); Vasculature (red).
(F) Localization of H2087-LCC1 cells (arrows) in the renal tubule (top-left) or glomeruli (top-right) of the kidney, within alveolar walls of the lung (bottom-left), or capillaries in the brain (bottom-right) 3 months post-injection by H&E staining.
(G) Immunohistochemical staining of disseminated H2087-LCC1 cells from panel 1F with human vimentin (brown).
(H) Time-course analysis of HCC1954-LCC1 cell morphology (green) along capillaries (red) as they extravasate and colonize the brain.
(I) Quantification of HCC1954-LCC1 cell morphology in the brain 3 days and 14 days post-injection. Data are mean percentage of coopting cells per brain ± S.E.M. N = 3 mice per group, scoring representative serial sections of the entire brain for each mouse. P < 0.001 (***), Student’s t-test.
Scale bars, 10μm (D), 100μm (E), 20μm (F and G), and 50μm (H). See also Figure S1.
We were able to histologically detect and recover cancer cells from 20 of 26 (77%) LCC-injected mice that remained metastasis-free, but only from 2/14 (14%) of mice injected with the parental lines (Figure S1F). H2087-LCC1 and -LCC2 cells targeted both the lungs and the kidneys, suggesting that the latent phenotype of these cells was not strictly organ specific (Figure 1D-G). Cells isolated from rare macrometastatic tumors arising in LCC-injected mice (LCC-M lines) showed a latent phenotype upon reinjection, with no increase in macrometastatic activity, arguing that LCC cells stochastically form overt metastases (Figure S1G).
LCC cell localization in infiltrated organs
We chose H2087-LCC1, H2087-LCC2, HCC1954-LCC1, and the HCC1954-LCC2 line derived from LCC1 injected mice, for further analysis. Two weeks after inoculation, approximately 20% of disseminated GFP+ HCC1954 LCC cells in the brain were found in small clusters (>10 cells) and the rest as single cells (Figure S1H). After 3 months, more than 90% of the GFP+ DTCs were found as single cells and the rest as small clusters of approximately 20 cells, as determined by anti-GFP immunostaining (Figure 1D-E), H&E staining (Figure 1F), and human vimentin immunostaining (Figure 1G).
In the kidney, LCC cells were detected between the renal tubules, adjacent to capillaries, or within glomeruli (Figure 1F-G). A few LCC cells were observed within the adrenal gland cortex (Figure S1I). In the lungs, LCC cells were invariably found within alveolar walls (Figure 1G). In the brain and other organs, LCC cells were closely associated with the vasculature (Figure 1D). Aggressive brain metastatic cells rapidly spread over the abluminal surface of cerebral microcapillaries, and this spreading is required for colony outgrowth (Valiente et al., 2014). In contrast, LCC cells infiltrating the brain parenchyma spread along capillaries only transiently (days 1–3 after inoculation), and subsequently adopted a rounded morphology (Figure 1H-I).
LCC cells are prone to enter quiescence
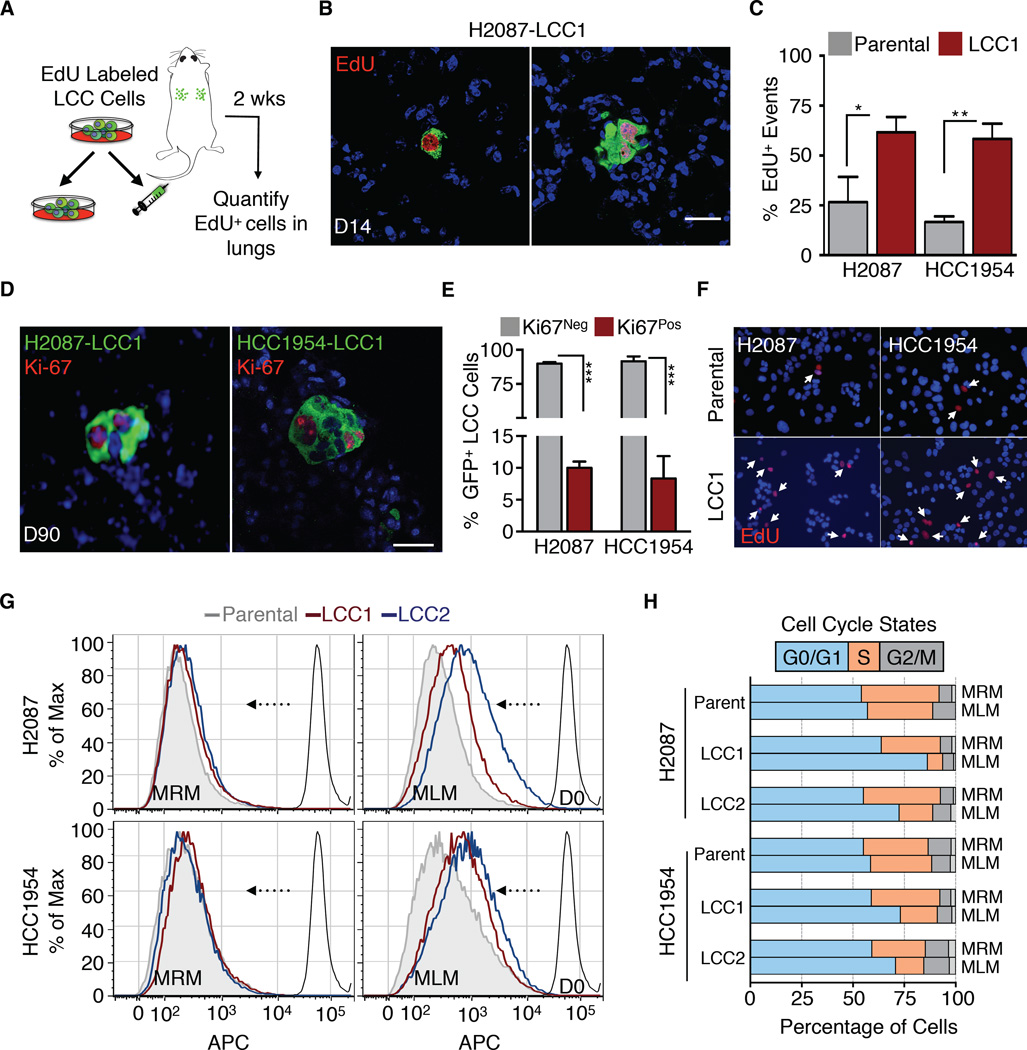
To monitor proliferation in the first days after inoculation, we labeled LCC cells with 5-ethynyl-2´-deoxyuridine (EdU) prior to intravenous injection into athymic mice (Figure 2A, S2A). Proliferation would dilute the amount of EdU retained in these cells. After 14 days, approximately 60% of LCC cells that reached the lungs still retained EdU versus 15–25% in the parental populations (Figure 2B-C), indicating that LCC cells entered quiescence more readily. Three months after inoculation, approximately 90% of H2087-LCC1 cells in lung and HCC1954-LCC1 cells in brain were negative for the proliferation marker Ki-67. Ki-67+ LCC cells were largely confined to cell clusters (Figure 2D-E).
Figure 2. LCC cells adopt a slow-cycling state in vitro and in vivo.
(A) Experimental design for EdU pulse-chase experiment.
(B) Immunofluorescence images of double-positive GFP+/EdU+ LCC cells (green, red) in lungs.
(C) Quantification of double-positive EdU+/GFP+ LCC cells versus parental counterparts in lungs harvested 2 weeks post-injection. Data are mean percentage of EdU+ cells per lung ± S.E.M. N = 3 mice per group, scoring representative serial sections of the entire lung for each mouse. P < 0.05 (*), P <0.01 (**), Mann-Whitney Test.
(D) Representative immunofluorescence images of proliferating Ki-67+ (red) H2087-LCC1 (in lung) and HCC1954-LCC1 cells (in brain) 3 months post-injection.
(E) Quantification of LCC cells from Figure 2D. Data are percentage of total cells that are Ki67+ per organ ± S.E.M. N = 5 per group, scoring representative serial sections of the entire organ of each mouse. P < 0.001 (***), Student’s t-test.
(F) Micrographs of the indicated cell lines in culture 4 days post EdU labeling. White arrows point to cells retaining EdU label.
(G) Retention of eFluor670 dye by indicated cell lines after 6 days in MRM or MLM culture conditions.
(H) Cell cycle analysis of the indicated cell lines by BrdU/APC after 3 days in MRM or MLM culture conditions.
Scale bars, 10μm (B and D), and 15μm (E). See also Figure S2.
To determine the effect of a mitogen-poor environment we cultured LCC cells in mitogen-low media (MLM, 2% serum) or regular mitogen-rich media (MRM, 10% serum). Under MLM conditions, LCC cells underwent a rapid decrease in proliferation, as determined by their ability to retain EdU in culture (Figure 2F), dye retention (Figure 2G) and CellTiter-Glo assays (Figure S2B), whereas the parental line showed little (H2087) or no decrease (HCC1954) in proliferation. LCC cells accumulated with a G0/G1 DNA content (Figure 2H) and no change in apoptosis marker (cleaved caspase-3) levels (Figure S2C). We performed genome-wide RNA-seq transcriptomic profiling under MRM and MLM conditions. Analysis of differentially expressed genes (Figure S2D) using signatures for specific cell cycle stages (Croft et al., 2014) showed increased expression of G0/G1-associated genes and lower expression of S phase associated genes in LCC cells under MLM conditions (Figure S2E).
In sum, LCC cells recapitulated key features of the latent metastatic state, including a propensity to enter proliferative quiescence, an ability to survive as latent entities in relevant organs for months, and a capacity to retain tumorigenic and metastasis-initiating potential. Although LCC cells suffered extensive clearing upon infiltrating target organs, as did the parental population from which they were derived, LCC cells were superior at seeding these organs with a minority of survivors to establish latent metastasis.
LCC cells molecularly cluster with stem/progenitor cells
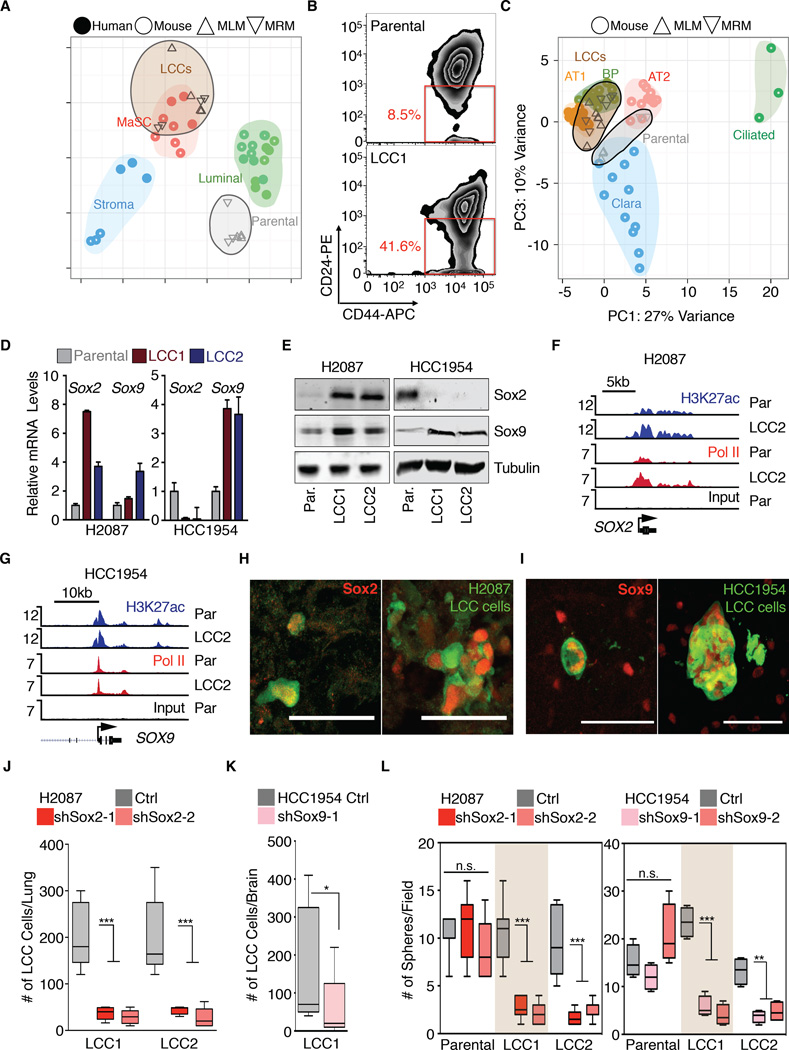
Gene Set Enrichment Analysis of differentially expressed genes in HCC1954-LCC cells versus to parental HCC1954 identified a mammary stem cell signature (MaSC) as a top-scoring gene set, under both MLM and MRM conditions (Figure S3A). Principal component analysis revealed clustering of HCC1954-LCC cells with human and mouse mammary stem and progenitor cells, away from mature luminal and stromal compartments (Figure 3A) (Lim et al., 2010). Moreover, HCC1954-LCC cells were enriched for the surface marker profile CD44Hi/CD24Lo, which is typical of human breast cancer stem cells (Figure 3B) (Al-Hajj et al., 2003).
Figure 3. LCC cells are enriched for stem cell-like characteristics.
(A) Principal Component Analysis (PCA) of HCC1954 derivatives and normal breast cell populations of human or mouse origin. HCC1954-LCC cells resemble mammary stem cell (MaSC) gene expression profiles.
(B) Flow cytometry analysis of shows marked enrichment in the CD44Hi/CD24Lo compartment, indicative of breast cancer stem cells in HCC1954 LCC1 cells.
(C) PCA plot of H2087 derivatives and mouse bronchiolar and alveolar cell lineages. H2087-LCC cells cluster with the stem-like alveolar type 1 (AT) and bipotent progenitor (BP) cells.
(D-E) Sox2 and Sox9 mRNA (D) and protein (E) expression in the indicated cell lines.
(F-G) Gene track view for H3K27ac and Pol II ChIP-seq data at the Sox2 locus (F) and Sox9 locus (G) in the indicated cell lines.
(H-I) Representative single cell and cell cluster images of Sox2 and Sox9 positive H2087-LCC1 in lung and HCC1954-LCC1 in brain.
(J-K) Sox2 or Sox9 depletion attenuates survival of H2087-LCC and HCC1954-LCC cells in lungs and brain of athymic mice respectively. Data are total number of LCC cells scored in the lung 3 months post-injection or brains 2 months post injection. N = 5–6 mice per group, scoring representative serial sections of the organ. P <0.001 (***), Mann-Whitney Test.
(L) Oncosphere-forming capacity upon Sox2 and Sox9 depletion in the indicated cell lines. Each panel depicts representative control or Sox2/9-depleted cell oncospheres. P < 0.01 (**), P < 0.001 (***), Student’s t-test.
Scale bars, 50μm (H-I). See also Figure S3.
Based on gene signatures of distinct cell types in the lung epithelium (Treutlein, 2014), H2087-LCC cells clustered with the stem-like alveolar type I and bipotent progenitor (BP) cells. Parental H2087 clustered with alveolar type II and Clara cell lineages under MRM and MLM conditions, respectively (Figure 3C). In limiting dilution assays, LCC cells showed a 4- to 15-fold higher probability of engrafting mice compared to parental cells (Figure S3B). These results suggested that our LCC isolation protocol enriched for cancer cells with stem/progenitor cell features.
Sox2 and Sox9 association with the LCC phenotype
Progenitor cell identity is determined by lineage-specific transcription factors. Notably, two master regulators of stem and progenitor cell identity, Sox2 (Arnold et al., 2011) and Sox9 (Guo et al., 2012), ranked high among transcription factors whose expression was prominently associated with the LCC phenotype. Sox2 was predominant in H2087-LCC cells whereas HCC1954-LCC cells showed high Sox9 expression and low Sox2 expression, both at the mRNA (Figures 3D, S3C-D) and the protein levels (Figure 3E). Sox2 and Sox9 regulate stem and progenitor cells in adult tissues (Sarkar and Hochedlinger, 2013). Sox2 is genetically amplified in squamous carcinomas and small cell lung cancers (Bass et al., 2009; Rudin et al., 2012). Sox9 is upregulated in brain tumors and basal cell carcinomas (Kordes and Hagel, 2006), and confers stem cell phenotype to breast cancer cells (Guo et al., 2012).
Histone H3 lysine 27 acetylation (H3K27ac) is a mark of transcriptionally active loci (Heintzman et al., 2009). Genome-wide analysis of H3K27ac marks by ChIP-seq revealed distinct patterns in LCC cells compared to the parental counterparts, with 11.9% and 15.6% of the H3K27ac peaks in LCC cells being absent in the H2087 and HCC1954 parental cell populations, respectively, and 7.1% and 24.0% of the H3K27ac peaks in parental cells being absent in the LCC cell populations, respectively (Figure S3E). Enrichment for H3K27ac and Pol II peaks was observed within the Sox2 locus in H2087 LCC cells (Figure 3F). Although H3K27ac and DNA polymerase II (Pol II) peaks on SOX9 were observed in HCC1954 LCC cells, we found no difference compared to parental populations (Figure 3G), suggesting that SOX9 mRNA up-regulation in these cells occurs at a post-transcriptional level.
We confirmed expression of Sox2 and Sox9 in LCC cells by immunofluorescence staining in vivo (Figure 3H-I). Sox expression was detected in single disseminated LCC cells and in rare proliferating clusters. LCC lesions that grew large accumulated cells with varying levels of Sox2 or Sox9 (see Figure S4A). Related, CD44Hi/CD24Lo mammary cell populations progressively drift towards a higher CD24 content as they divide (Liu et al., 2014). In agreement, sorting HCC1954-LCC2 for cells with a very low CD24 level enriched for Sox9 (Figure S3F).
Metastatic seeding by LCC cells requires Sox factors
We transduced H2087-LCC and HCC1954-LCC cells with shRNAs targeting Sox2 and Sox9 respectively (Figure S3G-H). The knockdown inhibited anchorage-independent growth of these cells in matrigel culture conditions as compared to controls (Figure S3I-J). We inoculated these LCC cells into athymic mice by tail vein injection, and monitored colonization of the lungs by whole-body BLI imaging weekly for 60 days. Histologic analysis of the lungs showed a marked decrease in the metastatic seeding capacity of the Sox-depleted cells compared to cells expressing a control vector (Figure 3J-K).
The ability of tumor-initiating cells to form colonies in suspension provides a test of their capacity to initiate tumor growth (Dontu et al., 2003). LCC cells formed the same number of oncospheres as their parental counterparts (Figure S3K), although LCC oncospheres were smaller (Figure S3L). Interestingly, Sox2 expression was enriched in H2087-LCC oncospheres, but not oncospheres formed by parental H2087 (Figure S3M). Knockdown of Sox2 or Sox9 inhibited oncosphere formation in LCC cells but not in their parental lines (Figure 3L, S3N). We concluded that LCC cells are distinct subpopulations of stem-like cancer cells characterized by high expression of Sox2 or Sox9 and a dependence on these transcription factors for growth under restrictive conditions in vitro and seeding of latent metastasis in mice.
Metastatic latency balanced by NK cell immune surveillance
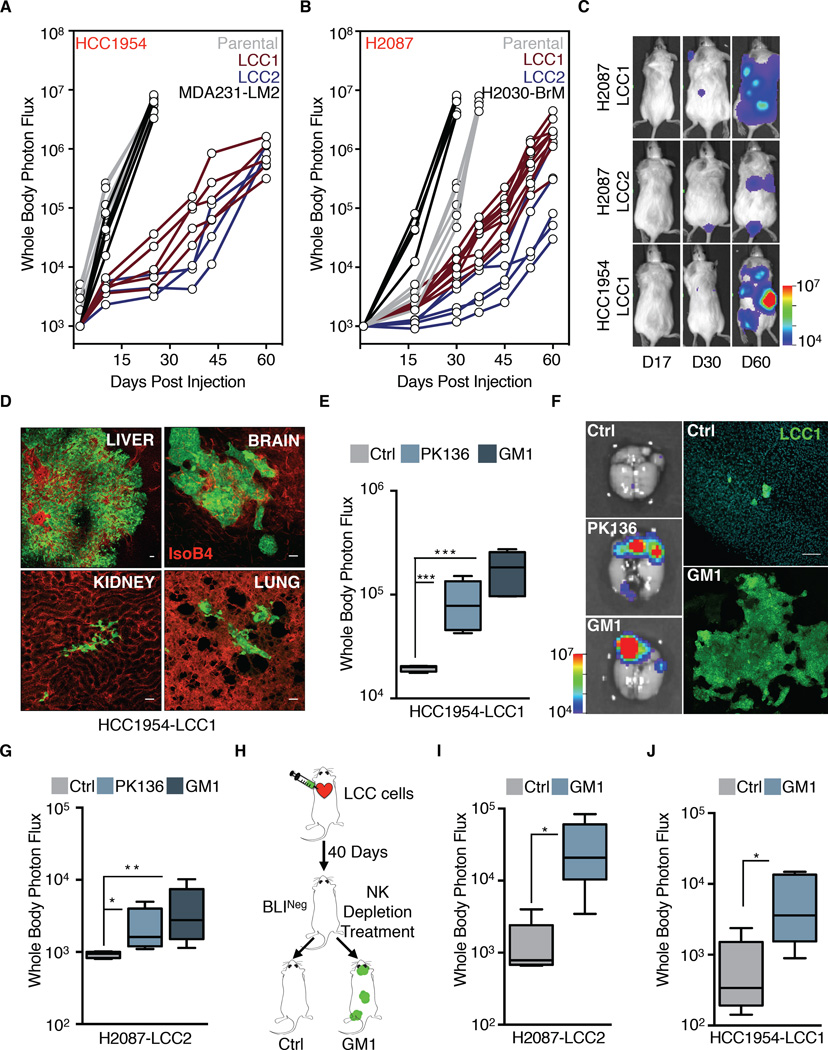
Although LCC cells were superior at seeding organs with latent disease, they still suffered massive attrition in athymic mice. The elimination of disseminated cancer cells may be due to metabolic and mechanical stresses (Goss and Chambers, 2010; Sosa et al., 2014), but also to immune surveillance (Dunn et al., 2004). Foxn1nu athymic mice retain functional NK cells and other components of the innate immune system, raising the possibility that immune surveillance restricts the expansion of LCC cells in these mice. To test this possibility, we inoculated LCC cells into NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice (NOD/SCID Gamma, NSG mice), which are defective for both adaptive and innate immune responses (Shultz et al., 2005). Strikingly, H2087-LCC and HCC1954-LCC cells formed overt metastasis with high penetrance (Figure 4A-B), in multiple organs (Figure 4C-D). These organs included the liver, a site normally rich in NK cells (Sojka et al., 2014). As the metastatic colonies expanded they accumulated cancer cells with varying levels of Sox2 and Sox9 (Figure S4A).
Figure 4. LCC cells are in equilibrium with the innate immune system.
(A-B) BLI tracking of NSG mice injected with the indicated cell lines. Each line represents one mouse.
(C) Representative images of BLI signal in NSG mice injected with the indicated LCC cell lines over the indicated time period.
(D) Metastatic outgrowth of HCC1954-LCC1 (green) in the liver, brain, kidney and lungs of NSG mice 2 months post-injection. Vasculature: isolectin-B4 (red).
(E-F) Depletion of NK cells by either anti-NK1.1 (PK136) or anti-asialo-GM1 antibody regimen allows outgrowth of HCC1954-LCC1 cells injected in athymic nude mice. Outgrowth quantified by whole body photon flux at 2 months post-injection, with whisker plots representing minimum and maximum values. N = 5–7 mice per group. P <0.001 (***), Mann-Whitney Test. Representative ex vivo brain BLI images of mice and brain tissue sections show massive outgrowth of HCC1954-LCC1 cells (green) upon NK depletion (F).
(G) NK cell depletion regimens also allow for outgrowth of H2087-LCC2 cells in athymic nude mice. Whole body photon flux at 2 months post-injection, with whisker plots representing minimum and maximum values. N = 5–7 mice per group. P <0.05 (*), P <0.01 (**), Mann-Whitney Test.
(H) NK depletion scheme in mice injected with LCC cells.
(I-J) NK cell depletion regimen 40 days post extravasation allows for outgrowth of the indicated cell lines in athymic nude mice. Outgrowth quantified by whole body photon flux, with whisker plots representing minimum and maximum values. N = 5 mice per group. P <0.05 (*). Mann-Whitney Test.
Scale bars, 100μm (D and F). See also Figure S4.
To test NK cells as candidate inhibitors of LCC expansion, we depleted athymic mice of NK cells by administration of polyclonal anti-asialo-GM1 antibody or anti-NK1.1 monoclonal antibody PK136 (Kasai et al., 1981; Sun and Lanier, 2008) (Figure S4B). Both NK cell depletion strategies resulted in permissive outgrowth of HCC1954-LCC1 cells, as shown by a marked increase in whole-body BLI signal, and an increased metastatic burden in the brain (Figures 4E-F, S4C). Similarly, NK cell depletion increased bone metastasis by H2087-LCC2 (Figure 4G).
To investigate the role of NK cells in immunocompetent mouse models we used lung adenocarcinoma cell lines derived from tumor-bearing KrasG12D/p53Del mice (Winslow et al., 2011). The 482T1 (T-Met) cell line readily metastasizes to the liver when injected into the spleen of syngeneic B6129SF1 mice, whereas the 368T1 (T-nonMet) line does not. NK cell depletion with anti-asialo-GM1 antibody resulted in a 100-fold increase in the liver metastatic activity of T-nonMet cells, reaching a level that was comparable to that of T-Met cells (Figure S4D-E). In a second model, we used 4T07 cells, a non-metastatic cell line derived from a spontaneous BALB/c mouse mammary tumor (Aslakson and Miller, 1992). NK cell depletion of recipient BALB/c mice significantly increased the overall metastatic activity of 4T07 (Figure S4F). Notably, treatment of athymic mice with anti-asialo-GM1 antibody 40 days after inoculation of H2087-LCC or HCC1954-LCC cells triggered an increase in metastatic burden in bones, lungs and brain compared to controls (Figure 4H-J).
These results demonstrated the ability of latent LCC cells to stochastically initiate outgrowth when NK cell surveillance in lifted. Collectively, the evidence suggests that LCC cells can proliferate after infiltrating distant organs, but NK cell immune surveillance prevents the accumulation of progeny, sparing LCC cells that entered quiescence. Latent LCC cells remain competent to initiate metastatic outgrowth, and abruptly manifest this competence if NK cell surveillance ends.
Downregulation of NK cell activators in quiescent LCC cells
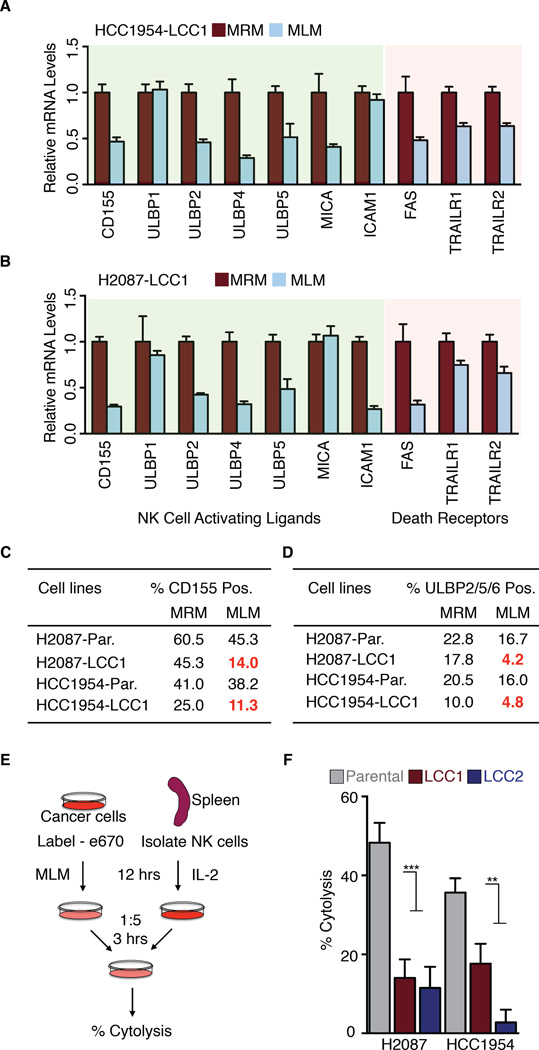
A balance of activating and inhibitory signals regulates the ability of NK cells to target cancer cells (Vesely et al., 2011; Wu and Lanier, 2003). Oncogenic transformation leads to loss of NK cell inhibitory receptors and upregulation of NK cell activating ligands. During tumor progression, cancer cells evolve to escape NK cell mediated recognition (Dunn et al., 2004; Ljunggren and Malmberg, 2007; O'Sullivan et al., 2012). As LCC cells survived NK cell surveillance in athymic mice, we queried LCC gene expression profiles for potential mechanisms of immune evasion. We examined gene expression signatures associated with immune recognition by macrophages, T, B, or NK cells. The expression of NK cytotoxicity signatures was specifically decreased in LCC cells relative to parental cells (Figure S5A-B). Moreover, quiescence (MLM) conditions induced further changes in the expression of NK cell ligands in LCC cells (Figure S5C-D). The specific genes (Figure 5A-B) include several mediators of anti-tumor responses: UL16-binding proteins (ULBP; also known as retinoic acid early transcript, RAET) ULBP2/RAET1H, ULBP3/RAET1N, and ULBP5/RAET1G, which bind to the NK activating receptor NKG2D/CD314 (Lanier, 2015), and PVR/CD155, a ligand for the cancer cell killing NK cell receptor CD226/DNAM-1 (DNAX accessory molecule-1) (Martinet and Smyth, 2015). The pro-apoptotic cytokine receptors FAS and TRAILR, which are critical for NK cell-mediated target killing (Bradley et al., 1998; Takeda et al., 2001), were also downregulated in LCC cells under quiescence conditions (Figure 5A-B). We confirmed the down-regulation of CD155 and ULBPs by flow cytometry analysis (Figure 5C-D, S5E-F).
Figure 5. LCC cells evade NK cell mediated immune surveillance.
(A-B) Genes important for NK cell recognition and cytotoxicity are downregulated in the indicated cell lines when grown in MLM conditions.
(C-D) Cell surface expression of NK cell activating ligands CD155 and ULBP 2/5/6 in the indicated cell lines under MRM and MLM culture conditions.
(E) Schematic of in vitro NK cell cytotoxicity experiment. Labeled cancer cells were incubated with activated primary NK cells at a 1:5 target:effector ratio for 3 hours, followed by quantification of remaining intact cancer cells.
(F) H2087 and HCC1954-LCC cells are more resistant to NK-cell mediated cytotoxicity compared to parental counterparts. Data are mean percentage of cytolysis ± S.E.M. for three replicates. P <0.01 (**), P <0.001 (***), Student’s t-test. See also Figure S5.
To determine whether LCC cells entering quiescence are intrinsically resistant to the cytotoxic action of NK cells, we incubated parental and LCC cells in low-mitogen media and then added freshly isolated, IL-2 activated mouse spleen NK cells to the cultures (Figure 5E). Compared to parental populations, LCC cells showed resistance to cytolysis when incubated with NK cells (Figure 5F). Thus, LCC cells that enter quiescence undergo a striking downregulation of NK cell activators, and acquire resistance to NK cell mediated killing.
Attenuated WNT signaling is associated with LCC quiescence
To identify what primes LCC cells to enter this immune evasive quiescent state, we applied signaling pathway classifier analysis to the transcriptomic data sets (Zhang et al., 2009). Under quiescence conditions, H2087-LCC cell isolates showed a reduction in WNT, MYC and NF-kB signaling, and an increase in TGF-β signaling (Figure 6A). WNT signaling was also attenuated in HCC1954 LCC cells under these conditions (Figure S6A). WNT signaling up-regulates MYC (He et al., 1998), and TGF-β causes Myc down-regulation (Massague, 2012). A high ratio of phospho-p38 to phospho-ERK kinases, previously described in dormant cancer cell models (Sosa et al., 2014), was present in HCC1954-LCC but not in H2087-LCC cells (Figure S6B).
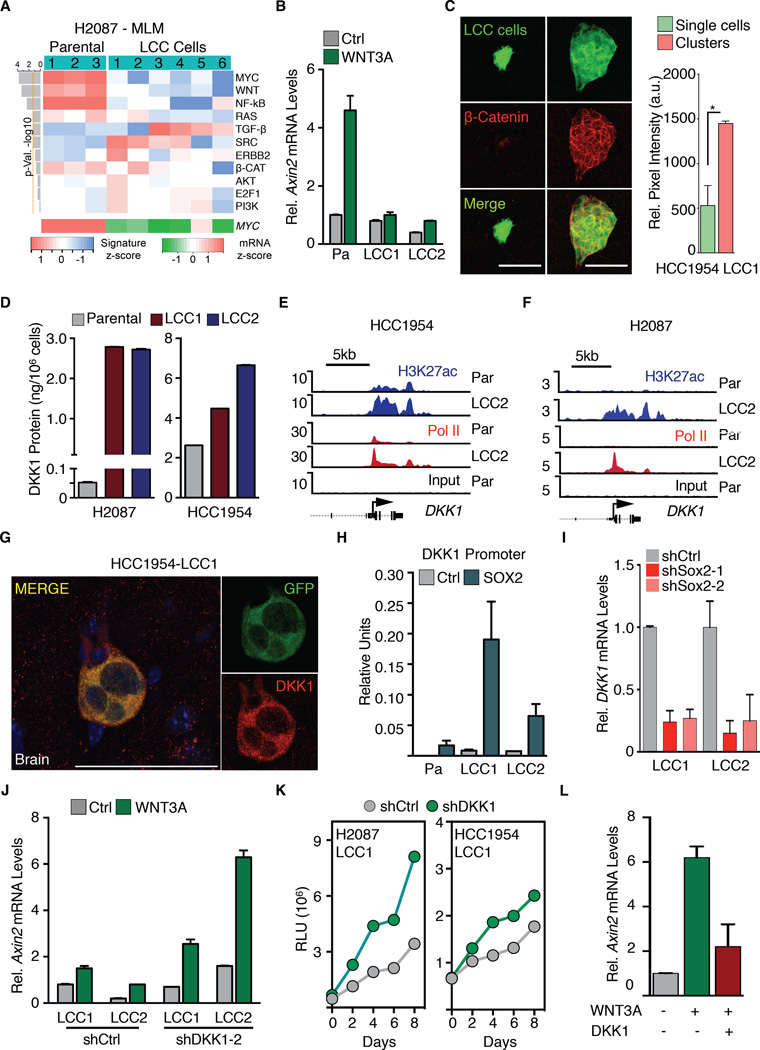
Figure 6. Attenuated WNT signaling enforces quiescence in LCC cells.
(A) Signaling pathway response signature analysis of H2087 derivatives under MLM conditions shows marked reduction in WNT and MYC pathway activity (upper panel). MYC mRNA z score of these derivatives was scored from RNA-seq data (lower panel).
(B) H2087-LCC cells are less responsive to WNT pathway activation by WNT3A stimulation as measured by Axin2 expression.
(C) Immunofluorescence images showing active β-Catenin expression (red) in clusters compared to single cells. Quantification of relative pixel intensity of β-Catenin is shown. N = 5 representative sections per condition. P < 0.05 (*), Student’s t-test.
(D) DKK1 protein expression levels (ELISA) in H2087 and HCC1954-LCC derivatives relative to parental. Data are mean amount of secreted DKK1 protein ± S.E.M. N = 3 technical replicates per group.
(E-F) Gene track view for H3K27ac and Pol II ChIP-seq data at the DKK1 locus in HCC1954 and H2087 parental and LCC cells. DKK1 gene body is represented at the bottom.
(G) HCC1954-LCC1 cells (green) detected in the brain parenchyma 1 month post-injection in athymic nude mice express DKK1 (red).
(H) Sox2 binding to the DKK1 promoter assayed by Sox2 ChIP followed by qRTPCR analysis of the DKK1 promoter in H2087 derivatives. Error bars ± S.D.
(I) Dkk1 mRNA expression is diminished in H2087-LCC cells depleted of Sox2.
(J) WNT pathway activation by WNT3a stimulation in H2087-LCC cells depleted of DKK1. Relative mRNA expression of Axin2 used as marker for WNT pathway activation.
(K) Tracking of cell proliferation in the indicated cell lines cultured under MLM conditions by CellTiter-Glo assay.
(L) Responsiveness of H2087 Parental cells to WNT-signaling induction by WNT3A stimulation in the presence or absence of recombinant human DKK1.
Scale bars, 50μm (C and G). See also Figure S6.
The drop in WNT signaling in LCC cells that entered quiescence was intriguing. WNT is a potent mitogen for stem and progenitor cells, and is implicated in the metastatic outgrowth of lung adenocarcinoma (Nguyen et al., 2009) and breast cancer stem cells (Malanchi et al., 2012; Oskarsson et al., 2011). H2087-LCC cells in MLM conditions showed resistance to pathway activation by WNT3A addition, as determined by Axin2 expression (Figure 6B) and TCF transcriptional reporter activation (Figure S6C) (Fuerer and Nusse, 2010). We assessed WNT activity in LCC cells in vivo by immunostaining for active β-catenin, a marker for canonical WNT pathway activation. We detected higher levels of active β-catenin in proliferating LCC cell clusters than in single disseminated LCC cells in the brain (Figure 6C). Thus, LCC cells have a propensity to resist WNT pathway activation.
Autocrine DKK1 expression in LCC cells
These results raised the possibility that LCC cells entering quiescence express a WNT inhibitor. Indeed, we observed that the WNT inhibitor dickkopf-related protein 1 (DKK1) was highly expressed in LCC cell lines (Figure 6D, S6D). ChIP-seq analysis showed a clear increase in H3K27ac and Pol II peaks at the DKK1 locus in both LCC models, compared to the parental lines (Figure 6E-F). Immunofluorescence staining demonstrated DKK1 expression in LCC cells in vivo (Figure 6G).
We extended these results with breast cancer patient derived xenografts (PDXs). Surgical orthotopic implantation of the hormone receptor-negative PDX models HCI-001 and HCI-002, and the HER2+ PDX model HCI-008 gives rise to tumors with no visible metastasis (DeRose et al., 2011). PDX tumors expressed human vimentin, which we used as a surrogate marker to identify DTCs in the lungs, brain and kidneys of mice bearing these tumors. 14%-51% of disseminated cancer cells were positive for both human vimentin and DKK1 (Figure S6E-F), demonstrating an association of DKK1 with DTCs from orthotopically implanted PDX models.
DKK1 is a direct transcriptional target of Sox2 in mesenchymal stem cells, with Sox2 binding 75 bp upstream of the transcriptional start site (Park et al., 2012; Seo et al., 2013). Indeed, H2087-LCC1 cells showed Sox2 binding to this promoter region (Figure 6H). Stable shRNA knockdown of Sox2 in LCC cells decreased the expression of DKK1 (Figure 6I). DKK1 knockdown (Figure S6G-H) re-sensitized LCC cells to WNT pathway activation (Figure 6J). CellTiter-Glo assays and dye retention assays (Figures 6K, S6I) showed that DKK1 knockdown stimulated the proliferation of LCC cells under low-mitogen conditions. In complementary experiments, incubation of parental H2087 cells with recombinant DKK1 inhibited WNT3A response and proliferation in LCC cells (Figures 6L, S6J).
Autocrine DKK1 enforces a quiescent, immune evasive state in latent metastasis
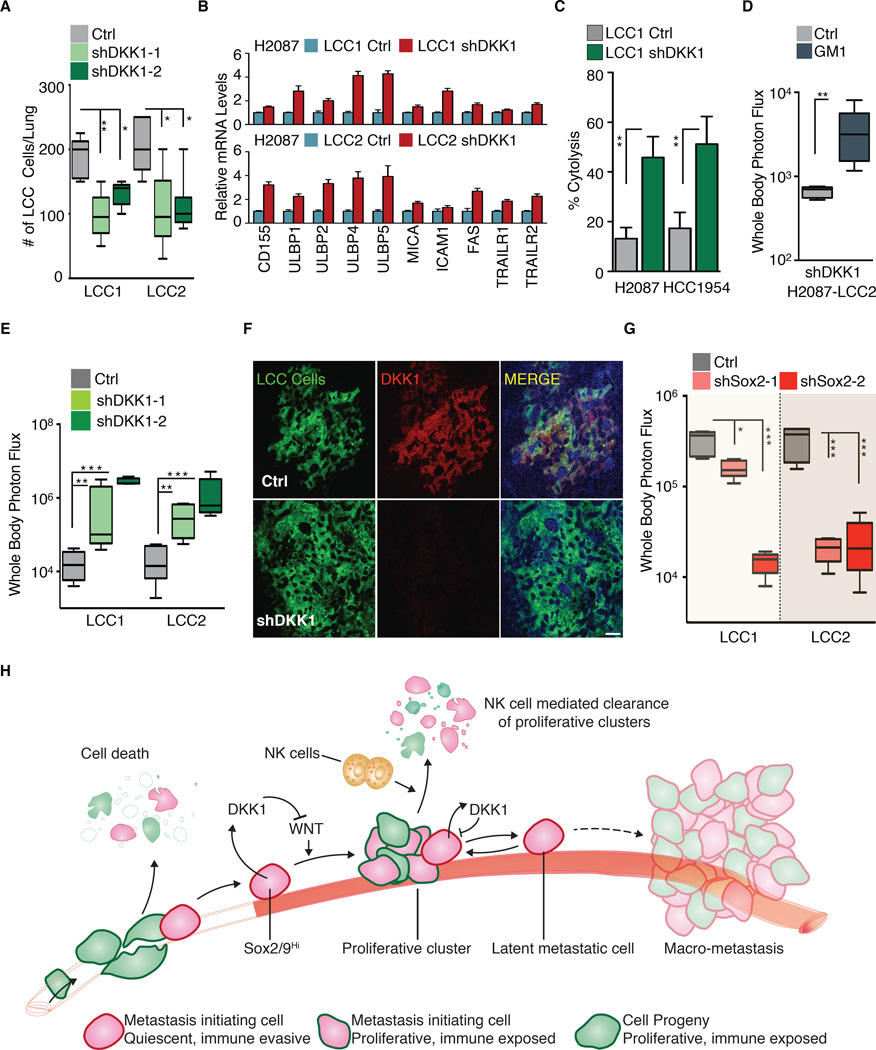
The preceding results suggested that autocrine DKK1 primes LCC cells to enter quiescence, which leads to downregulation of NK cell activators. One prediction of this model is that autocrine DKK1 is required to protect LCC cells from elimination in athymic mice. In line with this prediction, shRNA-mediated DKK1 depletion in H2087-LCC cells significantly decreased the accumulation of LCC cells in the lungs of inoculated athymic mice (Figure 7A). Ki-67+ stromal cells surrounded the cancer cells in bronchiolar regions under these conditions. Many of these stromal cells were positive for the leukocyte common antigen CD45, and included Ly6B.2+ neutrophils, and F4/80+ inflammatory monocytes and macrophages, suggesting that LCC cell proliferation triggers lymphocyte infiltration for cancer cell clearance (Figure S7A-B). DKK1 knockdown in H2087-LCC cells increased the expression of NK cell activating ligands ULBP1, 2, 4 and 5, and death signal receptors (Figure 7B). Conversely, addition of a high concentration of WNT3A to H2087-LCC1 cells increased the expression of ULBP1 and ICAM1 (Figure S7B). DKK1-depleted cells were more susceptible to NK cell mediated cytotoxicity (Figure 7C). Moreover, depletion of NK cells in athymic mice rescued the formation of macrometastases by DKK1-knockdown H2087-LCC cells (Figure 7D, S7C).
Figure 7. DKK1 enforces a quiescent immune evasive state.
(A) Growth of H2087-LCC cells depleted of DKK1 in athymic nude mice. Data are total number of LCC cells detected per lung after 3 months post-tail vein injection ± S.E.M. N = 5–6 mice per group, scoring representative serial sections of the entire lung for each mouse.
(B) Relative mRNA expression of NK cell activating ligands and death receptors in H2087-LCC cells upon DKK1 depletion. Relative quantification is normalized to LCC1 or LCC2 control.
(C) H2087 and HCC1954-LCC cells depleted of DKK1 are more susceptible to NK-cell mediated cytotoxicity compared to their controls. Data are mean percentage of cytolysis ± S.E.M. for three replicates. P <0.01 (**), Student’s t-test.
(D) Depletion of NK cells by anti-asialo-GM1 in athymic nude mice injected with shDKK1 bearing H2087-LCC2 cells. Whole body photon flux, 2 months post-injection. N = 5–6 mice per group. P <0.01 (**), Mann-Whitney Test.
(E) Growth of H2087-LCC cells depleted of DKK1 in NSG mice. Whole body photon flux, 2 months post-injection. N = 5–6 mice per group. P <0.01 (**), P <0.001 (***), Mann-Whitney Test.
(F) Representative immunofluorescence images of metastatic lesions in the lungs of NSG mice in Figure 7E.
(G) Growth of H2087-LCC cells depleted of Sox2 in NSG mice. Whole body photon flux, 2 months post-injection. N = 5–6 mice per group. P <0.05 (*), P <0.001 (***), Mann-Whitney Test.
(H) A model summarizing the central tenets of latency recapitulated in the present latency competent cancer cell models. A majority of disseminated cancer cells including LCC cells suffer massive attrition in circulation or upon extravasation due to mechanical and metabolic stress and immune surveillance. LCC cells associate with the vasculature, are enriched for Sox2/Sox9 stem cell-like programs, stochastically enter proliferative quiescence, and are superior at seeding organs. LCC cells can proliferate in response to stimulatory microenvironmental cues after infiltrating distant organs, but NK cell dependent immune surveillance prevents accumulation of their progeny, sparing only LCC cells that entered quiescence stochastically. Cells enriched for DKK1 are able to attenuate the proliferative response to local WNT signals, enter quiescence. Quiescent cells downregulate the expression of cell surface NK sensors to evade immune surveillance. Surviving latent metastatic cells may evolve, accumulating traits for eventual outbreak to form macrometastases (refer to Discussion).
Scale bars, 50μm (F). See also Figure S7.
In contrast to the deleterious effect of DKK1 knockdown on the viability of LCC cells in athymic mice, DKK1 knockdown increased the metastatic growth of H2087 and HCC1954 LCC cells in NSG mice (Figure 7E-F, S7D). Conversely, overexpressing DKK1 in H2087-parental cells decelerated their growth in NSG mice (Figure S7E-F). As a corollary, depletion of Sox2 in H2087-LCC or Sox9 in HCC1954-LCC cells inhibited the ability of these cells to form metastasis in NSG mice (Figure 7G, S7G). No significant effect was observed upon Sox2 and Sox9 depletion in parental populations (Figure S7H). These data confirmed the essential role of Sox transcription factors for metastatic growth of LCC cells in the presence or absence of NK cells, and the ability of NK cells to restrain LCC outgrowth but spare LCC cells that enter quiescence.
DISCUSSION
Our results show that cancer cell populations selected from lung and breast cancer cell lines for their competence to establish latent metastasis have high expression of stem/progenitor markers. They also have a capacity to enter immune evasive quiescence while retaining metastasis-initiating powers. These cells, which we operationally call latency competent cancer cells, suffer extensive attrition upon infiltrating target organs, as their unselected parental populations do. However, LCC cells can stochastically enter a self-imposed quiescent state, which leads to downregulation of NK cell ligands for evasion of immune surveillance. NK cells kill dividing LCC cells but spare quiescent LCC cells. As a result, cancer cells in these models can persist long-term as latent metastasis seeds in different organs (Figure 7H).
Stem-like nature of latency competent cancer cells
A defining trait of LCC cells is their stem-like phenotype. At the molecular level, breast cancer and lung cancer LCC cells resemble mammary stem cells or lung alveolar bi-potent progenitors, respectively. LCC cells express Sox2 or Sox9, transcription factors that play critical roles in stem cell identity and pluripotency. Sox2 and Sox9 enforce stemness in lung and breast adenocarcinomas, respectively (Arnold et al., 2011; Guo et al., 2012). We show that expression of Sox2 and Sox9 is essential for the survival and metastasis-initiating properties of LCC cells in multiple host tissues. Adult stem cells can exist in an actively dividing pool for homeostatic tissue renewal, and a slow-cycling pool responsible for tissue regeneration upon injury (Li and Clevers, 2010; Lien and Fuchs, 2014). LCC cells may be segmented into these two phenotypes as well. After infiltrating target organs, proliferating LCC cells are eliminated by NK cells. However, by entering quiescence, a minority of LCC cells can evade NK cell surveillance (Figure 7H).
Self-imposed quiescence
Growth inhibitory signals from the host microenvironment and the perivascular niche, including TGF-β and BMP, can contribute to metastatic dormancy (Sosa et al., 2014). However, disseminated cancer cells are likely exposed to mitogenic signals during the course of latency periods, as perivascular niches in the host organs support tissue homeostasis and regeneration though stromal WNT and other proliferative signals. Our results demonstrate an innate ability of LCC cells to self-impose a slow cycling state in this context. This ability is based on the expression of the WNT inhibitor DKK1, which prevents activation of β-catenin and LCC cell proliferation. DKK1 is a direct transcriptional target of Sox2 in LCC cells. We show that autocrine DKK1 helps disseminated LCC cells enter quiescence. In the model, this self-imposed quiescence counterbalances the action of stromal WNT signals to preserve the viability of residual LCC cells in the host tissue (Figure 7H).
Quiescence-associated immune evasion
Proliferative quiescence protects LCC cells from NK-mediated killing. On entering quiescence LCC cells downregulate cell surface ULBP activators of NK cell-mediated cytotoxicity and receptors for cell death signals. In mice with an active innate immune system, LCC cells are mostly found as quiescent single cells. Rare LCC cell clusters observed under these conditions contain actively proliferating cells, but may be fated to undergo NK-mediated clearance. Depletion of NK cells by different means in different mouse models led to aggressive metastatic outgrowth of LCC cells, arguing that quiescent LCC cells stochastically enter the cell cycle, and the proliferative clusters will progress to macrometastases if immune surveillance is relaxed. The ability to undergo periodic bursts of proliferation and elimination, with a small fraction of the progeny entering quiescence and surviving after each round, would allow disseminated cancer cells to evolve accumulate additional metastatic traits, such as active immunosuppression and organ-specific colonization traits, for an eventual macrometastatic outbreak (Figure 7H). Metastatic evolution would be less likely in a permanently quiescent cancer cell population.
Therapeutic implications
Our observations in latent metastasis are in line with the role of immune surveillance in restricting malignant cell proliferation in other contexts. Clinically, the presence of NK cell infiltrates in primary colorectal, gastric and lung cancer correlates with better patient survival outcomes, consistent with an ability of NK to keep metastases in check (Jin et al., 2014; Villegas et al., 2002). Because NK cells have the ability to limit LCC cell outgrowth, a drop in NK cytotoxicity index in disease-free cancer patients might serve as a prognostic indicator of disease relapse. The possibility of treating residual disease by inducing proliferation in order to re-sensitize cancer cells to adjuvant cytotoxic chemotherapy would entail the risk of triggering metastasis while trying to prevent it. Our findings raise the possibility of selectively reactivating NK cell ligands in quiescent metastatic cells in order to trigger the immunologic elimination of latent metastasis.
EXPERIMENTAL PROCEDURES
Animal Studies
All animal experiments were done in accordance with the guidelines provided by the MSKCC Institutional Animal Care and Use Committee. Athymic (NCI/Charles River/Harlan), BALB/c, B6129SF1 or NOD SCID gamma (Jackson Laboratory) female mice from 4–6 weeks of age were used for in vivo studies. For experimental metastasis assays, 1.0×105 cells were resuspended in 100ul 1X PBS and intracardially injected into the right ventricle with a 26G tuberculin syringe. Lung colonization assays were performed by injecting 0.5/1.0×105 cells into the lateral tail vein of mice with a 28G insulin syringe. We detected metastatic burden by non-invasive bioluminescence imaging of experimental animals using an IVIS Spectrum (PerkinElmer). Bioluminescence signal was measured using the ROI tool in Living Image 4.4 software (PerkinElmer).
LCC Isolation and Culture
1.0×105 cells expressing a lentiviral TK-GFP-Luciferase (TGL) construct in combination with antibiotic resistance marker blasticidin (H2087) or puromycin (HCC1954) in a volume of 100 μl was intracardially injected into the left cardiac ventricle of anesthetized (ketamine 100 mg/kg, xylazine 10 mg/kg) 4–6 week old athymic mice. Cancer cell colonization was tracked by retro-orbital injection of D-luciferin (150 mg/kg) and imaged with an IVIS Spectrum (PerkinElmer). Bioluminescence (BLI) analysis was performed with Living Image 4.4 software.
Organs from BLI-negative mice were resected under sterile conditions, and mechanically dissociated using a gentleMACS dissociator (Miltenyl Biotec) and placed in culture medium containing a 1:1 mixture of DMEM/Ham’s F12 supplemented with 0.125% collagenase III and 0.1% hyaluronidase. Minced samples were incubated at 37°C for 1 hr, with gentle rocking to produce single cell suspensions. After collagenase treatment, cells were briefly centrifuged, resuspended in 0.25% trypsin, and incubated for a further 15 min at 37°C. Cells were then resuspended in their respective culture conditions and allowed to grow to confluence on a 15cm dish. Cancer cells from these BLI-negative organs were selected for by adding respective antibiotics to the media.
Supplementary Material
Acknowledgments
We thank P. Bos, T. O’Sullivan, and J. Sun for insightful discussions and advice on NK cell depletion experiments; A. Welm for breast PDX models; S. Monette for review of pathology slides; T. Jacks and F. Giancotti for cell lines; and A. Obenauf, K. Ganesh, A. Laughney and E. Er for critical reading of the manuscript. We acknowledge the support of the MSKCC Genomics Core Facility and Molecular Cytology Core Facility. This work was supported by NIH grants P01-CA094060 and P01-CA129243, and DOD Innovator Award W81XWH-12-1-0074 (J.M.), and NIH grant P30-CA008748 (MSKCC). S.M. is supported by an American Cancer Society post-doctoral fellowship. X.J. is a Susan G. Komen Fellow.
Footnotes
AUTHOR CONTRIBUTIONS
S.M. and J.M. conceived the project and designed the experiments. S.M. and D.G.M. performed most experiments. X.J. and L.H. assisted with experiments. E.D. provided assistance with PDX models. H.B. performed ChIP-seq experiments. X.J., D.G.M., and Y.Z. performed computational analysis. S.M., D.G.M. and J.M. wrote the paper. All authors discussed results and revised the manuscript.
ACCESSION NUMBERS
The raw RNA-seq and ChIP-seq data for this manuscript are available at GEO under the accession number GSE72956.
Authors declare no financial interests in connection with this work.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, Hochedlinger K. Sox2(+) adult stem and progenitor cells are important for tissue regeneration and survival of mice. Cell Stem Cell. 2011;9:317–329. doi: 10.1016/j.stem.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M, Zeytun A, Rafi-Janajreh A, Nagarkatti PS, Nagarkatti M. Role of spontaneous and interleukin-2-induced natural killer cell activity in the cytotoxicity and rejection of Fas+ and Fas− tumor cells. Blood. 1998;92:4248–4255. [PubMed] [Google Scholar]
- Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, Schewe DM, Aguirre-Ghiso JA. TGF-beta2 dictates disseminated tumour cell fate in target organs through TGF-beta-RIII and p38alpha/beta signalling. Nat. Cell. Biol. 2013;15:1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N. Engl. J. Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Gene. Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchnowska R, Dziadziuszko R, Czartoryska-Arlukowicz B, Radecka B, Szostakiewicz B, Sosinska-Mielcarek K, Karpinska A, Staroslawska E, Kubiatowski T, Szczylik C. Risk factors for brain relapse in HER2-positive metastatic breast cancer patients. Breast Cancer Res. Tr. 2009;117:297–303. doi: 10.1007/s10549-008-0275-z. [DOI] [PubMed] [Google Scholar]
- Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Eyles J, Puaux AL, Wang X, Toh B, Prakash C, Hong M, Tan TG, Zheng L, Ong LC, Jin Y, et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J. Clinical Invest. 2010;120:2030–2039. doi: 10.1172/JCI42002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PloS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, Shen R, Brogi E, Brivanlou AH, Giancotti FG. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int. J. Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J. Cell. Biochem. Suppl. 1996;24:1–11. doi: 10.1002/jcb.240630502. [DOI] [PubMed] [Google Scholar]
- Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat. Rev. Cancer. 2010;10:871–877. doi: 10.1038/nrc2933. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Deng Y, Hao JW, Li Y, Liu B, Yu Y, Shi FD, Zhou QH. NK cell phenotypic modulation in lung cancer environment. PloS One. 2014;9:e109976. doi: 10.1371/journal.pone.0109976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai M, Yoneda T, Habu S, Maruyama Y, Okumura K, Tokunaga T. In vivo effect of anti-asialo GM1 antibody on natural killer activity. Nature. 1981;291:334–335. doi: 10.1038/291334a0. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat. Rev. Immunol. 2015;15:73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Okuda H, Xing F, Pandey PR, Watabe M, Hirota S, Pai SK, Liu W, Fukuda K, Chambers C, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J. Exp. Med. 2011;208:2641–2655. doi: 10.1084/jem.20110840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordes U, Hagel C. Expression of SOX9 and SOX10 in central neuroepithelial tumor. J. Neuro-Oncol. 2006;80:151–155. doi: 10.1007/s11060-006-9180-7. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 2015;3:575–582. doi: 10.1158/2326-6066.CIR-15-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Fuchs E. Wnt some lose some: transcriptional governance of stem cells by Wnt/beta-catenin signaling. Gene. Dev. 2014;28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljunggren HG, Malmberg KJ. Prospects for the use of NK cells in immunotherapy of human cancer. Nat. Rev. Immunol. 2007;7:329–339. doi: 10.1038/nri2073. [DOI] [PubMed] [Google Scholar]
- MacKie RM, Reid R, Junor B. Fatal melanoma transferred in a donated kidney 16 years after melanoma surgery. N. Engl. J. Med. 2003;348:567–568. doi: 10.1056/NEJM200302063480620. [DOI] [PubMed] [Google Scholar]
- Maeda R, Yoshida J, Hishida T, Aokage K, Nishimura M, Nishiwaki Y, Nagai K. Late recurrence of non-small cell lung cancer more than 5 years after complete resection: incidence and clinical implications in patient follow-up. Chest. 2010;138:145–150. doi: 10.1378/chest.09-2361. [DOI] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- Martinet L, Smyth MJ. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- Massague J, Obenauf AC. Metastatic colonization by circulating tumor cells. Nature. 2016 doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFβ signaling in context. Nat. Rev. Mol. Cell. Biol. 2012;18:5521–5525. [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan T, Saddawi-Konefka R, Vermi W, Koebel CM, Arthur C, White JM, Uppaluri R, Andrews DM, Ngiow SF, Teng MW, et al. Cancer immunoediting by the innate immune system in the absence of adaptive immunity. J. Exp. Med. 2012;209:1869–1882. doi: 10.1084/jem.20112738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massague J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SB, Seo KW, So AY, Seo MS, Yu KR, Kang SK, Kang KS. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2012;19:534–545. doi: 10.1038/cdd.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelleitier M, Montplaisir S. The nude mouse: a model of deficient T-cell function. Methods Achiev. Exp. Pathol. 1975;7:149–166. [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012;44:1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Basu-Roy U, Gunaratne PH, Coarfa C, Lim DS, Basilico C, Mansukhani A. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Rep. 2013;3:2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, Zhong C, Chase JM, Rothman PB, Yu J, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer. 2014;14:611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Lanier LL. Cutting edge: viral infection breaks NK cell tolerance to "missing self". J. Immunol. 2008;181:7453–7457. doi: 10.4049/jimmunol.181.11.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Smyth MJ, Cretney E, Hayakawa Y, Yamaguchi N, Yagita H, Okumura K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in NK cell-mediated and IFN-gamma-dependent suppression of subcutaneous tumor growth. Cell. Immunol. 2001;214:194–200. doi: 10.1006/cimm.2001.1896. [DOI] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- Villegas FR, Coca S, Villarrubia VG, Jimenez R, Chillon MJ, Jareno J, Zuil M, Callol L. Prognostic significance of tumor infiltrating natural killer cells subset CD57 in patients with squamous cell lung cancer. Lung Cancer. 2002;35:23–28. doi: 10.1016/s0169-5002(01)00292-6. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lanier LL. Natural killer cells and cancer. Adv. Cancer Res. 2003;90:127–156. doi: 10.1016/s0065-230x(03)90004-2. [DOI] [PubMed] [Google Scholar]
- Xiao D, Craig JC, Chapman JR, Dominguez-Gil B, Tong A, Wong G. Donor cancer transmission in kidney transplantation: a systematic review. Am. J. Transplant. 2013;13:2645–2652. doi: 10.1111/ajt.12430. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.