Abstract
To deepen our understanding of early rectal transmission of HIV-1, we studied virus-host interactions in the rectal mucosa using simian immunodeficiency virus (SIV)-Indian rhesus macaque model and mRNA deep sequencing. We found that rectal mucosa actively responded to SIV as early as 3 days post-rectal inoculation (dpi) and mobilized more robust responses at 6 and 10 dpi. Our results suggests that the failure of the host to contain virus replication at the portal of entry is attributable to both a high-level expression of lymphocyte chemoattractant, proinflammatory and immune activation genes, which can recruit and activate viral susceptible target cells into mucosa; and a high-level expression of SIV accessory genes, which are known to be able to counter and evade host restriction factors and innate immune responses. This study provides new insights into the mechanism of rectal transmission.
Keywords: Simian Immunodeficiency Virus (SIV), rectal transmission, mucosal response, mRNA-seq
Introduction
Anal-rectal mucosa is an important portal for HIV-1 entry in both men and women, and especially among men who have sex with men (MSM). MSM have a higher risk of contracting HIV-1 infection in both developed and developing countries than any other population (CDC, 2011; van Griensven et al., 2010). It has been estimated that one-half to two-thirds of MSM participate in anal intercourse (Mansergh et al., 2001; McGowan, 2008), and up to 40% of heterosexual men and 35% of women engage in anal intercourse (Kingsley et al., 1987; van Griensven et al., 2010). While the single columnar epithelial mucosal barrier and abundant HIV-1-susceptible target cells in the rectum may explain the increased rate of acquisition, the actual mechanisms have not been determined and the current knowledge about the earliest virus-host interactions in anorectal transmission is limited. It has been demonstrated that both lamina propria and epithelial cells of rhesus small intestines at three weeks post SIV infection have robust transcriptional responses (Mohan et al., 2012, 2013), however, the host responses in days post infection via rectal route, especially at the portal of entry, remain unknown. To better understand the earliest events of HIV-1 rectal transmission, we studied early virus-host interactions in rectal mucosa of rhesus macaques intra-rectally inoculated with simian immunodeficiency virus (SIV), the best available model of human rectal transmission of HIV-1 (Keele et al., 2009). Our study revealed profound changes in rectal mucosal gene expression as early as 3 days post-inoculation (dpi) using next-generation RNA sequencing (mRNA-seq), a time point before systemic viral dissemination, as compared with uninfected macaques. Interestingly at 3 dpi, chemokine CCL13/MCP-4 and CCL20/MIP-3α genes, which are well-known chemoattractants for T cells, monocytes and plasmacytoid dendritic cells, were up-regulated. At 6 and 10 dpi, a greater number of genes were significantly altered in expression, including proinflammatory, immune activation, chemoattractant for lymphocytes, and interferon-stimulated genes (ISGs) targeting various steps of the viral replication life cycle. Despite the strong mucosal anti-viral responses, the virus still subdued the host. Our data suggest that the key tactic the virus used to evade the mucosal host antiviral responses may be through high-level expression of SIV accessory genes and inducing chemoattractant, proinflammatory as well as immune activation genes. Together, this first comprehensive genome wide study of early virus-host interaction in rectum provides new insights into the mechanism of rectal transmission and identifies potential targets for future studies
Results
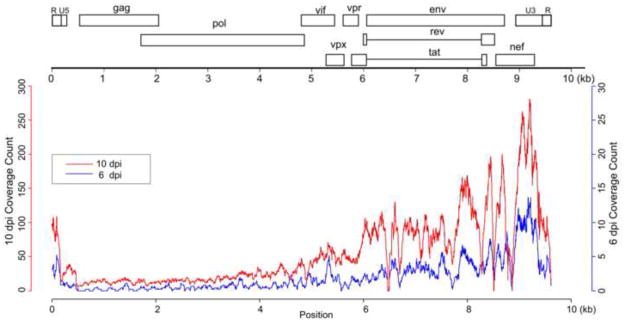
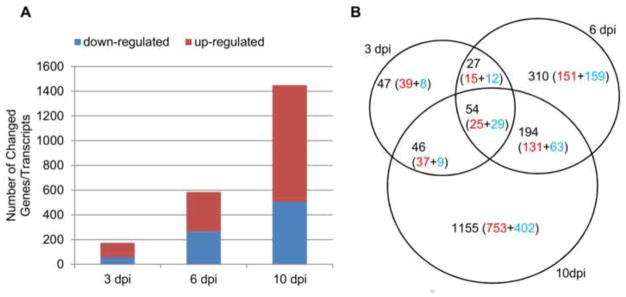
To elucidate the virus-host interaction in rectal mucosae during early rectal SIV transmission, mRNA-seq was used to simultaneously measure host and virus gene expression. Host genes with differential expression (adjusted p-value < 0.05, fold change > 2) were identified (Supplemental Table 1) and used for analysis. At 3 dpi, while SIV viral RNAs were detectable in the rectum by both quantitative RT-PCR and in situ hybridization (ISH), but not in distal lymph nodes and blood (manuscript in preparation), SIV viral RNAs were not detected in the rectum by RNA-seq, indicating SIV was only replicating locally and at low-level in this time point and RNA-seq is less sensitive to detect low-levels of viral RNA than RT-PCR and ISH. At 6 and 10 dpi, SIV vRNA was detected in rectum, blood and distal lymph nodes of all the rhesus macaques by RT-PCR and ISH (manuscript in preparation); furthermore SIV viral RNAs in the rectum were also detected by RNA-seq, and the read counts at 10 dpi are higher than 6 dpi (Fig. 5). The mRNA-seq analyses showed that host rectal mucosa responded to SIV as early as 3 dpi, with 116 (66.7%) genes up-regulated and 58 (33.3%) genes down-regulated (Fig. 1A). At day 6, there were 585 differentially expressed genes (DEGs), among which 320 (54.7%) were up-regulated and 265 (45.3%) were down-regulated. The largest number of DEGs occurred at 10 dpi, with 943 (65.1%) up-regulated and 506 (34.9%) down-regulated. Both unique and shared DEGs between each time point were observed, with 47 (3 dpi), 310 (6 dpi) and 1,155 (10 dpi) unique genes (Fig. 1B and supplementary Fig. 1). Notably, there was a clear preponderance of unique and shared DEGs during the course of infection, reflecting the enhanced host’s responses to infection over time. To better understand the biological functions of the altered genes and their associated signaling pathways in the context of early rectal SIV transmission, DEGs were further classified (Fig. 2). There were 37.9% (66/174), 26.8% (157/585) and 46.7% (677/1449) of DEGs classifiable at 3, 6 and 10 dpi, respectively.
Fig. 5.
Transcriptome analysis of SIV replication in rhesus macaques. Coverage of mRNA-seq reads at each position of the SIV genome at 6 dpi and 10 dpi.
Fig. 1.
Differentially expressed genes/transcripts in rectal mucosae of SIV-infected macaques at 3, 6 and 10 dpi. (A) Number of up- and down-related genes at each time point as compared with uninfected control macaques. (B) Uniquely and commonly altered genes at each time point. Up-regulated genes are depicted in red and down-regulated genes in blue.
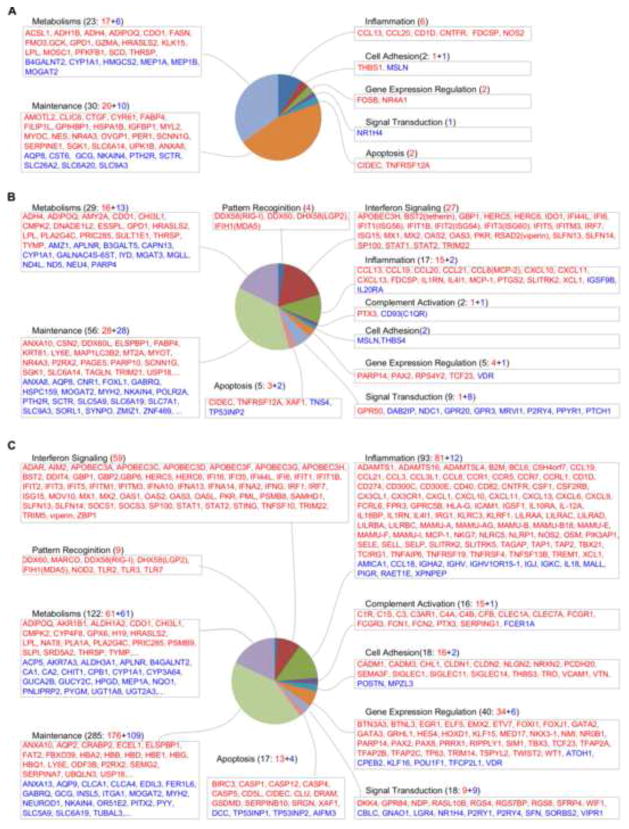
Fig. 2.
Functional categorization of differentially expressed genes in rectal mucosae at 3 (A), 6 (B) and 10 dpi (C). Only the top 20 up- and down-regulated genes in metabolisms and maintenance categories are listed in (B) and (C). Up-regulated genes are shown in red and down-regulated genes in blue.
Host rectal mucosal responses at 3 dpi
As early as 3 dpi, the rectal mucosae of SIV-infected macaques exhibited dramatic changes in gene expression as compared with uninfected control macaques (Fig. 2A). The DEGs encoded proteins involved in apoptosis (CIDEC and TNFRSF12A), cell adhesion (MSLN and THBS1), gene expression (FOSB and NR4A1), inflammation, cell metabolism and maintenance. Interestingly, chemokine CCL13/MCP-4 and CCL20/MIP-3α, which are well-known chemoattractants for T cells, monocytes and plasmacytoid dendritic cells, were up-regulated at 3 dpi. In addition to the genes encoding immune cell attractants, immune activation genes (NOS2, CD1D, CNTFR and FDCSP) were also up-regulated. Notably, FDCSP, which encodes a secreted protein in activated follicular dendritic cells (FDCs), was further up-regulated 3.7- and 10.7-fold at 6 dpi and 10 dpi, respectively, suggesting activation and proliferation of FDCs in rectal tertiary follicular aggregates.
Genes involved in cell metabolism and maintenance accounted for most of the DEGs at 3 dpi. These genes encode for proteins involved in the transportation of water (AQP8), iron (NKAIN4), chloride (CLIC6), sodium (SCNN1G) and other solutes (SLC6A20, SLC9A3, SLC26A2 and SLC6A14) across the cell membrane and protein modification (SGK1). Several DEGs encode for proteins involved in metabolism of lipids (MOGAT2, CYP1A1, THRSP, SCD, HMGCS2, LPL, ACSL1, FASN, ADIPOQ), carbohydrates (GCK, B4GALNT2, PFKFB1), proteins (MEP1B, MEP1A, KLK15, GZMA), alcohol (ADH4, ADH1B) and others (CDO1, FMO3, MOSC1, GPD1).
More active rectal responses at 6 and 10 dpi
Consistent with the elevated viral load and systemic dissemination of SIV at 6 and 10 dpi, there was significantly increased number of DEGs as compared with 3 dpi. Although the genes involved in cell metabolisms and maintenance still accounted for most of the DEGs, we will mainly discuss the immunity-related genes below.
The activation of pattern recognition receptors
Innate immune responses are triggered by host’s pattern recognition receptors (PRRs) bound to pathogen-associated molecular patterns (PAMPs). Four PRR genes (RIG-I, LGP2, MDA5 and DDX60) were up-regulated at 6 dpi, of which RIG-1 and MDA5 were up-regulated 7- and 4.6-fold, respectively, and LGP2, a protein homologous to RIG-I and MDA5, was up-regulated 3.7-fold (Fig. 2B and Fig. 3). Surprisingly, at 10 dpi, in addition to PRRs for virus recognition (RIG-I, MDA5, LGP2, TLR3 and TLR7), PRRs recognizing bacterial patterns (MARCO, NOD2 and TLR2) were also up-regulated (Fig. 2C and Fig. 3). In addition to viral RNA recognition, genes involved in viral DNA recognition (DAI, IFI16 and AIM2) were up-regulated (Fig. 2C and Fig. 3), indicating activation of viral DNA pattern recognition by the presence of SIV DNA in rectal mucosa.
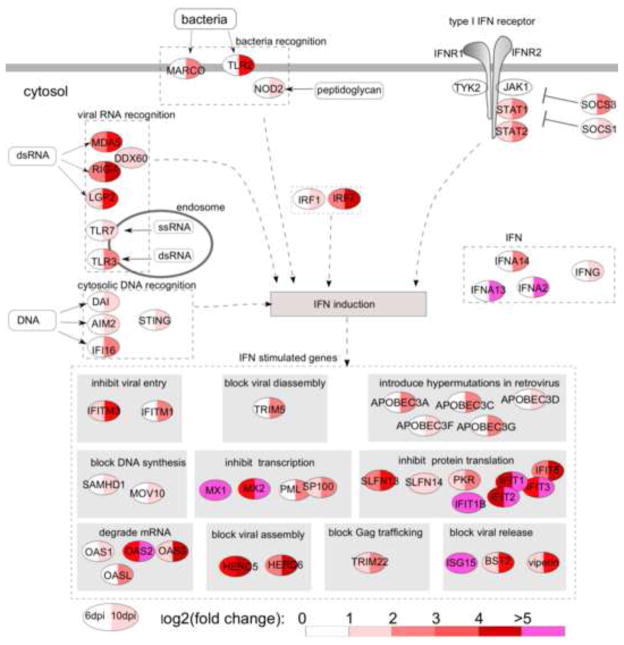
Fig. 3.
Differentially expressed genes related to IFN signaling in rectal mucosae at 6 and 10 dpi. IFN-stimulated genes are classified based on their functions. The levels of up-regulation are color-coded based on log2 (fold change).
Antiviral and IFN signaling activation
Among the DEGs, IFN signaling activation stood out at both 6 and 10 dpi (Fig. 2B and 2C), most of which were IFN-stimulated genes (ISGs) with anti-viral activities. To better understand the functions of these ISGs, we illustrated them in the context of IFN signaling pathways (Fig. 3). At 10 dpi, which was around the peak of viral replication in the rectum, the number of DEGs and the magnitude of up-regulation were significantly increased as compared with 6 dpi.
Interestingly, the genes of anti-viral ISGs were significantly up-regulated, targeting almost every step of virus replication life cycle, i.e., inhibition of viral entry (IFITM1 and IFITM3); blockade of viral disassembly (TRIM5); introduction of hypermutations (APOBEC3A, APOBEC3C, APOBEC3D, APOBEC3F and APOBEC3G); blockade of viral DNA synthesis (SAMHD1 and MOV10); inhibition of viral transcription (MX1, MX2, PML and SP100); degradation of mRNA (OAS1, OAS2, OAS3 and OASL); inhibition of protein translation (SLFN13, SLFN14, PKR and IFIT protein family); blockade of viral Gag protein trafficking (TRIM22); blockade of viral assembly (HERC5 and HERC6); and blockade of viral release (ISG15, BST2/tetherin and viperin). In addition to direct inhibition of the virus, several ISGs involved in viral suppression through antigen presentation (PSMB8) and unknown mechanisms (IFI44L, IFI6, IFI35, TNFSF10 and DDIT4) were also up-regulated.
Inflammation, immune activation and cell trafficking
In addition to active IFN responses, the expression of genes related to the mucosal proinflammatory responses, immune activation and immune cell trafficking was also significantly up-regulated (Fig. 2). At 6 and 10 dpi, CXCL11, the major ligand of CXCR3, was the most up-regulated cytokine, with greater than 100-fold up-regulation at 10 dpi. Other CXCR3 ligands (CXCL10 and CXCL9) were among the most up-regulated DEGs. DEGs encoding CCR5 ligands (CCL3/MIP-1α, CCL3L1, CCL5/RANTES and CCL8/MCP-2) were also up-regulated. Other important chemoattractant genes for T cells (CXCL1, CCR1, CCL13 and CCL8), myeloid dendritic cells (CCL19 and CCL21), monocytes (CCR1, CCL2, CCL13 and CCL8), and neutrophils (CXCL1, FPR3) were significantly up-regulated. Moreover, adhesion/homing receptor L-selectin (SELL/CD62L) and receptor CCR7, which control the migration of memory T cells to inflamed tissues, were up-regulated, suggesting the augmentation of T cell migration to the rectal mucosa. Notably, among the DEGs, IL18 was down-regulated, and IL18 inhibitor (IL18BP) and the immune suppressive molecule CD274/PD-L1 were up-regulated.
Complement and Fc receptor activation
In line with increased inflammatory responses and immune activation, complement and Fc receptor genes (PTX3 and CD93/C1QR) were altered in expression at 6 dpi, and many more genes in this category were altered in expression at 10 dpi, such as components of the complement classical pathway (C1R, C1S, PTX3, CLEC1A), lectin pathway (CLEC1A, FCN1, FCN2), alternative pathway (CFB), or common factors shared by these pathways (C3 and C4), and Fc receptors for IgE (FCER1A) and IgG (FCGR1, FCGR3). All of the above DEGs were up-regulated except FCER1A (-2.5-fold).
Antigen presentation
Interestingly, DEGs highlight the activation of MHC class I antigen presentation. TAP1, TAP2, PSMB8/LMP7/β5i and PSMB9/LMP2/β1i, members of the antigen-processing machinery (APM), were up-regulated. PSMB8 and PSMB9, both induced by IFN-γ, are integral components of the immunoproteasome where peptides are generated and transferred to the endoplasmic reticulum to form peptide-MHC class I complexes. Consistent with enhanced peptide processing and transportation, MHC class I α-chain (MAMU-A, MAMU-AG, MAMU-B, MAMU-E, MAMU-F, MAMU-I) and β-chain (B2M) genes were also up-regulated.
Mapping the DEGs to rectal mucosa in vivo
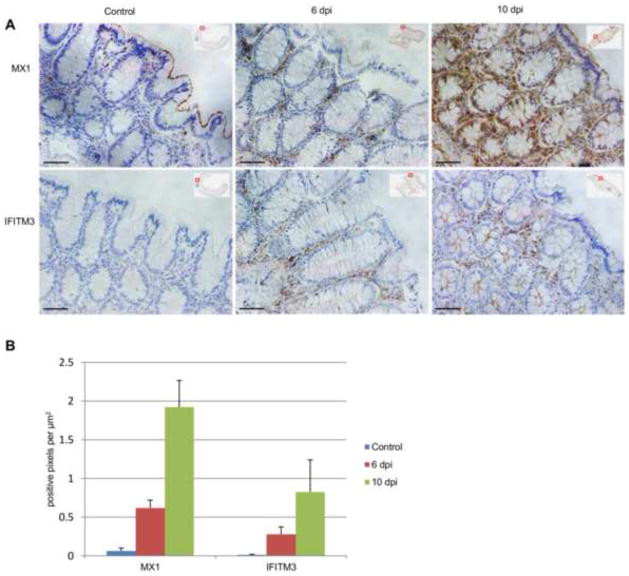
To validate the elevated expression of DEGs and determine their spatial distribution in the rectum, we performed immunohistochemical staining (IHCS) of IFITM3, a recently identified anti-viral factor, and Mx1, one of the most up-regulated genes during early SIV infection. Consistent with the mRNA-seq data, IFITM3 and Mx1 proteins were significantly elevated in the lamina propria at 6 dpi (13- and 25-fold, respectively) and 10 dpi (28- and 63-fold, respectively) (Fig. 4) as compared with the rectal mucosa of SIV-uninfected macaques.
Fig. 4.
Expression of MX1 and IFITM3 proteins in rectal mucosa. (A) Representative IHCS of MX1 and IFITM3 in uninfected control and SIV-infected rhesus macaques. (B) Quantification of MX1 and IFITM3 levels in rectal mucosa of all rhesus macaques in this study. Bar=100μm
SIV genome-wide expression
To better understand the genome-wide transcription pattern of SIV in vivo in early rectal infection, we mapped virus mRNA-seq reads to the SIVmac251 reference genome and calculated the read coverage at each nucleotide position. There was no significant difference between 3 dpi and the uninfected control for read mapping, indicating that mRNA-seq approach is less sensitive for detecting viral replication than qRT-PCR and ISH. As shown in Fig. 5, SIV mRNA was readily detectable at 6 dpi and increased further at 10 dpi using mRNA-seq, which is consistent with our qRT-PCR and ISH data (unpublished data). The mRNA levels of different SIV genes were not equivalent: genes encoding Env and accessory proteins (Vif, Vpu, Vpr, Vpx and Nef) were much higher than those of Gag and Pol in expression. For read coverage, there were larger peaks and valleys in the Env and Nef than the Gag and Pol regions. While the peaks reflected high levels of Env and Nef mRNAs, the valleys indicated that, in the transmitted/founder viruses in rhesus rectal mucosa, there were sequence polymorphism in env and nef genes, mismatched to SIVmac251 reference genome.
Discussion
The anorectal mucosa is a major entry site in HIV-1 transmission, especially among men who have sex with men (MSM). To shed light on the mechanism of HIV-1 rectal transmission, we employed next-generation mRNA-seq and studied virus-host interactions in the rectal mucosa during very early infection.
We found that there are robust host rectal mucosal responses to early rectal SIV transmission. At 3 dpi when SIV replication was still restricted within the portal of entry and before systemic dissemination, inflammation-related genes (CCL13, CCL20, CD1D, CNTFR, FDCSP, and NOS2) were significantly up-regulated. CCL13 is known for chemotaxis of monocytes, T cells, natural killer (NK) cells and immature dendritic cells, and CCL20 for T cells, B cells and immature dendritic cells. Strikingly, we found that the chemoattractant genes CCL13 and CCL20 were up-regulated at both 3 and 6 dpi. Up-regulated CCL20 and CCL13 gene expression in the rectal mucosa suggests that these chemokines may play a key role in recruiting virus-susceptible target cells to the rectal mucosa in the very early stage of transmission.
The early immune responses to rectal SIV exposure are initiated by PRRs that recognize SIV PAMPs. Viral RNA pattern recognition genes (RIG-I, LGP2, MDA5, DDX60, TLR3, and TLR7) were up-regulated at 6 and 10 dpi. RIG-I is a key PRR that triggers IFN expression after detection of double-stranded RNA with a 5′ triphosphate (5′ppp-dsRNA) (Kowalinski et al., 2011). A recent report suggested that LGP2 acts upstream of RIG-I and MDA5 to facilitate their recognition of viral dsRNA (Satoh et al., 2010). DDX60 was recently reported to be an antiviral factor that interacts with RIG-I, MDA5 and LGP2 and regulates RIG-I- and MDA5-mediated type I IFN activation after viral infection (Miyashita et al., 2011). Moreover, genes involved in viral DNA recognition (DAI, IFI16 and AIM2) were significantly up-regulated at 10 dpi, reflecting the presence of viral DNA in the cytoplasm after reverse transcription of SIV genomes, and previous studies showed that the presence of DNA in cytoplasm is a danger signal and can trigger defense responses in eukaryotic cells (Barbalat et al., 2011; Thompson et al., 2011). Surprisingly, in addition to recognition of viruses, receptors recognizing bacteria (MARCO, NOD2 and TLR2) were also up-regulated at 10 dpi, which suggests that the rectal mucosal barrier may be compromised, allowing bacteria and/or their products cross over. This speculation is consistent with our previous finding of increased intestinal epithelial cell apoptosis during early SIV infection (Li et al., 2008).
Our results revealed that after sensing PAMPs, the host’s rectal mucosa mounted two major responses: 1) activation of IFN signaling and antiviral responses; and 2) inflammation, cell trafficking and immune activation. As shown in Fig. 3, the IFN signaling pathway (STAT1, STAT2, SOCS1, SOCS3, IFN-α, IFN-γ, IRF1 and IRF7) was significantly activated, and antiviral IFN-stimulated genes (ISGs) were induced at both 6 and 10 dpi. IRF1 plays a crucial role in regulating MAVS-dependent signaling from peroxisomes (Dixit et al., 2010) and induces the expression of ISGs in the absence of IFN signaling, thereby demonstrating broad-spectrum anti-viral activity (Schoggins et al., 2011). IRF7 can induce the expression of ISGs in the absence of IFN; therefore, the cell can produce rapid ISG expression before IFN is produced (Schmid et al., 2010), which may explain the high levels of ISGs as early as 6 days after SIV infection. Suppressor of cytokine signaling (SOCS) genes, which negatively regulate type I IFN signaling, were also up-regulated. It has been demonstrated that HIV-1 can exploit this mechanism by inducing SOCS3 expression in macrophages, resulting in the failure of IFN-β to suppress viral replication (Akhtar et al., 2010). Cytosolic DNAs are sensed, triggering IFN-β expression through STING (Barber, 2011), which was also up-regulated at 10 dpi.
As shown in Fig. 3, The products of up-regulated ISGs possess broad antiviral activities targeting each step of the viral life cycle, which we will discuss in categories based on their mechanisms: 1) Inhibition of viral entry. IFITM1 and IFITM3 were up-regulated. It is now clear that IFITM1, 2 and 3 have anti-influenza activity (Brass et al., 2009), and a recent study showed that IFITM3 inhibits HIV-1 replication in vitro (Schoggins et al., 2011); 2) Blocking of virus disassembly. TRIM5 was up-regulated. TRIM5 can inhibit HIV/SIV replication by prematurely accelerating viral capsid disassembly (Song et al., 2005; Stremlau et al., 2006). In addition to disruption of viral disassembly, binding of TRIM5 to the HIV-1 capsid catalyzes the synthesis of free ubiquitin chains, leading to AP-1 and NF-κB activation (Iwasaki, 2012); 3) Hypermutation of viral genomes. Many apolipoprotein B editing catalytic subunit-like 3 family (APOBEC3) genes (A, C, D, F, G and H), which have cytidine deaminase (CDA) domains and can induce G-to-A hypermutations in viral DNA during reverse transcription, thereby impairing virus infectivity, were significantly up-regulated at 10 dpi (Bishop et al., 2004). APOBEC3G and APOBEC3F restrict the replication of HIV-1 by introducing hypermutations in the viral genome (Zheng et al., 2004). APOBEC3C is an SIV inhibitor (Yu et al., 2004) and was the most up-regulated (6-fold) member of the APOBEC3 family at 10 dpi. A recent study showed that APOBEC3A inhibits early phases of HIV-1 infection by decreasing viral DNA accumulation (Berger et al., 2011); 4) Blocking of reverse transcription. SAM domain HD domain-containing protein 1 (SAMHD1) can block reverse transcription of HIV-1 by depleting the intracellular pool of deoxynucleoside triphosphates (dNTPs) (Lahouassa et al., 2012). MOV10 can be packaged into HIV-1 virions and inhibit reverse transcription after viral entry into host cells (Abudu et al., 2012); 5) Inhibition of viral transcription. Mx1, Mx2, PML and Sp100 were up-regulated. After assembly into multimers, GTPases Mx1 and Mx2 inhibit transcription and interfere with viral particle assembly. Sp100 is a major component of nuclear domain 10 (ND10), which harbors multiple functions, including viral inhibition by transcriptional repression (Tavalai and Stamminger, 2009); 6) Degradation of viral RNA. OAS1, OAS2, OAS3 and OASL, which can synthesize oligo-A, in turn binding and activating RNase L and leading to mRNA degradation, were significantly up-regulated (Ishibashi et al., 2010); 7) Inhibition of translation. PKR/EIF2AK2, IFIT1/ISG56, IFIT1B, ITIT2, IFIT3, and IFIT5 were significantly up-regulated at both 6 and 10 dpi. PKR/EIF2AK2 is induced by IFN-β and activated by dsRNA (Der et al., 1998). Once activated, PKR inhibits all protein synthesis in the cell via phosphorylation of the translation initiation factor eIF2α. IFIT1/ISG56, IFIT1B, ITIT2, IFIT3, and IFIT5 are functionally related, containing IFN-stimulated response elements (ISREs) for gene transcription after IFN activation and can also be triggered by PAMPs before the induction of type I IFNs (Diamond and Farzan, 2013). A recent study demonstrated that IFIT1 and IFIT3 possess antiviral functions (Guo et al., 2000). IFIT1 can inhibit translation by binding to eukaryotic initiation factor 3 (eIF3) and also prevents viral replication by targeting viral 5′ triphosphate-RNAs (PPP-RNA) for destruction (Pichlmair et al., 2011). To escape the inhibition of IFIT1, influenza virus has evolved to express large quantities of short ppp-RNA to avoid restriction (Umbach et al., 2010). It will be interesting to investigate whether HIV and SIV possess a similar mechanism to escape inhibition by IFIT proteins; 8) Blocking of Gag trafficking. TRIM22, which can inhibit HIV-1 replication by blocking intracellular trafficking of HIV-1 Gag to the cell surface, was up-regulated (Barr et al., 2008); 9) Blocking of virus assembly. USP18 and HERC5 were significantly up-regulated. HERC5 is the major enzyme mediating the conjugation of ISG15 to more than 100 proteins in IFN-stimulated cells. A recent study showed that human HERC5 arrests HIV-1 assembly at the plasma membrane via conjugation of ISG15 to Gag protein (Woods et al., 2011). After SIV infection, the HERC5 gene was up-regulated 8.8- and 29-fold at 6 dpi and 10 dpi, respectively, which may help the host cell contain the release of SIV; 10) Blocking of virus release. Tetherin/BST-2, ISG15, and viperin were significantly up-regulated at both 6 and 10 dpi. Tetherin/BST-2 is an IFN-α-induced protein localized to the cell surface and is critical for the antiretroviral activity of type I IFN in vivo (Liberatore and Bieniasz, 2011). Tetherin/BST-2 efficiently inhibits retroviral release by trapping virions on the cell surface (Neil et al., 2008). ISG15 gene was one of the most up-regulated genes, and expression of ISG15 inhibits the ubiquitination of HIV Gag protein and other host proteins, resulting in impaired viral budding from the host cell (Okumura et al., 2006). Moreover, ISG15 plays a critical role in the host’s response against other viral infections (Lenschow, 2010).
It should be noted that, some of the above discussed viral restriction factors were shown to inhibit viruses other than SIV and HIV. Their inhibition mechanisms on SIV and HIV might be different, and therefore need to be further studied. As the top up-regulated genes at 6 and 10dpi, the IFIT protein family members very likely contribute to SIV inhibition. And other factors may also have more mechanisms of viral restriction than we currently know, such as MX2 gene, which was up-regulated 75-fold 10dpi and was recently shown to inhibit HIV-1 DNA integration (Liu et al., 2013). It will be interesting to study the kinetics of expression levels and patterns of the restriction factors in various cell types to further elucidate interactions between host and virus.
Besides the well-studied anti-viral factors, our study also revealed DEGs with similar levels of up-regulation as top altered restriction factors, for example IFI44L, IFI6, PLA2G4C, USP18, LY6E etc. (Tab. S1). Previous high through-put screening showed that IFI6 inhibits yellow fever virus and IFI44L inhibits hepatitis C virus (HCV), whereas LY6E significantly enhances yellow fever virus (YFV) infection(Schoggins et al., 2011). And PLA2G4C and USP18 were both reported to facilitate HCV replication(Randall et al., 2006; Xu et al., 2012). While the effects of up-regulated levels of IFI44L and IFI6 on SIV replication in vivo need to be further investigated, PLA2G4C, USP18 and LY6E, together with some DEGs identified in this study, might represent positive forces from host that facilitate SIV replication.
Despite potent antiviral responses presented in rhesus rectal mucosa, SIV still finds ways to overcome the restrictions and expand locally and disseminate systemically. The possible mechanisms may be the followings. Firstly, SIV preferentially expresses abundant accessory genes during very early rectal infection as revealed by mRNA-seq (Fig. 5). The SIV accessory gene products are known to antagonize host intrinsic and innate anti-viral molecules. The high levels of accessory gene products (Vif, Vpu, Vpr, Vpx and Nef) antagonize the restriction from host cells and facilitate viral replication. SIV Vpx protein binds SAMHD1 and recruits the ubiquitin ligase complex, resulting in SAMHD1 degradation in the proteasome (Blanco-Melo et al., 2012), and also counteracts APOBEC3A by inducing its degradation (Berger et al., 2011). Vif binds APOBEC3 proteins and results in its degradation in the proteasome (Blanco-Melo et al., 2012). HIV-1 Vpu and SIV Nef bind tetherin/BST-2, resulting in its sequestration and degradation (Blanco-Melo et al., 2012). Vpr and Vif interact with cullin-associated ubiquitin complex and target IRF3 for degradation, resulting in the inhibition of IFNβ synthesis (Okumura et al., 2008). Secondly, some altered host genes in expression revealed in this study might facilitate SIV replication, just like the factors identified to enhance the replication of other viruses (Randall et al., 2006; Xu et al., 2012), PLA2G4C and USP18 are potential factors in this category. Thirdly, the significantly up-regulated genes related to the mucosal proinflammatory responses, immune activation and immune cell trafficking may facilitate SIV replication by attracting CCR5+ CD4+ T cells to the rectal mucosa(Fig. 2).
Collectively, our study revealed for the first time robust responses in rhesus rectal mucosa during very early rectal transmission of SIV, including pattern recognition, complement, inflammation, IFN signaling and innate antiviral responses. The DEGs we identified in this study include all the previous known anti-HIV factors, and potential HIV-1 restriction factor candidates. Furthermore, our study revealed genome-wide SIV expression in vivo in early rectal infection. Together, this first comprehensive genome wide study of early host and SIV interaction in rectum provides new insights into the mechanism of rectal transmission and identifies potential targets for future study to validate and block this route of transmission.
Materials and Methods
Rhesus macaques and rectal SIV inoculation
Thirteen adult male rhesus macaques (Macacca mulatta) of Indian origin housed at BIOQUAL, Inc. in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care standards were used in this study. All animals were tested negative for HIV-2, SIV, type D retrovirus, and SRV-1 infections. Infectious SIVmac251 swarms, provided by Dr. Nancy Miller, were propagated in PBMC of rhesus macaques. Ten macaques were intra-rectally inoculated with SIVmac251 (3.1 × 104 TCID50) and were euthanized at 3 days (3 macaques), 6 days (4 macaques) or 10 days (3 macaques) post-inoculation. Three macaques without virus inoculation served as uninfected controls. Rectal tissues were frozen in liquid nitrogen immediately after dissection and stored at -80°C until analysis.
RNA extraction and next-generation sequencing
At necropsy, the anus and rectum were cut open, divided into six strips with 1.5 cm in length numbered from anus upward, and the third strip was further cut into 1.5 cm × 0.5 cm pieces for fixation or snap frozen for RNA extraction. For RNA extraction in this study, one or two pieces of frozen rectum sample from strip 3 were used depending on yield of RNA extracted. Total RNA was extracted according to our previously published protocol (Li et al., 2009b). Briefly, rectal tissues were homogenized with a power homogenizer in TRIzol (cat #15596-018, Invitrogen) and further purified using the RNeasy Mini Kit (cat #74104, Qiagen). RNA quality was verified on a Bioanalyzer 2100 (Agilent, Palo Alto, CA). A total of 10 μg RNA from each monkey was used for downstream preparation for deep-sequencing. mRNA-seq libraries were generated using the Sample Prep Kit (cat #FC-122-1001, Illumina Inc.) according to the manufacturer’s instructions. Briefly, mRNAs were purified from total RNA using oligo-dT magnetic beads, fragmented, and reverse transcribed into cDNAs, which went through an end-repair process followed by ligation to adapters. A single DNA fragment was enriched by PCR and used for sequencing on an Illumina Genome Analyzer IIx (GA IIx) sequencer at the Genomics Core Research Facility (GCRF) at the University of Nebraska-Lincoln (UNL). Each of the 13 samples sequenced had approximately 30 million 76-nucleotide single-end reads, and the sequencing results passed quality control. The raw data were deposited in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under accession number SRX270636.
Transcriptome data Analysis
Reads from each sample were mapped to the rhesus macaque genome (Mmul_051212) and SIVmac251 full-length reference sequences using GSNAP program (Wu and Nacu, 2010) with default parameters, and the resulting SAM files were converted to BAM files using SAMtools (Li et al., 2009a). Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al., 2013) was used for visualization and mapping quality checking. For rhesus transcriptome analysis, Java Picard utilities were used to count the number of reads mapped to each gene annotated in the rhesus macaque reference genome. The read count data were then statistically analyzed using the DESeq package, which implements an extended negative binomial distribution model (Anders and Huber, 2010). Genes with adjusted p-value (Benjamini-Hochberg method) lower than 0.05 and a fold change greater than two were defined as differentially expressed genes (DEGs), which were further analyzed using IPA (Ingenuity Systems, www.ingenuity.com), DAVID (http://david.abcc.ncifcrf.gov/) and literature search for gene categorization and pathway activation. To analyze the level of SIV mRNA, reads mapped to the SIVmac251 reference sequence were counted base-by-base using SAMtools. The count data were normalized based on total reads in each sample, averaged and plotted to show the different levels of reads coverage at 6 dpi and 10 dpi.
Immunohistochemical staining and quantitative image analysis
Rectal tissues were dissected from euthanized monkeys and immediately fixed in SafeFix II (cat# 23-042-600, Fisher Scientific) and embedded in paraffin. Antibodies against IFITM3 (1:5,000 dilution; cat# 3776-1, Epitomics Inc.) and MX1 (1:3,000 dilution; cat# ab95926, Abcam) were used for Immunohistochemical staining (IHCS) by following procedures described previously (Li et al., 2005). The stained slides were digitized using ScanScope (Aperio, Vista, CA). For quantitative image analysis, the images of slides were imported into ImageScope, and regions of laminar propria were selected using ImageScope drawing tools. IFITM3+ or MX1+ cells were quantified using a positive pixel count algorithm in the Spectrum Plus analysis program (version 9.1). The parameters of the algorithm were manually tuned to accurately match the IFITM3+ or MX1+ markup image over the background DAB stain. Once the parameters were set, the algorithm was applied automatically to all images to measure IFITM3+ or MX1+ positive signals by area.
Supplementary Material
Heat map of DEGs in rectal mucosae at 3 dpi, 6 dpi and 10 dpi. Altered genes in expression are color-coded based on log2 (fold change) as compared with un-infected macaques.
Mucosal responses in rhesus monkeys during early SIV rectal transmission were studied
Rectal mucosal responses were detected as early as 3 days post infection
More robust responses were mobilized at 6 and 10 days post infection.
Genes whose expression may contribute to rectal transmission are identified
Acknowledgments
The authors would like to thank all of the macaque care and veterinary staff at Bioqual, Inc. We thank Dr. Yuannan Xia and Mei Chen at the Genomics Core Research Facility, University of Nebraska-Lincoln for mRNA-seq services. We thank Andrew J. Demers for his critical reading of this manuscript.
This work was supported by NIH grant DK087625 (to Li, Q.) and by Preclinical Research & Development Branch, VRP, DAIDS, NIAID, NIH, Task Order under N01-AI-30018 (to Li, Q). Yue Li is supported by the Fogarty International Center grant at the University of Nebraska-Lincoln (D43 TW001429).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abudu A, Wang X, Dang Y, Zhou T, Xiang SH, Zheng YH. Identification of molecular determinants from Moloney leukemia virus 10 homolog (MOV10) protein for virion packaging and anti-HIV-1 activity. The Journal of biological chemistry. 2012;287:1220–1228. doi: 10.1074/jbc.M111.309831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar LN, Qin H, Muldowney MT, Yanagisawa LL, Kutsch O, Clements JE, Benveniste EN. Suppressor of cytokine signaling 3 inhibits antiviral IFN-beta signaling to enhance HIV-1 replication in macrophages. J Immunol. 2010;185:2393–2404. doi: 10.4049/jimmunol.0903563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annual review of immunology. 2011;29:185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011;243:99–108. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, Bouaziz S, Muriaux D, Darlix JL, Cimarelli A. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7:e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Current biology: CB. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D, Venkatesh S, Bieniasz PD. Intrinsic Cellular Defenses against Human Immunodeficiency Viruses. Immunity. 2012;37:399–411. doi: 10.1016/j.immuni.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. HIV Surveillance in Men Who Have Sex with Men. 2011 Retrieved May 8, 2013, from http://www.cdc.gov/hiv/pdf/statistics_surveillance_MSM_2011.pdf.
- Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. The EMBO journal. 2000;19:6891–6899. doi: 10.1093/emboj/19.24.6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Wakita T, Esumi M. 2′,5′-Oligoadenylate synthetase-like gene highly induced by hepatitis C virus infection in human liver is inhibitory to viral replication in vitro. Biochemical and biophysical research communications. 2010;392:397–402. doi: 10.1016/j.bbrc.2010.01.034. [DOI] [PubMed] [Google Scholar]
- Iwasaki A. Innate Immune Recognition of HIV-1. Immunity. 2012;37:389–398. doi: 10.1016/j.immuni.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. The Journal of experimental medicine. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley LA, Detels R, Kaslow R, Polk BF, Rinaldo CR, Jr, Chmiel J, Detre K, Kelsey SF, Odaka N, Ostrow D, et al. Risk factors for seroconversion to human immunodeficiency virus among male homosexuals. Results from the Multicenter AIDS Cohort Study. Lancet. 1987;1:345–349. doi: 10.1016/s0140-6736(87)91725-9. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenschow DJ. Antiviral Properties of ISG15. Viruses. 2010;2:2154–2168. doi: 10.3390/v2102154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009a;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- Li Q, Smith AJ, Schacker TW, Carlis JV, Duan L, Reilly CS, Haase AT. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J Immunol. 2009b;183:1975–1982. doi: 10.4049/jimmunol.0803222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QS, Estes JD, Duan L, Jessurun J, Pambuccian S, Forster C, Wietgrefe S, Zupancic M, Schacker T, Reilly C, Carlis JV, Haase AT. Simian immunodeficiency virus-induced intestinal cell apoptosis is the underlying mechanism of the regenerative enteropathy of early infection. Journal of Infectious Diseases. 2008;197:420–429. doi: 10.1086/525046. [DOI] [PubMed] [Google Scholar]
- Liberatore RA, Bieniasz PD. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci U S A. 2011;108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, Cen S, Guo F, Liang C. The Interferon-Inducible MxB Protein Inhibits HIV-1 Infection. Cell Host Microbe. 2013;14:398–410. doi: 10.1016/j.chom.2013.08.015. [DOI] [PubMed] [Google Scholar]
- Mansergh G, Colfax GN, Marks G, Rader M, Guzman R, Buchbinder S. The Circuit Party Men’s Health Survey: findings and implications for gay and bisexual men. American journal of public health. 2001;91:953–958. doi: 10.2105/ajph.91.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan I. Rectal microbicides: a new focus for HIV prevention. Sexually transmitted infections. 2008;84:413–417. doi: 10.1136/sti.2008.031328. [DOI] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Molecular and cellular biology. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal lamina propria yields greater molecular insight into mechanisms underlying SIV induced immune dysfunction. PloS one. 2012;7:e34561. doi: 10.1371/journal.pone.0034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Kaushal D, Aye PP, Alvarez X, Veazey RS, Lackner AA. Focused examination of the intestinal epithelium reveals transcriptional signatures consistent with disturbances in enterocyte maturation and differentiation during the course of SIV infection. PloS one. 2013;8:e60122. doi: 10.1371/journal.pone.0060122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Okumura A, Alce T, Lubyova B, Ezelle H, Strebel K, Pitha PM. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology. 2008;373:85–97. doi: 10.1016/j.virol.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, Heathcote J, Rice CM, Edwards AM, McGilvray ID. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology. 2006;131:1584–1591. doi: 10.1053/j.gastro.2006.08.043. [DOI] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S, Mordstein M, Kochs G, Garcia-Sastre A, Tenoever BR. Transcription factor redundancy ensures induction of the antiviral state. The Journal of biological chemistry. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005;79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N, Stamminger T. Interplay between Herpesvirus Infection and Host Defense by PML Nuclear Bodies. Viruses. 2009;1:1240–1264. doi: 10.3390/v1031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach JL, Yen HL, Poon LL, Cullen BR. Influenza A virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio. 2010;1 doi: 10.1128/mBio.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griensven F, de Lind van Wijngaarden JW. A review of the epidemiology of HIV infection and prevention responses among MSM in Asia. AIDS. 2010;24(Suppl 3):S30–40. doi: 10.1097/01.aids.0000390087.22565.b4. [DOI] [PubMed] [Google Scholar]
- Woods MW, Kelly JN, Hattlmann CJ, Tong JG, Xu LS, Coleman MD, Quest GR, Smiley JR, Barr SD. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology. 2011;8:95. doi: 10.1186/1742-4690-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Pei R, Guo M, Han Q, Lai J, Wang Y, Wu C, Zhou Y, Lu M, Chen X. Cytosolic phospholipase A2 gamma is involved in hepatitis C virus replication and assembly. J Virol. 2012;86:13025–13037. doi: 10.1128/JVI.01785-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. The Journal of biological chemistry. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heat map of DEGs in rectal mucosae at 3 dpi, 6 dpi and 10 dpi. Altered genes in expression are color-coded based on log2 (fold change) as compared with un-infected macaques.