Abstract
We report genomic analysis of 300 meningiomas, the most common primary brain tumors, leading to the discovery of mutations in TRAF7, a proapoptotic E3 ubiquitin ligase, in nearly one-fourth of all meningiomas. Mutations in TRAF7commonly occurred with a recurrent mutation (K409Q) in KLF4, a transcription factor known for its role in inducing pluripotency, or with AKT1E17K, a mutation known to activate the PI3K pathway. SMO mutations, which activate Hedgehog signaling, were identified in ~5% of non-NF2 mutant meningiomas. These non-NF2 meningiomas were clinically distinctive—nearly always benign, with chromosomal stability, and originating from the medial skull base. In contrast, meningiomas with mutant NF2 and/or chromosome 22 loss were more likely to be atypical, showing genomic instability, and localizing to the cerebral and cerebellar hemispheres. Collectively, these findings identify distinct meningioma subtypes, suggesting avenues for targeted therapeutics.
Meningiomas, arising from the meninges of the central nervous system, are the most common primary brain tumors, with a prevalence of ~170,000 cases in the United States (1). Although most are histologically classified as benign (grade I), about 10% represent atypical (grade II) or anaplastic (grade III) forms. Meningiomas frequently invade surrounding brain and critical neurovascular structures, often causing neurological deficits and requiring surgical intervention. Loss of Neurofibromin 2 (merlin, NF2) is found in 40 to 60% of sporadic meningiomas (2), but the genetic architecture of the remainder remains obscure, limiting options for the development of rational therapies.
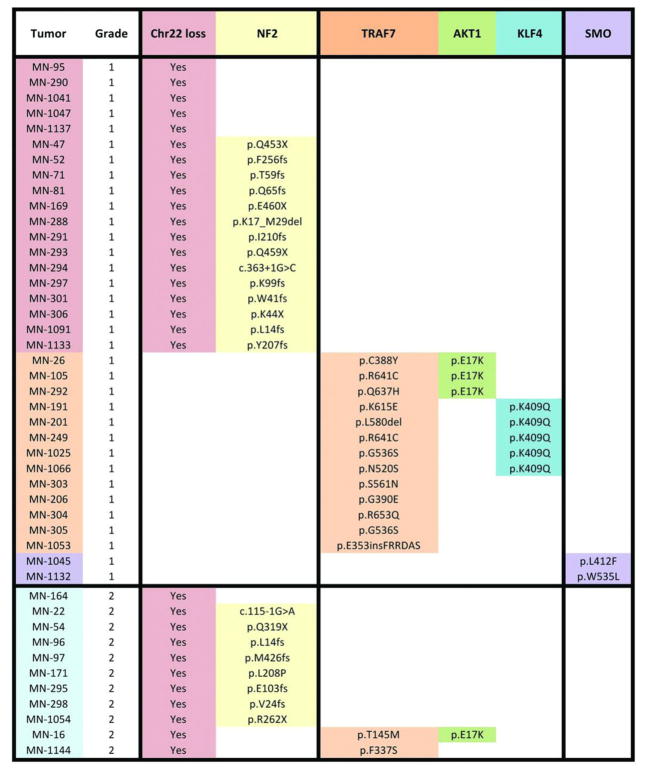
To comprehensively characterize the genomics of meningioma and to gain further insight into molecular mechanisms of tumor formation, we performed genome-wide genotyping and exome sequencing (average depth of coverage 255-fold) of 50 previously nonirradiated grade I (n = 39) and grade II (n = 11) meningiomas and matched normal DNA (3) (table S1). For the meningiomas in which matching blood samples were available (n = 39), the mean number of protein-altering somatic mutations was 7.2 (range 1 to 15), a considerably smaller number compared with malignant tumors (table S2). We next searched for genes with significantly more somatic mutations than expected by chance (fig. S1). Besides NF2, we identified increased mutation burden in TNF receptor-associated factor 7 (TRAF7), Krupple-like factor 4 (KLF4), v-akt murine thymoma viral oncogene homolog 1 (AKT1), and Smoothened, frizzled family receptor (SMO) (as a group, referred to as non-NF2 mutant hereafter) (Fig. 1). Mutations in these genes were mutually exclusive of NF2 mutations. In addition, we identified single mutations in genes previously reported to play a role in other neoplasias, including CREBBP, PIK3CA (R108H variant), PIK3R1 (deletion p.306-307), and BRCA1 as well as two SMARCB1 mutations, which coexisted with NF2 loss and have previously been reported in meningiomas (4) (table S3).
Fig. 1.
Exome sequencing identifies meningioma subgroups based on mutually exclusive mutation profiles.
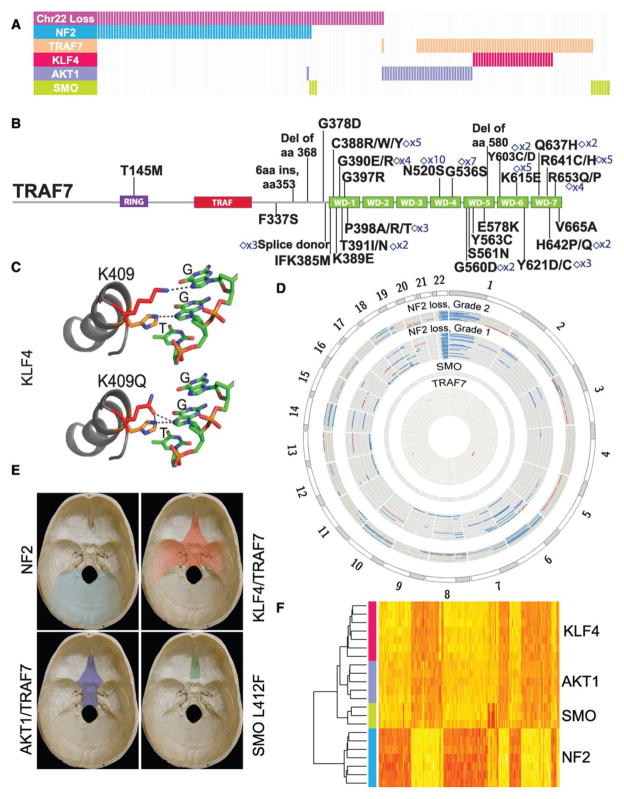
We next performed targeted resequencing of these top five genes, along with chromosome 22 copy-number analysis, in an independent set of 250 unradiated meningiomas (204 grade I and 46 high-grade meningiomas) (fig. S2). In the combined analysis of 300 meningiomas, we identified coding mutations in one of these five genes and/or evidence for chromosome 22 loss in 237 (79%) (Fig. 2A and table S3). NF2 mutations were present in 108 (36%). TRAF7 mutations, which were always exclusive of NF2 mutations [mutual exclusivity P value (Pme) = 2.55 × 10−17 (5)], were observed in nearly one-fourth of the meningiomas examined (n = 72). TRAF7 is a proapoptotic N-terminal RING and zinc finger domain protein with E3 ubiquitin ligase activity that contains seven WD40 repeats in its C terminus (6). TRAF7 interacts with several molecules, such as MEKK3, through these WD40 repeats, affects multiple signaling pathways, including NF-κB, and targets ubiquitination of proteins including c-FLIP, an antiapoptotic molecule (7). It is notable that 67 of the 72 TRAF7 mutations, including 15 recurrent mutations, all map to the WD40 domains (Fig. 2B).
Fig. 2.
Genomic architecture of meningiomas. (A) NF2, TRAF7, and SMO coding mutations along with recurrent AKT1E17K and KLF4K409Q variants reveal meningioma subtypes with mutually exclusive profiles. Analysis for chromosome 22 copy number is also shown. Each bar represents a grade I meningioma sample; 191 samples are depicted. (B) TRAF7 mutations, which are identified in 72 of 300 meningiomas analyzed, are clustered within its WD40 domains. The count of recurrent mutations, which are denoted by diamonds, is indicated. (C) The recurrent KLF4K409Q mutation is located within the first zinc finger domain, which makes direct DNA contact. (D) Circos plot of large-scale genomic abnormalities identified (blue: deletion, red: amplification). Whereas all NF2/chr22loss meningiomas (outer circles, n = 41, including n = 30 with coding NF2 mutations) show chromosome 22 loss, which is typically associated with further chromosomal abnormalities in grade II tumors (n = 11, including n = 8 with coding NF2 mutations), genomic stability is a hallmark of grade I non-NF2 tumors (inner circles, n = 36). (E) Along the skull base, NF2/chr22loss meningiomas originate from the lateral and posterior regions, whereas the vast majority of anterior and medial meningiomas are non-NF2 mutant. (F) Unsupervised hierarchical clustering of gene expression profiles defines two major benign meningioma subgroups, those with NF2/chr22loss and non-NF2 mutant tumors. Each subgroup reveals differential H3K27ac and gene expression profiles (figs. S10 to S14 and tables S5 to S8).
In the transcription factor KLF4, we identified a recurrent K409Q mutation, which almost always co-occurred with TRAF7 mutations [n = 31; co-occurrence P value (Pco) = 2.50 × 10−20] and were exclusive of NF2 mutations (Pme = 3.77 × 10−7). KLF4 is expressed in meningiomas (fig. S3). KLF4 regulates differentiation of several cell types and is best known as one of four genes that together promote reprogramming of differentiated somatic cells into pluripotent stem cells (8). Deletion of the KLF4 DNA binding domain blocks differentiation and induces self-renewal in hematopoietic cells (9). The recurrently mutated KLF4 residue, K409, lies within the first zinc finger and makes direct DNA contact in the major groove of the DNA binding motif (9) (Fig. 2C and fig. S4).
The known neoplasia-related recurrent mutation, AKT1E17K, was identified in 38 meningiomas. Although the AKT1E17K mutation co-occurred with TRAF7 mutations in 25 of the 38 tumors (Pco = 3.90 × 10−9), it was exclusive of the KLF4K409Q (Pme = 1.18 × 10−2) and NF2 mutations, except in one case (Pme = 2.70 × 10−7). The AKT1E17K mutation has been shown to activate PI3K/AKT signaling (10) and was readily detectable by immunohistochemistry using an antibody specific for this mutation (fig. S3).
Finally, in 11 tumors, we identified mutations in SMO, which is expressed in meningiomas (fig. S5). These mutations include a recurrent L412F variant in seven meningiomas and a previously reported W535L mutation, which has been shown to result in activation of Hedgehog signaling in basal cell carcinoma (11). Eight of these SMO mutations were mutually exclusive of mutations in the other four genes (Pme = 1.24 × 10−2).
We next evaluated chromosomal instability. Chromosome 22 loss, observed in 149 tumors, was the most common event and was strongly associated with the presence of coding NF2 mutations (Pco = 1.32 × 10−47). These were also significantly associated with higher grade meningiomas [P = 5.90 × 10−5; odds ratio (OR) = 3.54]. Higher-grade tumors also showed an increased number of large-scale chromosomal abnormalities (Fig. 2D and fig. S6) (6.9 versus 1.7 events per tumor) and an increased rate of NF2 mutations (P = 0.03; OR = 1.96) and were observed more frequently in males than females (P = 6.45 × 10−4; OR = 2.93).
Given these observations pointing to distinct tumor subtypes based on mutation profiles, we examined whether the mutation spectrum correlated with anatomical distribution and histological subtype. We initially grouped cerebral meningiomas into those originating along the skull base or those present in the cerebral hemispheres (Fig. 2E, fig. S7, and table S4). Interestingly, tumors with NF2 mutations and/or chromosome 22 loss (NF2/chr22loss) were predominantly found in the hemispheres (P = 9.22 × 10−14; OR = 6.74) with nearly all posterior cerebral (parieto-occipital), cerebellar, or spinal meningiomas being NF2/chr22loss tumors (fig. S8). For the meningiomas originating from the skull base, we observed a difference between those originating from medial versus lateral regions. The vast majority of non-NF2 meningiomas were medial (P = 4.36 × 10−8; medial versus lateral OR = 8.80), whereas the lateral and posterior skull base meningiomas had NF2/chr22loss (P = 1.55 × 10−12; OR = 23.11). Meningiomas with only the recurrent SMO L412F mutation (n = 5) all localized to the medial anterior skull base, near the midline. This is particularly interesting because mutations in Hedgehog signaling result in holoprosencephaly, the midline failure of embryonic forebrain to divide into two hemispheres (12).
Mutational profiles also were correlated with histological diagnoses. For example, all of the meningiomas with a “secretory” component (n = 12), which follow a more aggressive clinical course owing to increased brain swelling, carried both TRAF7 andKLF4 mutations (Pco = 6.02 × 10−12) (fig. S9).
Consistent with these clinical observations, unsupervised hierarchical clustering of meningiomas based on gene expression and chromatin immunoprecipitation for H3K27 acetylation followed by sequencing (H3K27ac ChIP-seq) analyses confirmed clustering into NF2/chr22loss versus non-NF2 mutant subgroups (Fig. 2F and figs. S10 and S11) and revealed several molecules whose acetylation and expression was specific to a subtype (tables S5 and S6). For these differentially expressed genes, there was a strong correlation between expression and ChIP-seq data (fig. S12). Among the non-NF2 meningiomas, SMO mutants were clearly defined by increased expression and activation of the Hedgehog pathway (fig. S13 and tables S7 and S8).
These results clearly identify meningioma subgroups, distinguishing them based on their mutually exclusive distribution of mutations, distinct potential for chromosomal instability and malignancy, anatomical location, histological appearance, gene expression, and H3K27ac profile. Our results show that the mutational profile of a meningioma can largely be predicted based on its anatomical position, which in turn may predict likely drug response (e.g., Hedgehog inhibitors for midline tumors). This may prove relevant for surgically unresectable, recurrent, or invasive meningiomas and could spare patients surgery or irradiation, an independent risk factor for progression of these generally benign tumors.
Supplementary Material
Acknowledgments
We are grateful to the patients and their families who have contributed to this study. This study was supported by the Gregory M. Kiez and Mehmet Kutman Foundation. R.P.L. is an investigator of the Howard Hughes Institute. V.E.C. is supported by NIH T32GM07205. All somatic mutations identified through exome sequencing of meningiomas are reported in the supplementary materials and submitted to the Catalogue of Somatic Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic, submission ID COSP30702). Yale University has filed a provisional patent application based on the results of this study.
Footnotes
References and Notes
- 1.Wiemels J, Wrensch M, Claus EB. J Neurooncol. 2010;99:307. doi: 10.1007/s11060-010-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riemenschneider MJ, Perry A, Reifenberger G. Lancet Neurology. 2006;5:1045. doi: 10.1016/S1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- 3.Materials and methods are available as supporting material on Science online.
- 4.Schmitz U, et al. Br J Cancer. 2001;84:199. doi: 10.1054/bjoc.2000.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui Q. PloS One. 2010;5 doi: 10.1371/journal.pone.0013180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu LG, Li LY, Shu HB. J Biol Chem. 2004;279:17278. doi: 10.1074/jbc.C400063200. [DOI] [PubMed] [Google Scholar]
- 7.Bouwmeester T, et al. Nat Cell Biol. 2004;6:97. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, et al. Cell. 2007;131:861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Schuetz A, et al. Cell Mol Life Sci. 2011;68:3121. doi: 10.1007/s00018-010-0618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpten JD, et al. Nature. 2007;448:439. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 11.Xie J, et al. Nature. 1998;391:90. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 12.Roessler E, et al. Nature Genetics. 1996;14:357. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.