Abstract
Objective
To investigate the heterogeneity in clinical course among those with pediatric acute liver failure (PALF) of indeterminate disease etiology.
Study design
We studied participants enrolled in the PALF registry study with “indeterminate disease” (IND) final diagnosis. Growth mixture modeling (GMM) was used to analyze participants’ INR, total bilirubin and clinical encephalopathy (HE) trajectories in the first 7 days following enrollment. Participants with at least 3 values for one or more of the measurements were included. We examined the association between the resulting latent subgroup classification with participants’ characteristics and disease outcomes. Data from PALF participants with specified etiologies (non-IND) were utilized to investigate the potential diagnostic value of the latent subgroups.
Results
In this sample of 380 IND participants, 115 (30%) experienced mild and quickly improving disease trajectories and another 48 (13%) started with severe disease, but improved by day 7. The majority of participants (216, 57%) had disease trajectories that worsened over time. The identified patterns of disease trajectories are predictive of outcome (p<.001). The trajectory patterns are associated with the underlying disease etiology (p<.001) for the 488 non-IND participants.
Conclusions
The clinical courses of indeterminate PALF participants exhibit distinct trajectory patterns which have important prognostic and potentially diagnostic value.
Keywords: classification, disease trajectory, growth mixture model, liver transplantation, prognosis
Pediatric acute liver failure (PALF) is a life-threatening clinical syndrome in which children without previous history of liver disease suffer from rapid loss of liver function. The disease may progress quickly and lead to severe impairment of hepatic function within days or weeks, as evidenced in many children by jaundice, coagulation abnormalities and hepatic encephalopathy (HE). The outcomes of PALF are poor, and one-half of patients die or receive liver transplantation (LTx)(1).
Medical management of PALF is largely supportive in the absence of a condition known to respond to specific therapy (e.g., acute acetaminophen toxicity, herpes simplex virus). Liver transplantation becomes an option once liver function deteriorates to such an extent that recovery is judged to be unlikely. Due to the rapid progression of PALF in some patients, a timely decision to proceed to LTx is needed to interrupt damaging sequelae associated with PALF such as cerebral edema and renal injury. Yet, it is undesirable for a patient to undergo LTx if survival with the native liver would have occurred.
Reliable prognostic tools are needed to predict the outcomes of PALF and to guide the LTx decision. The King’s College Hospital Criteria (KCHC)(2) is the only predictive model for acute liver failure developed in the pre-LTx era when patient outcomes were limited to survival or death. Patients who met KCHC in the initial report had a high likelihood of death with a positive predictive value (PPV) of 97% for those with non-acetaminophen acute liver failure(2). However, when KCHC were recently applied to a cohort of PALF study participants consisting of those who died or survived with their native liver to 21 days, the PPV of KCHC fell to 33%(3).
Etiologies of PALF are diverse and include drug toxicity such as acetaminophen overdose (APAP), autoimmune liver disease, metabolic disease, and viral hepatitis(1). Etiology is an important factor in determining outcome. For example, patients with acute APAP toxicity or herpes simplex hepatitis would be expected to have a relatively good or poor prognosis, respectively, given the known pathobiology and treatment for these conditions. Yet, individual patients with APAP toxicity do die and those with herpes simplex can survive, suggesting factors other than etiology play a role in determining outcome.
The 40% to 50% of cases of PALF with an indeterminate cause of PALF present a formidable challenge in predicting outcome as underlying causes or treatment strategies are not known(4, 5). Patients with indeterminate PALF were more likely to receive LTx than are patients with PALF with specified etiologies (4). Importantly, those with an indeterminate diagnosis have inherent heterogeneity likely involving the unknown underlying etiology, pathobiology and outcomes.
The goal of this analysis was to determine whether PALF dynamics, as measured by trajectories of disease markers, could aid in the prognosis following PALF of indeterminate etiology. Hence, this analysis attempts to determine if disease trajectory over up to 7 days of observation can aid with determining who should undergo LTx and who may be able to wait for signs of spontaneous improvement.
Methods
The Pediatric Acute Liver Failure (PALF) study group is a multicenter collaborative study formed in 1999 to investigate the diagnosis, etiology, prognosis, and management of PALF(1). The first phase of the PALF study was an ancillary to the adult ALF study group, which was sponsored by the National Institutes of Health-National Institute of Diabetes, Digestive and Kidney disease (NIH-NIDDK). During this initial phase, the study included 22 pediatric sites and a Data Coordinating Center (DCC) at the University of Texas Southwestern Medical School (UTSW). The PALF study transitioned to its second phase 2005, when the pediatric consortium received independent funding from NIH-NIDDK. The second phase of the PALF study consisted of 20 sites and a DCC at the University of Pittsburgh. There were 986 participants enrolled in the PALF study between 1999 and 2010. The inclusion/exclusion criteria and primary aims were identical for the 2 phases of the PALF study.
The PALF study group created a registry database including demographic, clinical, laboratory and outcome data among pediatric participants with acute liver failure. Inclusion criteria were less than 18 years of age, no evidence of chronic liver disease, biochemical evidence of acute liver injury, and coagulopathy not corrected by vitamin K. Patients could be recruited to the study if the they had INR≥ 1.5 (or prothrombin time≥ 15 seconds) in the presence of clinical hepatic encephalopathy (HE) or INR≥ 2 (or prothrombin time ≥ 20 seconds) regardless of presence or absence of HE(1).
The study was observational because patient management, including the decision regarding liver transplantation, was determined by treating clinicians who followed the local standard of care. The PALF study did not have any treatment protocols outside of a clinical trial of NAC for non-APAP caused PALF(6). Clinical measurements and laboratory test results were recorded daily for up to seven consecutive days following enrollment. In phase 1, the earliest outcome (hospital discharge, death, liver transplantation, survival without transplantation) 21 days following enrollment was recorded. Any of these outcomes that occurred up to 1 year following enrollment was recorded in phase 2. The daily maximum of the HE grade was recorded.
The site principal investigator determined a primary etiology at the time of study enrollment and a final diagnosis at the time of the outcome event. An indeterminate etiology was assigned if the participant could not be classified into any specific etiology.
Statistical Analyses
To explore the heterogeneity in the clinical course among participants with an indeterminate final etiology, participants were classified into latent subgroups based on the dynamic trajectories of several key clinical and laboratory measurements using growth mixture modeling (GMM), a multilevel random effect modeling framework (7–10). The GMM assumes that the heterogeneous study population, exemplified by the indeterminate cohort, is comprised of homogeneous latent subgroups that can be identified by similar dynamic trajectories of data elements. Each latent subgroup features its own set of parameters which defines a pattern of changing clinical course for those in the same subgroup. Therefore, the GMM serves a powerful tool for clustering subjects into unobserved subgroups and for estimating the dynamic disease trajectories within different subgroups. Subject-specific random effect terms were used to account for the within subject correlation across study days.
The GMM parameters can be estimated via maximum likelihood methods. The maximum likelihood estimators accommodate the “missing at random” (MAR) mechanism, allowing use of a participant’s data even if his/her measures were not available for each of the 7 days of data collection or until an outcome was reached. The MAR assumption allows the probability of data missing to depend on observed data.
Rigorous model selection procedures were conducted to determine the number of subgroups and the shape of the trajectories. For model selection, we considered statistical measures (i.e., the Bayesian information criteria [BIC], entropy, the Lo, Mendell and Rubin likelihood ratio test [LMR-LRT], and the bootstrap likelihood ratio test [BLRT]), and the clinical meaningfulness of the resulting classifications(9, 10). The BIC is a penalized-likelihood model selection criterion that accounts for both the model fit and the number of parameters, whereby models with smaller BIC values are often preferred. Entropy is a measure of the classification quality, where entropy values close to 1 suggest good discrimination among the latent subgroups(11). Entropy values of 0.8 or higher implies adequate separation among latent subgroups. LMR-LRT and BLRT are hypothesis testing procedures, for which significant test results suggest that the K-subgroup GMM is better than the (K-1)-subgroup GMM. Here, K is an integer that denotes the number of latent subgroups. These measures have their respective strengths and limitations and may not always agree on the best GMM to select in terms of the optimal number of latent subgroups and other model parameters. Thus, clinical judgment regarding potential latent subgroups was also used for model selection. A GMM is regarded as clinically meaningful if the trajectory patterns in different latent subgroups exhibit clinically important differences and if there are adequate proportions of patients in each of the resulting latent subgroups.
Based on results of univariable GMM analysis and a priori clinical input, the three measures included in the multivariable GMM model were international normalized ratio (INR), total bilirubin and HE. Albumin was considered but not included in the final GMM model, on the basis of univariable analysis results. Total bilirubin, INR, albumin and HE are recognized biomarkers for patients with liver failure. For the proposed model to have general clinical relevance, variables considered in the univariate analysis should be rapidly available and believed to reflect the dynamic changes in liver function. For example, INR, total bilirubin, and albumin are components of the PELD score, and HE is regarded as a key feature of acute liver failure. INR was loge transformed due to its skewness. HE categories were none, I–II, and III–IV. HE grade may be recorded as not assessable when the participant was on a respirator or in an induced coma. Because these participants may have had severe disease and poor outcomes, the missing HE may be informative. Hence, for such participants HE was imputed as HE grade III–IV if the subject ever had an HE grade above 0 in the first 7 days following enrollment and subsequently was determined “not assessable” or on a ventilator. If HE was never greater than 0 prior to being non-assessable, then non-assessable HE was considered to be missing. All GMM analyses were conducted in Mplus, Version 6.12. A minimum of three records for one or more measurements was required for inclusion.
Following classification into a subgroup as defined by GMM, participant characteristics and outcomes were compared across subgroups. Continuous variables (eg, age) were summarized using medians, 25th and 75th percentiles and categorical data (eg, 21-day outcome) was summarized via frequencies and percentages. For formal statistical comparisons, the Kruskal-Wallis test for continuous variables and the Pearson’s chi-square test or its exact version was used for categorical variables as appropriate. Because the GMM differentiated subgroups based on INR, total bilirubin, and HE, statistical tests were not performed to determine whether trajectory groups differed significantly with respect to these three variables. These analyses were conducted using SAS 9.4. (SAS Institute, NC).
The estimated GMM parameters can be used to classify a new patients with PALF into one of the identified subgroups. Specifically, we can calculate the posterior probabilities of the latent subgroups based on the estimated GMM parameters and the patient’s INR, total bilirubin and HE trajectories. Posterior probabilities are the estimated probabilities that a participant belongs to different latent subgroups on the basis of his/her observed data. The participant can be classified into the subgroup with the largest posterior probability. The 488 participants with specified etiology were subsequently classified into the subgroups identified among those with indeterminate etiology to investigate the diagnostic and prognostic value of the subgroup classification. The exact chi-squared test was used to determine the statistical significance of the association between subgroup indices and diagnostic categories, and the association between subgroup indices and 21 day outcomes.
Results
Among the 986 participants enrolled in the 2 phases of the PALF registry were 437 (44%) with an indeterminate final etiology. There were 57 subjects with fewer than 3 values for all three measurements (INR, total bilirubin, and HE) excluded from analyses, leaving 380 indeterminate participants included. When compared with the patients included in the analysis, the 57 excluded patients had higher INR (p<.001) and worse HE (p<.001) but similar total bilirubin (p=0.5) at enrollment. Among the 380 patients with IND, the median age was 3.5 years (25th to 75th percentiles 1.0 to 9.2 years), 211/380 (56%) were male, the median INR was 2.7 (25th to 75th percentiles 2.1 to 3.9), and the HE grade was greater than 0 for 198 (54%) participants (Table I).
Table 1.
Patient characteristics at enrollment for the N=380 participants with indeterminate diagnosis (IND), overall and by identified latent subgroup index.
| characteristics | All N=380 |
Subgroup1 N=59 (15.5%) |
Subgroup2 N=57 (15%) |
Subgroup3 N=48 (12.6%) |
Subgroup4 N=130 (34.2%) |
Subgroup5 N=86 (22.6%) |
pA |
|---|---|---|---|---|---|---|---|
| Age (yrs)** | N=380 3.5 (1.0,9.2) |
N=59 1.7 (0.9,6.3) |
N=57 4.2 (1.6,11.0) |
N=48 3.1 (0.5,9.4) |
N=130 3.3 (0.6,9.3) |
N=86 5.7 (1.5,10.0) |
0.1 |
|
| |||||||
| INR** | N=303 2.7 (2.1,3.9) |
N=46 2.3 (1.8,3.7) |
N=47 2.2 (1.8,2.7) |
N=41 3.6 (2.7,7.0) |
N=95 2.6 (2.1,3.5) |
N=74 3.1 (2.3,5.1) |
N/A |
|
| |||||||
| Total bilirubin** | N=312 13.7 (5.1,19.8) |
N=51 1.8 (0.9,2.5) |
N=51 8.4 (5.4,11.4) |
N=41 19.1 (9.8,25.4) |
N=100 17.6 (12.5,20.2) |
N=69 16.5 (13.4, 23.6) |
N/A |
|
| |||||||
| HE* | N=367 | N=56 | N=54 | N=47 | N=126 | N=84 | N/A |
| None | 169(46%) | 21(38%) | 28(52%) | 14(30%) | 58(46%) | 48(57%) | |
| I–II | 157(43%) | 28(50%) | 15(28%) | 25(53%) | 61(48%) | 28(33%) | |
| III–IV | 41(11%) | 7(13%) | 11(20%) | 8(17%) | 7(6%) | 8(10%) | |
|
| |||||||
| Jaundice* | N=150 | N=35 | N=20 | N=15 | N=48 | N=32 | <.001 |
| Yes | 102(68%) | 5(14%) | 14(70%) | 9(60%) | 45(94%) | 29(91%) | |
|
| |||||||
| Ascites* | N=378 | N=59 | N=57 | N=48 | N=128 | N=86 | 0.002 |
| Yes | 71(19%) | 3(5%) | 5(9%) | 11(23%) | 33(26%) | 19(22%) | |
|
| |||||||
| ALT (U/L)** | N=297 1897 (793,3156) |
N=49 3615 (2051,6545) |
N=46 2209 (879,3278) |
N=41 1390 (896,3893) |
N=95 1350 (461,2372) |
N=66 1992 (1074,2980) |
<.001 |
|
| |||||||
| Albumin** | N=323 2.9 (2.5,3.2) |
N=55 2.8 (2.3,3.3) |
N=51 3.0 (2.6,3.2) |
N=42 2.8 (2.6,3.2) |
N=103 2.7 (2.4,3.1) |
N=72 2.9 (2.6,3.4) |
0.1 |
summarized by freq (%);
summarized by median (25th, and 75th percentiles).
The p-values reflect the difference across the 5 subgroups. Statistical tests were not conducted for the three elements (INR, total bilirubin and HE) used for subgroup classification. Statistical tests were conducted for Age, Jaundice, Ascites, ALT and Albumin, which were not used for subgroup classification in the GMM model.
Based on extensive model selection procedures, we selected a linear-trajectory GMM model with K=5 latent subgroups. The decision to use 5 subgroups was based on the following: (1) the GMM model with K latent subgroups is statistically better than that with K-1 subgroups for K=2, 3, 4, 5 (BLRT p-value <0.05), and the GMM model with 6 subgroups does not have statistical improvement over that with 5 subgroups (BLRT p-value=0.2). Therefore, K=5 is the best choice according to hypothesis testing. (2) the model with K=5 had lower BIC than other choices of K between 2 and 6, and entropy for K=5 (0.892) was exceeded only when K=2 (0.929), for which BIC was substantially higher (14256 which was second highest of BICs examined vs. 13252). (3) The five subgroups exhibit clinically meaningful differences in the identified trajectory patterns, and there were adequate proportions of patients in all 5 latent subgroups.
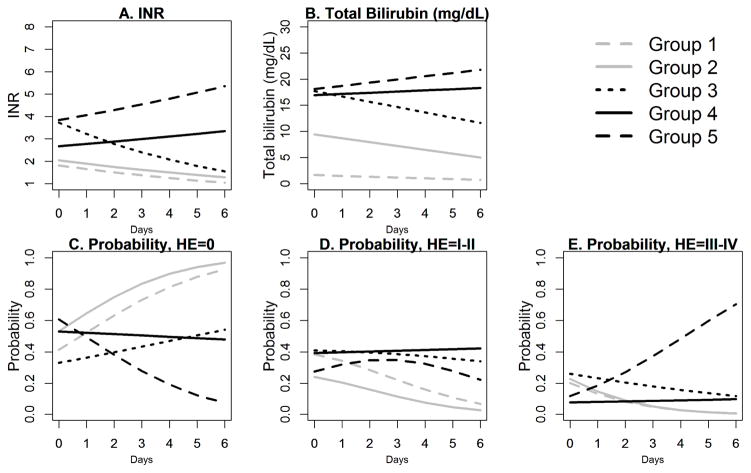
In the selected model with 5 latent subgroups, the entropy value was 0.89, suggesting clear delineation among the subgroups. Figure 1 presents the estimated disease trajectories in the first 7 days, where the day of enrollment is denoted by Day 0. The identified subgroups showed different patterns of evolution in disease trajectories of INR, total bilirubin and HE. Subgroups 1 and 2 feature low and decreasing INR and steady or decreasing total bilirubin trajectories. They are differentiated by total bilirubin which is higher in subgroup 2 than in group 1 at time of enrollment and, though decreasing, remains higher in group 2 than group 1 at the end of day 6. The probabilities of having no HE increase with time for both subgroups and exceed 0.9 on day 6, and the probabilities of having moderate HE of I–II or high HE or III–IV both decrease with time (Figure, C-E). Groups 3 and 4 had higher INR and total bilirubin than groups 1 and 2 at enrollment with group 3 having higher values of each than group 4 at enrollment. The disease trajectories of the three measures among Group 3 participants tended to improve over time. On day 6, the estimated INR for group 3 is below 2, the estimated total bilirubin in group 3 is approximately 12 mg/dL, and the estimated probability of having no HE for group 3 is over 0.5 (Figure, A–C). By comparison, both the INR trajectory and the total bilirubin trajectory appear slightly increasing for group 4 (Figure, A–B). The probabilities of having mild HE of I–II or having high HE of III–IV appear relatively stable for Group 4. On day 4, the trajectory values in Group 4 are worse than those in Group 3 in all three measures. Group 5 appears to have the worst disease trajectories with high and increasing INR and total bilirubin. The probability of having HE grade of III–IV in group 5 approaches 0.7 at day 6 (Figure, E). The probability of HE grade of III or IV at day 6 is < 0.2 for all of the other subgroups.
Figure.
Estimated disease trajectories for the 5 latent subgroups in the first 7 days following enrollment. Day 0 corresponds to the day of enrollment.
The most common trajectory was subgroup 4 with 130 (34%) participants. There were similar numbers of participants in subgroups 1, 2 and 3 with 59 (16%), 57 (15%) and 48 (13%) participants, respectively. The remaining 86 (23%) participants were in subgroup 5.
Table II (available at www.jpeds.com) presents the mean of estimated posterior probabilities by estimated latent subgroup index. The results suggest that the GMM distinguishes between the 5 subgroups well. For example, the mean posterior probability of belonging to subgroups 1–2 is 0.01 for patients who were classified into subgroup 4; the mean posterior probability of belonging to subgroup 3 and 5 is 0.031 and 0.029, respectively, for a subgroup 4 patients. This result suggests that the GMM is able to classify patients with IND with a high degree of certainty.
Table 2.
Mean of estimated posterior probabilities (row) by latent subgroup index (column)* for the N=380 participants with indeterminate diagnosis (IND).
| Subgroup1 (N=59) | Subgroup2 (N=57) | Subgroup3 (N=48) | Subgroup4 (N=130) | Subgroup5 (N=86) | |
|---|---|---|---|---|---|
| Subgroup 1 | 0.961 | 0.005 | 0.001 | 0.001 | 0.000 |
| Subgroup 2 | 0.021 | 0.954 | 0.021 | 0.009 | 0.000 |
| Subgroup 3 | 0.015 | 0.029 | 0.935 | 0.031 | 0.044 |
| Subgroup 4 | 0.001 | 0.010 | 0.023 | 0.929 | 0.049 |
| Subgroup 5 | 0.002 | 0.002 | 0.020 | 0.029 | 0.907 |
For each participant, the GMM calculates his/her posterior probabilities of belonging to different latent subgroups. Then, the GMM classifies the participant into the subgroup with the largest posterior probability. The columns here represent the latent subgroup that the participants were classified into. Within each column, the rows represents the mean posterior probabilities of belonging to different subgroups.
Participants’ characteristics at study enrollment by subgroup are summarized in Table I. The proportions of participants with ascites were higher in groups 3–5 than groups 1 and 2 (p=0.002), where the proportions were < 10% for groups 1–2 but > 20% for groups 3–5. However, the numerical differences in ascites proportions may not be apparent enough for practical discriminations. ALT was highest in group 1, followed by group 2 and 5 (p<.001). The distributions of age and albumin were not significantly different across the latent subgroups.
The 21 day outcomes differ significantly among the disease trajectory subgroups (p<.001; Table III). For example, the proportions of subjects who were alive without LTx at day 21 were 95% for group 1 and 93% for group 2. For group 3, only 2/48 (4%) died within 21 days, however 14 (29%) underwent LTx mostly by day 7. Although the proportions of death and LTx by day 7 were similar between group 3 and 4, participants in group 4 were more likely to die (1/48, 2% vs 9/130, 7%) or undergo LTx (1/48, 2% vs 30/130, 23%) between day 8 and 21. Only 3/86 (3%) subjects in group 5 were alive without LTx at day 21. The majority (80%) of Group 5 participants underwent LTx by day 21 and 16% died.
Table 3.
Summary of 21-day outcomes among participants with indeterminate diagnosis (IND). Each column represents the distribution of the 21-day outcome, overall or within a certain subgroup. The distribution of the 21-day outcome are significantly different across the five latent subgroups (p<.001).
| All N=380 |
Subgroup1 N=59 |
Subgroup2 N=57 |
Subgroup3 N=48 |
Subgroup4 N=130 |
Subgroup5 N=86 |
|
|---|---|---|---|---|---|---|
| 21-day Outcome* | N=379 | N=58 | N=57 | N=48 | N=130 | N=86 |
| Death by day 7 | 14(4%) | 0(0%) | 2(4%) | 1(2%) | 3(2%) | 8(9%) |
| LTx by day 7 | 110(29%) | 0(0%) | 2(4%) | 13(27%) | 36(28%) | 59(69%) |
| Death between day 8–21 | 19(5%) | 3(5%) | 0(0%) | 1(2%) | 9(7%) | 6(7%) |
| LTx between day 8–21 | 41(11%) | 0(0%) | 0(0%) | 1(2%) | 30(23%) | 10(12%) |
| Alive with native liver at day 21 | 195(52%) | 55(95%) | 53(93%) | 32(67%) | 52(40%) | 3(3%) |
|
| ||||||
| Total death by day 21 | 33 (9%) | 3(5%) | 2(4%) | 2(4%) | 12(9%) | 14(16%) |
| Total LTx by day 21 | 151(40%) | 0(0%) | 2(4%) | 14(29%) | 66(51%) | 69(80%) |
Of the 549 PALF participants with specified etiologies (non-IND) there were 488 who had at least 3 values for one or more three measurements. To explore the association between the potential subgroup index and the underlying disease, these participants were classified into the previously identified latent subgroups (Table IV). There were 196/488 (40%) classified into either Group 4 or 5, the two groups with worsening trajectories, compared with 216/380 (55%) among the participants with IND. Only 5/107 (5%) participants with a final diagnosis of APAP were classified into Group 4 or 5. Among participants with a final diagnosis of viral hepatitis other than hepatitis A/B/C/E, such as herpes simplex, EBV and CMV, there were 36/54 (67%) who were assigned to group 4 or group 5. Eight (36%) of the 22 participants with hemophagocytic syndrome were classified into group 5. These results demonstrate that the subgroup indices are strongly associated with the underlying PALF etiology (p<.001), so may carry important diagnostic value for the indeterminate group.
Table 4.
The distribution of subgroup classification among the N=488 patients with identified disease etiologies (non-IND) and sufficient data on INR, total bilirubin and HE, overall and by disease etiology. Each row represents the distribution of subgroup classifications within a specific diagnostic category. The distribution of subgroups are significantly different across different diagnostic categories (p<.001).
| categories | N | Subgroup1 | Subgroup2 | Subgroup3 | Subgroup4 | Subgroup5 |
|---|---|---|---|---|---|---|
| All non-IND | 488 | 136(28%) | 95(19%) | 61(13%) | 135(28%) | 61(13%) |
| Diagnosis | ||||||
| Acetaminophen | 107 | 71(66%) | 20(19%) | 11(10%) | 2(2%) | 3(3%) |
| Drug-induced hepatitis | 21 | 5(24%) | 7(33%) | 2(10%) | 6(29%) | 1(5%) |
| Hemophagocytic Syndrome | 22 | 1(5%) | 5(23%) | 4(18%) | 4(18%) | 8(36%) |
| Hepatitis, A/B/C/E | 11 | 1(9%) | 7(64%) | 1(9%) | 2(18%) | 0(0%) |
| Other viral hepatitis | 54 | 11(20%) | 4(7%) | 3(6%) | 26(48%) | 10(19%) |
| Shock/ischemia | 31 | 12(39%) | 6(19%) | 7(23%) | 2(6%) | 4(13%) |
| Neonatal iron storage | 26 | 1(4%) | 2(8%) | 1(4%) | 15(58%) | 7(27%) |
| Veno-oclusive disease | 12 | 0(0%) | 3(25%) | 2(17%) | 7(58%) | 0(0%) |
| Wilsons disease | 31 | 0(0%) | 9(29%) | 6(19%) | 12(39%) | 4(13%) |
| Other Metabolic | 58 | 11(19%) | 14(24%) | 14(24%) | 15(26%) | 4(7%) |
| Autioimmune Hepatitis | 63 | 5(8%) | 12(19%) | 7(11%) | 30(48%) | 9(14%) |
| Other diagnosis | 31 | 11(35%) | 3(10%) | 2(6%) | 9(29%) | 6(19%) |
| Multiple | 21 | 7(33%) | 3(14%) | 1(5%) | 5(24%) | 5(24%) |
In addition, we summarized the 21-day outcomes among the 488 patients with non-IND PALF by their latent subgroup indices (Table V; available at www.jpeds.com). The results agree with those observed in Table III, and suggest that the subgroup indices also carry prognostic values for patients with PALF with a specified disease etiology (p<.001).
Table 5.
the distribution of 21-day outcomes among the N=488 non-IND patients by estimated latent subgroup index, where the p-value between the 21-day outcome and the latent subgroup index is <.001.
| characteristics | All N=488 |
Subgroup1 N=136 |
Subgroup2 N=95 |
Subgroup3 N=61 |
Subgroup4 N=135 |
Subgroup5 N=61 |
|---|---|---|---|---|---|---|
| 21-day Outcome* | N=488 | N=136 | N=95 | N=61 | N=135 | N=61 |
| Death by day 7 | 30(6%) | 2(1%) | 1(1%) | 4(7%) | 12(9%) | 11(18%) |
| LTx by day 7 | 56(11%) | 1(1%) | 5(5%) | 9(15%) | 18(13%) | 23(38%) |
| Death between day 8–21 | 29(6%) | 0(0%) | 3(3%) | 2(3%) | 16(12%) | 8(13%) |
| LTx between day 8–21 | 20(4%) | 0(0%) | 1(1%) | 0(0%) | 16(12%) | 3(5%) |
| Alive with native liver at day 21 | 353(72%) | 133(98%) | 85(89%) | 46(75%) | 73(54%) | 16(26%) |
|
| ||||||
| Total death by day 21 | 59(12%) | 2(1%) | 4(4%) | 6(10%) | 28(21%) | 19(31%) |
| Total LTx by day 21 | 76(16%) | 1(1%) | 6(6%) | 9(15%) | 34(25%) | 26(43%) |
Discussion
Pediatric acute liver failure is a dynamic clinical syndrome in which patients can be observed to improve, progress to a fatal outcome, or experience clinical deterioration followed by full recovery as a result of directed therapies or supportive care. The natural history of PALF is interrupted by LTx. Although LTx can be life-saving, it is also life-altering and should be avoided in a child who would have fully recovered without LTx. In this work, we employed growth mixture modeling (GMM) to identify clinically meaningful latent subgroups utilizing the dynamic clinical and biochemical trends among PALF participants of indeterminate disease etiology. The five latent subgroups feature distinct clinical courses, and the subgroups were shown to be significantly associated with 21 day outcomes. This work systematically studied the dynamic clinical courses of PALF and explored the predictive value of the clinical courses.
Medical and transplant decisions relative to both listing for, and accepting, an organ are addressed continuously by the clinician through the hospital course utilizing a complex array of factors (12). Distilling multiple clinical and biochemical measures into a practical model that can be used at the bedside for each decision interval remains a challenge. Previous predictive models for ALF in adults and children have included clinical and biochemical measures that are objective (e.g, INR, bilirubin, ammonia, age) or subjective (e.g., jaundice, encephalopathy) at the time of admission to hospital, the highest recorded or “peak” value(13, 14), or combinations of these data elements(2, 15–17). Measures obtained only at admission are limited by their inability to account for disease progression or improvement during the ensuing days. Utilization of a peak value is problematic given the uncertainty of when that value may be achieved. For example, a peak value may represent the initial result obtained, as we see for the INR value in groups 1, 2 and 3 in our model, and the INR may increase over time as in groups 4 and 5. A study from New Delhi, where the option of liver transplantation was not available, found dynamic changes in HE, INR, arterial ammonia and total bilirubin collected over 3 hospital days to be predictive of survival or death(18). The study involved adults with etiologies quite different from those in the PALF study, but it does point to the potential usefulness of dynamic changes within a manageable palette of variables to predict outcome.
The GMM model we constructed may serve as a useful prognostic tool for a new patient with PALF with an indeterminate or established disease etiology. Using clinical measures routinely obtained for patients with PALF, we were able to classify PALF participants into one of the five subgroups each with unique dynamic trajectories of INR, total bilirubin and HE that were associated with outcomes. In a patient with a clinical trajectory mirroring subgroup 1 or 2, a decision to proceed with LTx should be made with caution given the high likelihood of spontaneous survival in over 92% of patients with similar trajectories. Subjects in subgroup 3 had a trajectory of improvement in all 3 measured parameters that might suggest an opportunity for spontaneous recovery, yet LTx occurred in 14/48 (29%). Careful assessment of the need for LTx in patients with similar trajectories would appear to be worthwhile. Participants in subgroup 4 were found to have an INR and total bilirubin that increased over the 7 day observation period. Although the degree of increase in the laboratory values was relatively modest and HE was clinically mild (0–II) in the majority, participants in the subgroup experienced a high frequency of LTx (51%). Liver transplantation may be appropriate for patients classified into subgroups 4 or 5, although the high rates of LTx in the two groups preclude the possibility of observing that some of these participants may have improved if data were available beyond 7 days. We have included 4 virtual clinical illustrations (Appendix 2; available at www.jpeds.com) of how one might use the GMM at the bedside to assist in assessing the potential outcome for a patient with more the 3, but less than 7 days of data.
Our model demonstrates its potential usefulness for participants with either an IND or non-IND diagnosis. For our results to be used for clinical decision-making, however, it will be necessary to further validate our results. Moreover, like other prognostic tools, the model should be used in conjunction with, and not as a replacement for, clinical judgements regarding short-term disease status. The timing of availability of organs is also critical and needs to be taken into account.
The modeling procedure utilized the information for INR, total bilirubin and HE for up to 7 days. However, patients with fewer days of measurements can still be classified, just with less classification accuracy. In these cases, the reliability of the classification may depend on many things, such as the specific days of measurement (eg, the measurements on days 0, 3, 5 may provide richer information than the measurements on days 0, 1, 2, despite that the total number days of measurements is the same) and the variability of an individual trajectory. One way to assess the reliability of the classification in practice is to examine the posterior probabilities that the patient belongs to each of the five latent subgroups, which can be calculated based on his/her available trajectory data. The situation when one of the five posterior probabilities exceeds 0.8 would provide confidence about the classification. When more updated data become available, we suggest that the posterior probabilities be updated and the resulting subgroup classification be re-examined.
Ideally, determining the trajectory of a clinical condition would begin with the day of disease onset. For the majority of PALF cases, however, the day of onset cannot be precisely determined. Non-specific symptoms may have been present for hours, days or even weeks before a clinical suspicion prompted laboratory tests that met PALF study entry criteria. In our analysis, we used data in the first 7 days following enrollment to serve as a window into the clinical trajectory of what is considered to be a rapidly evolving condition. The study captured data at the earliest possible interval following consent the family of the critically ill child.
From the day of enrollment, the trajectory data were only available for 7 days in the first two phases of the PALF study. For participants with an outcome beyond the 7 days of data collection, it is possible the trajectory after the first 7 days would differ from the initial trajectory. We recognize that it may be necessary to extend comprehensive data analysis beyond 7 days. We speculate the long-term trajectories may change over time, reflecting different stages of a longer clinical course. Future studies will assess the GMM using detailed clinical and laboratory assessments beyond 7 days.
There are some limitations in this analysis. First, we have excluded patients with fewer than 3 measurements for all three trajectories, because their limited trajectory data would not enable a good classification. As a result, the PALF participants who experienced the outcome (death, LTx, or hospital discharge) quickly after enrollment were not included. Therefore, our results only apply to patients with PALF who remained alive without undergoing liver transplantation for three or more days. Secondly, because the clinical measurements are only available for up to 7 days, we adopted the linear trajectory assumption to model the trajectory patterns in the GMM. With the sample size, the linear trajectory model strikes a balance between the model fit quality and the discriminative ability of the classification. With wider time range and larger sample size, it may be worthwhile to consider more complicated trajectory models, such as the piecewise-linear trajectory model, which can accommodate richer trajectory patterns. Furthermore, we adopted the MAR assumption in our estimation procedures. There are two reasons to believe that MAR may be appropriate for this analysis. The first is that missing data were due to various reasons, including both positive (hospital discharge) and negative (death) outcomes, and blood volume considerations, which is a bigger issue for younger participants. The second reason is that the three measures we studied are quite informative for the disease outcomes of death, LTx and spontaneous recovery. The part of missingness that can be explained by the observed values of INR, total bilirubin and HE will not bias the model estimates. Note that we did not include the event time outcomes (e.g., time to death and LTx) in the statistical modeling, so that we can assess the prognostic value of the trajectories independently without assuming it in the modeling building procedure. A joint modeling procedure may enable us to achieve latent subgroup classifications using both the clinical trajectories and the event time outcomes.
In conclusion, this study examined the trajectories of three key clinical and laboratory measures among a large cohort of patients with PALF without a specified disease etiology. The GMM accounts for the heterogeneity in disease etiology and progression pattern, and furthermore entails a meaningful and rigorous classification according to participants’ disease trajectories. The results reported shed insight into the predictive value of the dynamic trajectory patterns, and show promise as a powerful and reliable prognostic tool for new patients with PALF with or without an identified disease etiology.
Acknowledgments
We thank Edward Doo, MD, and Averell H. Sherker, MD (the National Institutes of Health), for <>; and members of the Data Coordinating Center at the University of Pittsburgh.
Supported by the National Institutes of Health (2U01DK072146, UL1 RR024153, and UL1TR000005).
Abbreviations and Acronyms
- APAP
acetaminophen overdose
- BLRT
bootstrap likelihood ratio test
- GMM
growth mixture modeling
- HE
clinical encephalopathy
- IND
indeterminate final diagnosis
- KCHC
the King’s college hospital criteria
- LTx
liver transplantation
- PALF
pediatric acute liver failure
- non-IND
determined final diagnosis
Appendix 1
Additional members of the Pediatric Acute Liver Failure Study Group include:
Current Sites, Principal Investigators and Coordinators –Kathryn Bukauskas, RN, CCRC (Children’s Hospital of Pittsburgh of UPMC, Pittsburgh, Pennsylvania); Michael R. Narkewicz, MD, Michelle Hite, MA, CCRC (Children’s Hospital Colorado, Aurora, Colorado); Kathleen M. Loomes, MD, Elizabeth B. Rand, MD, David Piccoli, MD, Deborah Kawchak, MS, RD (Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania); Rene Romero, MD, Saul Karpen, MD, PhD, Liezl de la Cruz-Tracy, CCRC (Emory University, Atlanta, Georgia); Vicky Ng, MD, Kelsey Hunt, Clinical Research Coordinator (Hospital for Sick Children, Toronto, Ontario, Canada); Girish C. Subbarao, MD, Ann Klipsch, RN (Indiana University Riley Hospital, Indianapolis, Indiana); Estella M. Alonso, MD, Lisa Sorenson, PhD, Susan Kelly, RN, BSN, Dhey Delute, RN, CCRC, Katie Neighbors, MPH, CCRC (Lurie Children’s Hospital of Chicago, Chicago, Illinois); Philip J. Rosenthal, MD, Shannon Fleck, Clinical Research Coordinator (University of California San Francisco, San Francisco, California); Mike A. Leonis, MD, PhD, John Bucuvalas, MD, Tracie Horning, Clinical Research Coordinator (University of Cincinnati, Cincinnati, Ohio); Norberto Rodriguez Baez, MD, Shirley Montanye, RN, Clinical Research Coordinator, Margaret Cowie, Clinical Research Coordinator (University of Texas Southwestern, Dallas, Texas); Karen Murray, MD, Melissa Young, Clinical Research Coordinator, Heather Vendettuoli, Clinical Research Coordinator (University of Washington, Seattle, Washington); David A. Rudnick, MD, PhD, Ross W. Shepherd, MD, Kathy Harris, Clinical Research Coordinator (Washington University, St. Louis, Missouri).
Previous Sites, Principal Investigators and Coordinators –Saul J. Karpen, MD, PhD, Alejandro De La Torre, Clinical Research Coordinator (Baylor College of Medicine, Houston, Texas); Dominic Dell Olio, MD, Deirdre Kelly, MD, Carla Lloyd, Clinical Research Coordinator (Birmingham Children’s Hospital, Birmingham, United Kingdom); Steven J. Lobritto, MD, Sumerah Bakhsh, MPH, Clinical Research Coordinator (Columbia University, New York, New York); Maureen Jonas, MD, Scott A. Elifoson, MD, Roshan Raza, MBBS (Harvard Medical School, Boston, Massachusetts); Kathleen B. Schwarz, MD, Wikrom W. Karnsakul, MD, Mary Kay Alford, RN, MSN, CPNP (Johns Hopkins University, Baltimore, Maryland); Anil Dhawan, MD, Emer Fitzpatrick, MD (King’s College Hospital, London, United Kingdom); Nanda N. Kerkar, MD, Brandy Haydel, CCRC, Sreevidya Narayanappa, Clinical Research Coordinator (Mt. Sinai School of Medicine, New York, New York); M. James Lopez, MD, PhD, Victoria Shieck, RN, BSN (University of Michigan, Ann Arbor, Michigan).
Appendix 2. Detailed Illustration of the Subgroup Classification
We illustrate the subgroup classification for several virtual participants. The classification is conducted through a computer algorithm we developed, which utilizes the GMM results to calculate the posterior probabilities of the five latent subgroups. The participant should be classified into the subgroup with the largest posterior probability.
For future research, we plan to further validate the algorithm on a new cohort of PALF patients. After that, the algorithm will be transformed into a user-friendly web interface, where a clinician can input the observed trajectory data to obtain the estimated posterior probabilities.
Participant 1
In the following table, each row presents a measure, and the columns represent the value of the measure for 7 consecutive days since the day of enrollment. * means missing data.
| Day0 | Day1 | Day2 | Day3 | Day4 | Day5 | Day6 | |
|---|---|---|---|---|---|---|---|
| INR | * | 1.80 | 1.90 | 1.90 | 1.20 | 1.40 | 1.30 |
| Total Bilirubin (mg/dL) | * | 15.1 | * | 11.9 | 10.2 | 8.8 | 6.5 |
| Encephalopathy | I–II | 0 | 0 | 0 | 0 | 0 | 0 |
Using participant 1’s data for all 7 days, his/her estimated posterior probabilities for subgroups 1–5 are: 0, 0.97, 0.03, 0, and 0. The five posterior probabilities always sum up to 1, and the person was classified into the subgroup with the largest posterior probability. Therefore, this participant was classified into subgroup 2.
If we only use participant 1’s data up till Day 4, the posterior probabilities for subgroups 1–5 are: 0, 0.82, 0.17, 0.01, and 0. The person would still be classified into subgroup 2. However, the certainty of the classification may reduce when we have fewer data points.
The person’s 21 day outcome is alive with native liver.
Participant 2
| Day0 | Day1 | Day2 | Day3 | Day4 | Day5 | Day6 | |
|---|---|---|---|---|---|---|---|
| INR | 2.6 | 2.2 | 2.5 | 1.6 | 1.9 | * | * |
| Total Bilirubin (mg/dL) | 11.2 | 14.6 | 18.8 | 21.2 | 20.6 | * | * |
| Encephalopathy | 0 | I–II | III–IV | III–IV | III–IV | * | * |
This participant had improving INR but worsening total bilirubin and HE. Using all his/her data, her posterior probabilities for subgroups 1–5 are: 0, 0, 0, 0, 1. Thus, this participant was classified into subgroup 5.
If we only utilize the data from Day 0 to Day 3, the posterior probabilities are still 0, 0, 0, 0, 1. The person’s 21 day outcome is death.
Participant 3
| Day0 | Day1 | Day2 | Day3 | Day4 | Day5 | Day6 | |
|---|---|---|---|---|---|---|---|
| INR | 2.1 | 2.2 | 2.4 | 3.0 | 3.2 | 2.9 | 3.3 |
| Total Bilirubin (mg/dL) | 14.7 | 16.9 | 17 | 20.2 | 20.9 | 21.6 | 23.4 |
| Encephalopathy | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
On the basis of Day0–Day6 data, the estimated posterior probabilities for subgroups 1–5 are: 0, 0, 0, 1, 0. Thus, this is a subgroup 4 participant.
If we only use the data between Day 0 and Day 4, then the posterior probabilities for the 5 subgroups are: 0, 0, 0, 0.98, 0.02, still leading to subgroup 4.
This participant’s 21-day outcome is LTx.
Participant 4
| Day0 | Day1 | Day2 | Day3 | Day4 | Day5 | Day6 | |
|---|---|---|---|---|---|---|---|
| INR | 5.1 | 4.0 | 4.2 | 4.7 | 4.7 | 4.6 | 4.1 |
| Total Bilirubin (mg/dL) | 31.7 | 37 | 32.2 | 38.8 | 32.8 | 33.9 | 32.9 |
| Encephalopathy | 0 | I–II | I–II | I–II | * | I–II | I–II |
Using all the data from Day 0 to Day 6, the posterior probability estimates are: 0, 0, 0, 1, 0. Thus, the participant was classified into subgroup 4.
If we only use the data between Day 0 and Day 4, then the posterior probabilities are: 0, 0, 0, 0.41, 0.59. The probabilities for subgroup 4 and subgroup 5 are both not low. Therefore, the classification for this participant was not that certain when fewer data points were available. On Day 4, we can only say that this participant belongs to either subgroup 4 or subgroup 5.
This participant’s 21-day outcome is LTx.
Footnotes
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Squires RH, Jr, Shneider BL, Bucuvalas J, Alonso E, Sokol RJ, Narkewicz MR, et al. Acute liver failure in children: the first 348 patients in the pediatric acute liver failure study group. The Journal of pediatrics. 2006;148:652–8. doi: 10.1016/j.jpeds.2005.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Grady JG, Alexander GJ, Hayllar KM, Williams R. Early indicators of prognosis in fulminant hepatic failure. Gastroenterology. 1989;97:439–45. doi: 10.1016/0016-5085(89)90081-4. [DOI] [PubMed] [Google Scholar]
- 3.Sundaram V, Shneider BL, Dhawan A, Ng VL, Im K, Belle S, et al. King’s College Hospital Criteria for non-acetaminophen induced acute liver failure in an international cohort of children. The Journal of pediatrics. 2013;162:319–23e1. doi: 10.1016/j.jpeds.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narkewicz MR, Dell Olio D, Karpen SJ, Murray KF, Schwarz K, Yazigi N, et al. Pattern of diagnostic evaluation for the causes of pediatric acute liver failure: an opportunity for quality improvement. The Journal of pediatrics. 2009;155:801–6e1. doi: 10.1016/j.jpeds.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suchy FJ, Sokol RJ, Balistreri WF. Liver disease in children. 3. Cambridge ; New York: Cambridge University Press; 2007. p. xvii.p. 1030.p. 22. [Google Scholar]
- 6.Squires RH, Dhawan A, Alonso E, Narkewicz MR, Shneider BL, Rodriguez-Baez N, et al. Intravenous N-acetylcysteine in pediatric patients with nonacetaminophen acute liver failure: a placebocontrolled clinical trial. Hepatology. 2013;57:1542–9. doi: 10.1002/hep.26001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muthen B, Shedden K. Finite mixture modeling with mixture outcomes using the EM algorithm. Biometrics. 1999;55:463–9. doi: 10.1111/j.0006-341x.1999.00463.x. [DOI] [PubMed] [Google Scholar]
- 8.Muthen B, Muthen LK. Integrating person-centered and variable-centered analyses: growth mixture modeling with latent trajectory classes. Alcoholism, clinical and experimental research. 2000;24:882–91. [PubMed] [Google Scholar]
- 9.Jung T, Wickrama KAS. An introduction to latent class growth analysis and growth mixture modeling. Social and Personality Psychology Compass. 2008:2. [Google Scholar]
- 10.Ram N, Grimm KJ. Growth mixture modeling: A method for identifying differences in longitudinal change among unobserved groups. Int J Behav Dev. 2009;33:565–76. doi: 10.1177/0165025409343765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celeux G, Soromenho G. An entropy criterion for assessing the number of clusters in a mixture model. J Classif. 1996;13:195–212. [Google Scholar]
- 12.Alagoz O, Hsu H, Schaefer AJ, Roberts MS. Markov decision processes: a tool for sequential decision making under uncertainty. Medical decision making : an international journal of the Society for Medical Decision Making. 2010;30:474–83. doi: 10.1177/0272989X09353194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu E, MacKenzie T, Dobyns EL, Parikh CR, Karrer FM, Narkewicz MR, et al. Characterization of acute liver failure and development of a continuous risk of death staging system in children. Journal of hepatology. 2006;44:134–41. doi: 10.1016/j.jhep.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Lee WS, McKiernan P, Kelly DA. Etiology, outcome and prognostic indicators of childhood fulminant hepatic failure in the United kingdom. Journal of pediatric gastroenterology and nutrition. 2005;40:575–81. doi: 10.1097/01.mpg.0000158524.30294.e2. [DOI] [PubMed] [Google Scholar]
- 15.Lu BR, Gralla J, Liu E, Dobyns EL, Narkewicz MR, Sokol RJ. Evaluation of a scoring system for assessing prognosis in pediatric acute liver failure. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2008;6:1140–5. doi: 10.1016/j.cgh.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajanayagam J, Coman D, Cartwright D, Lewindon PJ. Pediatric acute liver failure: etiology, outcomes, and the role of serial pediatric end-stage liver disease scores. Pediatric transplantation. 2013;17:362–8. doi: 10.1111/petr.12083. [DOI] [PubMed] [Google Scholar]
- 17.Rajanayagam J, Frank E, Shepherd RW, Lewindon PJ. Artificial neural network is highly predictive of outcome in paediatric acute liver failure. Pediatric transplantation. 2013;17:535–42. doi: 10.1111/petr.12100. [DOI] [PubMed] [Google Scholar]
- 18.Kumar R, Shalimar, Sharma H, Goyal R, Kumar A, Khanal S, et al. Prospective derivation and validation of early dynamic model for predicting outcome in patients with acute liver failure. Gut. 2012;61:1068–75. doi: 10.1136/gutjnl-2011-301762. [DOI] [PubMed] [Google Scholar]