Abstract
BACKGROUND
Appendiceal cancer (AC) patients treated with cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC) often demonstrate an unpredictable variability in their survival outcomes. Biomarkers predictive of CRS/HIPEC efficacy could better guide treatment decisions. We hypothesized that variation in the transcriptional programming of AC tumors might distinguish molecular subtypes with differential outcomes after CRS/HIPEC.
STUDY DESIGN
Gene expression profiles of 2 AC cohorts were analyzed using Affymetrix whole-genome expression microarrays. Hierarchical clustering methods, Kaplan-Meier analysis, and Cox regression models were used to discover and validate prognostic molecular subtypes of AC. Gene set enrichment analysis was used to infer pathologic attributes of the molecular subtypes.
RESULTS
Unsupervised hierarchical clustering analysis of tumor expression profiles revealed a 139-gene cassette that distinguished 2 molecular subtypes (based on low vs high expression of the gene cassette) with statistically significant survival differences (disease-specific survival, p = 0.0075; progression-free survival, p = 0.0072). In a second AC cohort, the 139-gene cassette reproducibly partitioned tumors into subtypes with significant survival differences. Tumors showing high relative expression of the genes comprising the cassette associated with poor survival outcomes (disease-specific survival, p = 0.047; progression-free survival, p = 0.0079), and exhibited gene expression patterns enriched for oncogenic processes and pathways. The prognostic value of the molecular subtypes was specific for low-grade appendiceal tumors (disease-specific survival, p = 0.028; progression-free survival, p = 0.0016), and remained significant in the presence of conventional prognostic markers, including grade, surgical resection score, Eastern Cooperative Oncology Group status, and age.
CONCLUSIONS
The 139-gene cassette can have actionable clinical utility for identifying low-grade appendiceal tumor molecular subtypes predictive of therapeutic efficacy of CRS/HIPEC.
It is estimated that approximately 1% of all appendectomy specimens will contain a neoplasm.1 The most common cancers of the appendix are neuroendocrine tumors (carcinoid), benign mucoceles, and mucinous carcinoma. Appendiceal cancer (AC) is a rare disease, yet its incidence in the reported literature varies, depending on the histologic types included in the classification of appendiceal malignancies.2,3 In a Surveillance, Epidemiology, and End Results database retrospective analysis, that excluded low-grade carcinoid tumors, the annual age-adjusted incidence of appendiceal primaries was 0.12 cases per 1,000,000 of population. Appendiceal adenocarcinoma represented 66.5% of these patients.3 Extrapolating from the fact that the Surveillance, Epidemiology, and End Results program collects data from 14% of the US population, the annual incidence of appendiceal adenocarcinoma in the country should be around 300 to 400 cases, although estimates up to 3,500 cases annually in the United States have been made.4 The rate of appendiceal neoplasms is believed to have increased by >50% since the turn of the century.4
Although rare, AC is associated with considerable mortality due to the late stage at diagnosis and the low likelihood of it being found on screening colonoscopic examinations.5 Mucinous ACs rupture all too frequently, leading to peritoneal surface disease (PSD) or so-called “carcinomatosis.” Cytoreductive surgery (CRS) with hyperthermic intraperitoneal chemotherapy (HIPEC) is an established modality for the treatment of peritoneal dissemination from appendiceal tumors,6–13 as well as a variety of epithelial primaries. Survival after CRS/HIPEC for appendiceal neoplasms with PSD is multifactorial and often depends on tumor biology, volume of disease at presentation, completeness of CRS, and patient’s functional status and comorbidities.7–12
Patients with PSD from low-grade appendiceal (LGA) primaries have traditionally been considered the best candidates for CRS and HIPEC, primarily due to favorable biologic behavior characterized by a predominant pattern of late or noninvasive superficial spread into tissues, with minimal risk of hematogenous dissemination.8,9,12 However, even within the LGA group, clinical outcomes, such as progression-free survival (PFS) and disease-specific survival (DSS), show a significant and often unpredictable variability.7–12 This variability is greater when the studied cohorts include patients with high-grade appendiceal primaries, even when the extent of disease and completion of CRS are factored into the survival analysis.12
Two well-accepted light microscopy-based histologic classification systems have only partially stratified the polymorphic and often convoluted clinical spectrum of PSD from appendiceal primaries.8,13 Both systems use a combination of features, including presence of mucin and epithelium, cytologic atypia, degree of proliferation, architectural complexity, mitotic activity and parenchymal invasion. The Ronnett system13 identified 3 tiers of tumor histology with prognostic significance: disseminated peritoneal adenomucinosis, peritoneal mucinous carcinomatosis with intermediate or discordant features, and peritoneal mucinous carcinomatosis. On the other hand, the Bradley system8 combined the intermediate and the low categories together and came up with a 2-tier system: mucinous carcinoma peritonei low grade and high grade. Although the 2 histologic classification systems are each significant and reliable for predicting prognosis, even in the good prognostic subgroup, there is a substantial failure rate despite complete cytoreduction.
In previous work, we investigated the genome-wide gene expression profiles of a panel of peritoneal metastases comprising 26 appendiceal and 15 colorectal tumors.14 Through unsupervised clustering analysis, 3 distinct tumor subclusters of mixed histologies were identified. Kaplan-Meier analysis demonstrated that the tumor subclusters, composed of both appendiceal and colorectal tumors, were prognostic in nature, stratifying patients into significantly different survival groups with potential for clinical impact, particularly in the context of low-grade appendiceal disease.
In the current study, we sought to more fully characterize the transcriptome of AC, focusing on the global gene expression patterns and transcriptional subsets of genes that associate with patient prognosis. We hypothesized that the clinical diversity in patient outcomes could be resolved, in part, through the identification of differences in tumor transcriptional programming that distinguish more and less aggressive disease subtypes. In this work, we discovered and validated the existence of 2 distinct prognostic subtypes most applicable to LGA tumors, as differentiated by a 139-gene transcriptional cassette, and demonstrated the additive prognostic value of these novel subtypes in comparison with conventional predictive variables.
METHODS
Patients and clinical characteristics
Selection of AC cases was facilitated by an IRB-approved, prospectively maintained database of clinical and demographic information on all patients treated with CRS/HIPEC at Wake Forest Baptist Medical Center from 1992 to the present. In the original series,14 24 appendiceal tumor samples were collected for analysis from patients who received treatment at Wake Forest Baptist Medical Center from 2000 through 2006. Salient characteristics of this series were the following: 29% female, 71% male, mean age at surgery of 54 years, and mean clinical follow-up of 4.5 years. In the second series (the validation cohort), 39 samples with adequate ribonucleic acid (RNA) integrity from patients who received treatment from 2001 to 2011 were analyzed. Demographic characteristics were the following: 51% female, 49% male, mean age at surgery of 51 years, and mean clinical follow-up of 3.9 years.
Tumor specimens
Tumor samples from both patient series were collected according to a routine rapid tissue collection protocol developed by our Cancer Center’s Tumor Tissue and Pathology Shared Resource. Accordingly, immediately after resection, tumor specimens were identified grossly by a pathologist, snap frozen in liquid nitrogen, and maintained at −80°C. Tissue analysis, qualification, and processing were conducted as described previously.14 Tumor tissue content was verified by a surgical pathologist via a subsequent frozen section.
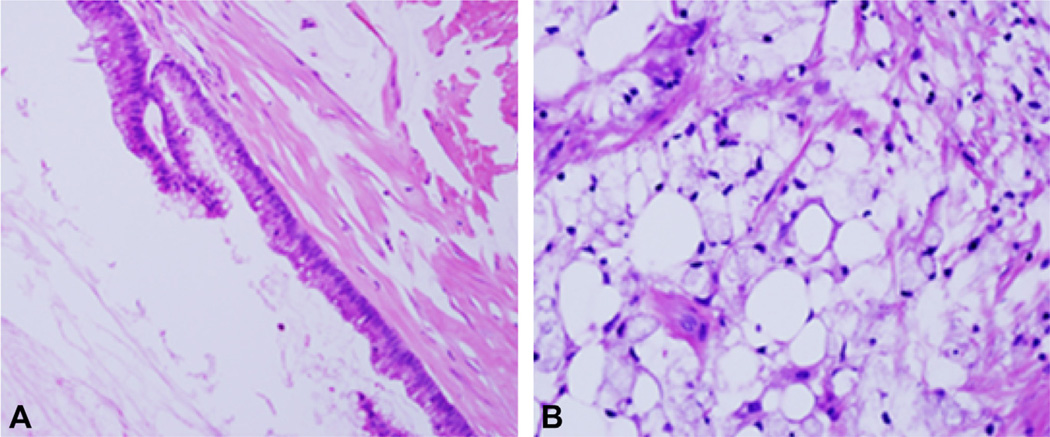
The associated diagnostic materials for all cases were reviewed by 2 pathologists. Tumors were divided into 2 groups (high grade and low grade), following the Bradley system.8 The peritoneal lesions were used for grading. Tumors that demonstrated any of the following were classified as high grade: high-grade nuclear atypia to include prominent nucleoli, hyperchromatic nuclei, markedly irregular nuclear membrane or dense coarse chromatin; architectural complexity, such as cribriform pattern, papillary formation, or extensive nuclear stratification; high cellularity; frequent mitosis; and signet ring cells. Examples of low and high-grade cancer are shown in Figure 1.
Figure 1.
Histology of peritoneal mucinous tumors. (A) Low-grade tumor showing simple bland appearing epithelium and abundant mucin. (B) High-grade tumor with signet-cell morphology and tissue invasion. Hematoxylin and eosin, 20× (A) and 40× (B) magnification.
Ribonucleic acid isolation and microarray analysis
Ribonucleic acid isolates from the original tumor series (n = 24) were processed as described previously.14 For the validation series (n = 39), total RNA was isolated using the Qiagen RNeasy kit according to the manufacturer’s instructions. Ribonucleic acid quality and quantity was assessed on an Agilent Bioanalyzer and samples with RNA Integrity values >6.0 were carried forward for microarray profiling. Ribonucleic acid samples of the original tumor series were profiled on the Affymetrix U133A GeneChip system as described previously.14 Ribonucleic acid samples of the validation tumor series were profiled on the Affymetrix GeneAtlas U219 array strips at our Cancer Center’s Cancer Genomics Shared Resource. Briefly, 250 ng total RNA was used to generate labeled in vitro transcription products (complementary RNA). The complementary RNA was purified, hybridized to the array, washed, and scanned according to GeneAtlas standard operating procedures. Raw signal-intensity data were normalized and log2 transformed using the Robust Multichip Averaging algorithm15 as implemented16 via the simpleaffy package from Bioconductor.17 Analysis of log-intensity distributions, pair-wise correlation, and internal Affymetrix controls was performed to evaluate and ensure data quality.
Unsupervised clustering
Hierarchical clustering of tumor gene expression profiles and data visualization by heatmap were carried out using Eisen’s Cluster (v2.11) and TreeView (v1.60) software.18 Logged signal intensity data were mean centered (on genes), and both genes and tumors were hierarchically clustered using average linkage clustering and uncentered Pearson correlation as the distance metric. Clustered data were visualized by TreeView using optimal color saturation settings.
Survival analyses
Disease-specific survival time was computed as the date of CRS/HIPEC to the last known date of clinical follow-up (censored) or death resulting from progressive disease (event). Progression-free survival time was computed as the date of CRS/HIPEC to the first subsequent clinical follow-up resulting in diagnosis of progressive disease or death due to progressive disease (event); or the last known date of clinical follow-up without evidence of disease progression (censored). The Kaplan-Meier survival analysis was used to estimate patient DSS and PFS survival rates. Log-rank p values <0.05 were considered to be statistically significant. Cox proportional hazards regression was used to assess the prognostic significance of the gene-based classification system in the presence of conventional clinical variables. These models were fit using a stepwise selection approach. SAS version 9.3 (SAS Institute) or SigmaPlot 12.0 (Systat Software Inc) software was used to conduct all survival analyses.
Gene set enrichment analysis
In the original tumor series of 24 patients, the 14 tumors comprising the poor prognosis subtype and the 10 tumors comprising the favorable prognosis subtype were subjected to differential expression analysis. For each microarray probe set, the mean log fold change (LFC) between the 2 subtypes was computed and used to rank order the probe sets with positive LFC reflecting overexpression in the favorable subtype, and negative LFC indicating overexpression in the poor prognosis subtype. In the case of multiple probe sets corresponding to the same gene symbol, the probe set with greatest absolute magnitude LFC was retained resulting in 13,211 probe sets with unique gene symbols. The rank-ordered list was then uploaded to the gene set enrichment analysis (GSEA) pre-ranked application in the GSEA software package (http://software.broadinstitute.org/gsea/index.jsp) and compared against the Molecular Signatures Database (MSigDB; containing 10,348 gene sets) as described.19 MSigDB gene sets corresponding to the Hallmark, Curated and Oncogenic Signatures collections were used with 1,000 permutations for significance testing.
RESULTS
Global gene expression patterns delineate prognostic tumor subtypes
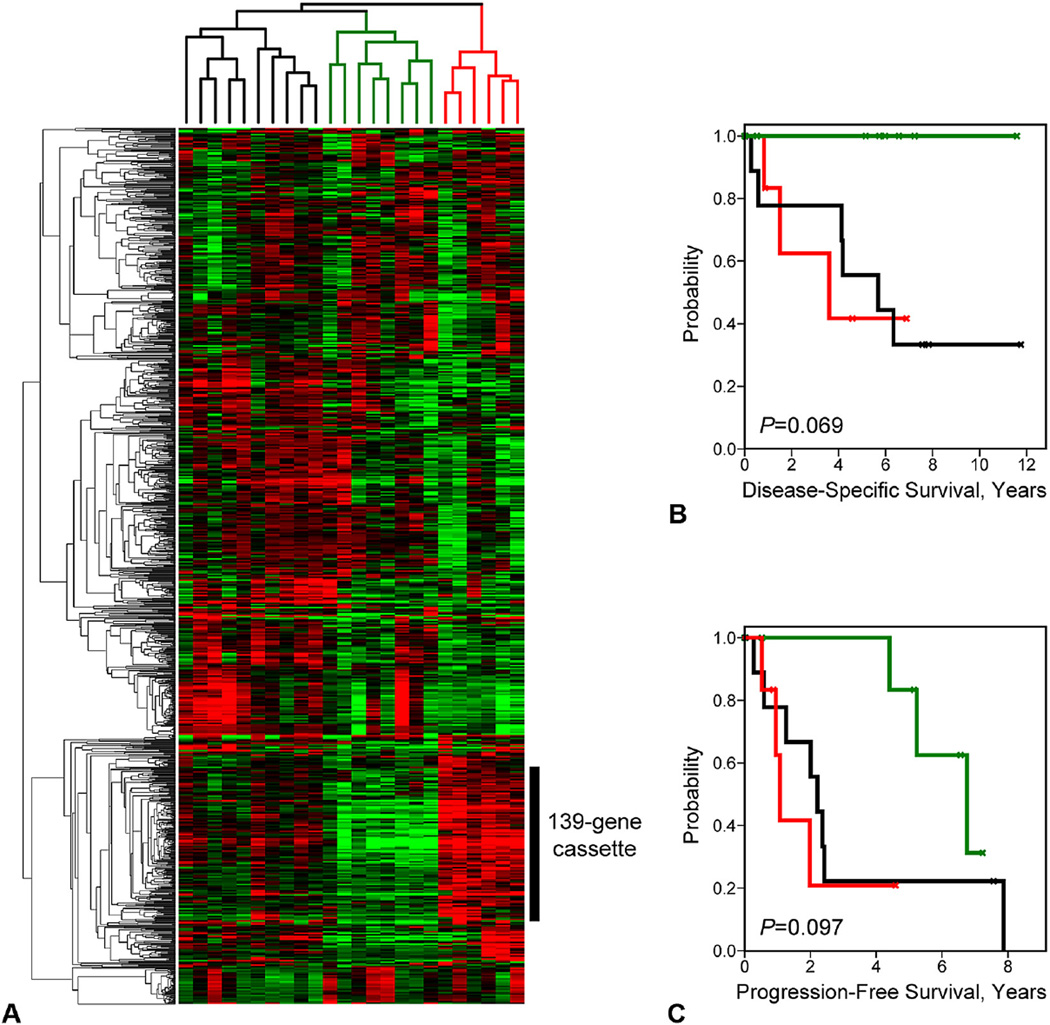
To investigate the relationship between gene expression patterns and patient prognosis, we analyzed comprehensive gene expression profiles of 24 ACs annotated for DSS and PFS. Unsupervised hierarchical clustering analysis of the tumors was performed using the set of genes that exhibited the greatest magnitude of variation in expression across the sample population, independent of patient outcomes. These genes, identified by 1,152 probe sets (Affymetrix U133A GeneChip), were defined as having log2 signal intensity distributions in the 90%-central range (from the 5th to 95th percentiles) that spanned >2.5 logs of gene expression. As indicated by the dendrogram in Figure 2A, hierarchical clustering partitioned the tumors into 3 primary branches (identified by black, green, and red labeling). We investigated the prognostic attributes of these tumor clusters by Kaplan-Meier analysis (Figs. 2B, C) and observed that the clusters stratified patients into moderately significantly different outcomes groups by both DSS (p = 0.069) and PFS (p = 0.097) analysis, despite the small number of patients represented in each cluster.
Figure 2.
Unsupervised hierarchical clustering discerns prognostic tumor subtypes. (A) Heatmap of clustered gene expression patterns (rows) and tumor expression profiles (columns) is shown. Red indicates above-mean expression; green indicates below-mean expression. Degree of color saturation reflects the magnitude of gene expression. (B, C) Kaplan-Meier survival curves corresponding by color to the tumor branches designated in (A) are shown for (B) disease-specific survival and (C) progression-free survival.
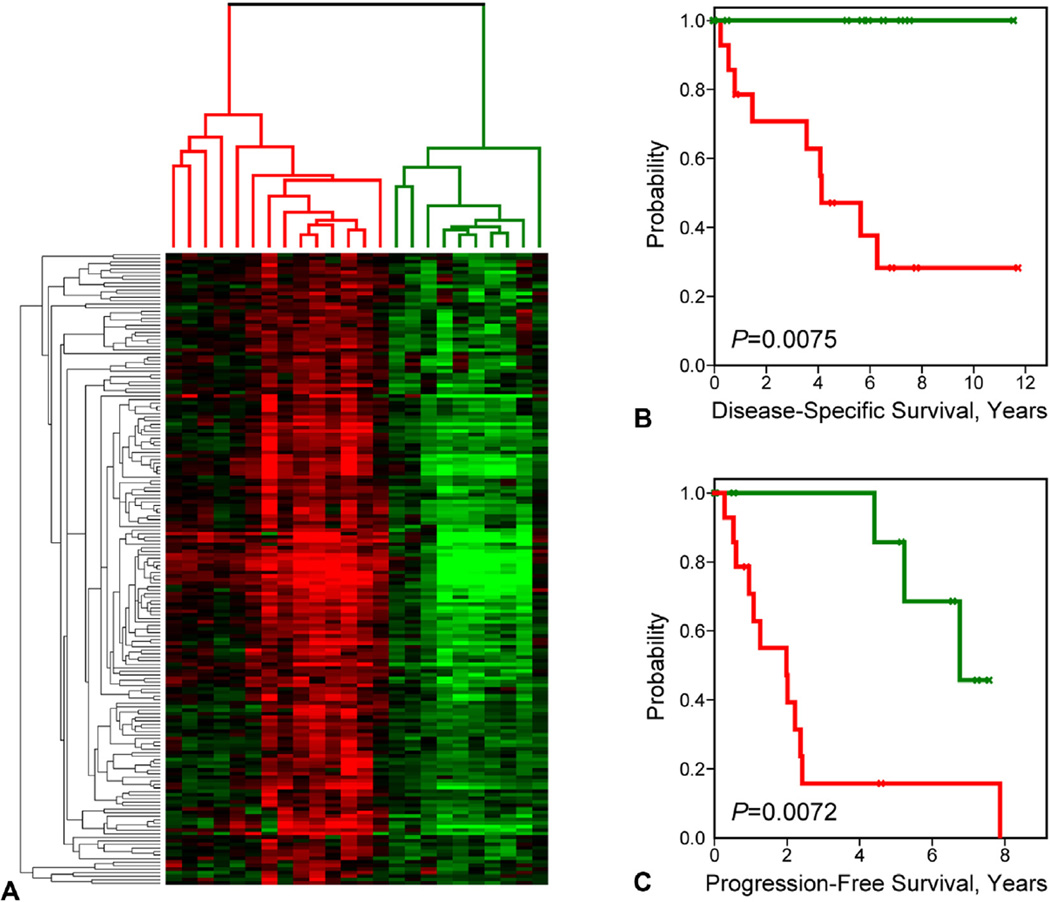
When correlating the gene clusters to the tumor branches, we observed that a highly correlated cluster of 139 genes (corresponding to 179 distinct probe sets; Fig. 2A, vertical black bar) was the dominant contributing factor in the formation of the 3 observed tumor clusters—stratifying the clusters according to intermediate, low, and high gene expression levels, respectively (see Appendix 1, online only, for gene list). Given the small size of the tumor cohort, and with an interest in determining the significance of a binary classification approach, we re-clustered the tumors based solely on the 139-gene cassette (Fig. 3A). As expected, the resulting dendrogram partitioned the AC samples into 2 primary branches, revealing molecular subtypes delineated on the basis of relatively high (red cluster) or low (green cluster) gene expression values. In subsequent Kaplan-Meier analysis, the 2 subtypes partitioned patients into highly significantly different survival groups by both DSS (p = 0.0075) and PFS (p = 0.0072) analysis, with high overall gene expression levels being associated with poor patient outcomes (Figs. 3B, C). We next addressed the possibility that in addition to the 139-gene cassette, other genes on the array might also be significantly associated with patient outcomes. We performed Cox regression analysis for DSS and PFS associations for all individual gene probe sets included on the microarray platform. After false discovery correction, no associations remained statistically significant, potentially owing to the small sample size of the cohort. However, on ranking the genes in decreasing order of statistical significance, we observed that among the top 100 most significant genes (of >22,000 probe sets) 65% belonged to our 139-gene cassette. This indicates that the cassette is highly enriched for the most significant survival-associated genes.
Figure 3.
Prognostic tumor clusters based on the 139-gene cassette. (A) Genes comprising the 139-gene cassette cluster the tumors into 2 primary branches (subtypes) based on relative high and low gene expression. (B, C) Kaplan-Meier survival curves corresponding by color to the tumor branches designated in (A) are shown for (B) disease-specific survival and (C) progression-free survival.
Prognostic subtypes are reproducible in an independent patient cohort
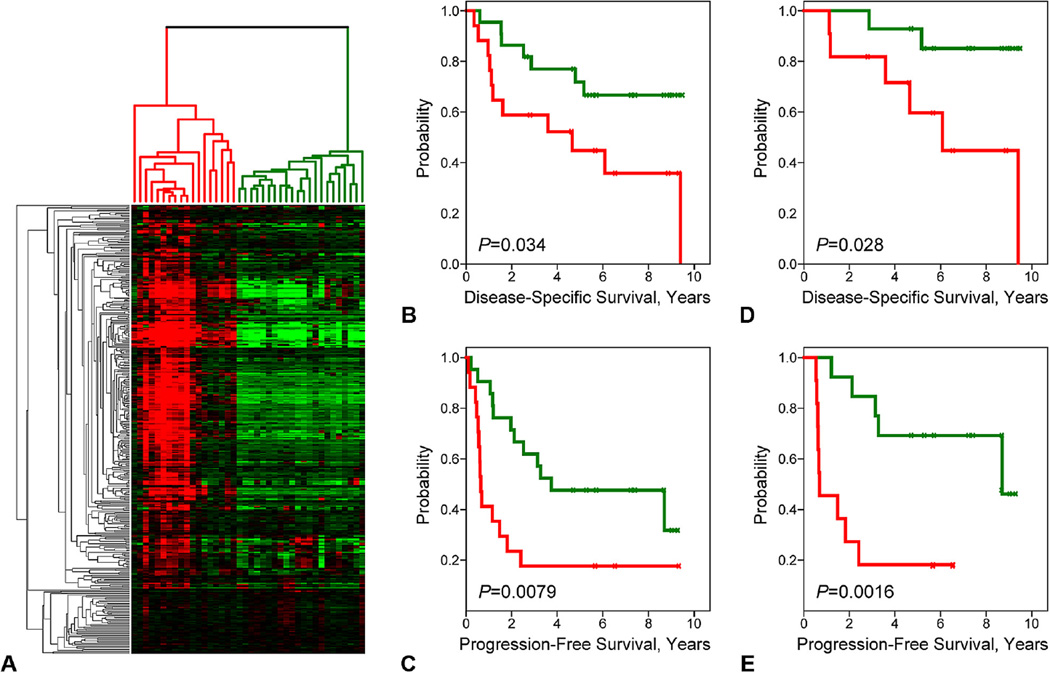
To confirm the prognostic value of the molecular subtypes discerned by the 139-gene cassette, we profiled a second series of appendiceal tumors (the validation cohort) from patients similarly treated at our Cancer Center (see Methods). In this series, 39 frozen tumor samples maintained by our Cancer Center’s Tumor Tissue Bank were profiled on Affymetrix U219 gene expression arrays. Probe sets matching the 139-gene cassette were identified (corresponding to 391 U219 probe sets representing 134 genes) and subsequently used to hierarchically cluster the new tumor samples. As displayed by the heatmap in Figure 4A, the majority of the U219 probe sets recapitulated the gene correlation structure observed in the original tumor series, resulting in the reproducible clustering of tumors into low and high gene-expressing subtypes. We next examined the prognostic significance of the molecular subtypes by Kaplan-Meier analysis (Figs. 4B, C). Again, the molecular subtypes stratified patients into significantly different outcomes groups by both DSS (p = 0.034) and PFS (p = 0.0079) with the high gene-expressing subtype associating with poor outcomes. These findings confirmed the reproducible nature of the prognostic attributes of the AC subtypes discerned by the 139-gene cassette.
Figure 4.
The prognostic subtypes are reproducible entities. (A) The 139-gene cassette was used to cluster an independent appendiceal tumor cohort. The resulting tumor subtypes were analyzed for survival associations. (B, C) Kaplan-Meier plots for (B) disease-specific survival and (C) progression-free survival are shown. (D, E) Kaplan-Meier plots for (D) disease-specific survival and (E) progression-free survival are shown for patients with low-grade disease only.
The prognostic power of the molecular subtypes is independent of conventional prognostic variables
In the original tumor series (n = 24), all but 2 of the cases were pathologically classified as LGA cancer. By contrast, 64% of cases in the second cohort were defined as LGA. Given the clinical importance of prognostication in the low-grade setting, we examined the ability of the molecular subtypes to distinguish outcomes among low-grade tumors. In LGA tumors, the molecular subtypes exhibited moderate to strong prognostic significance by both DSS (p = 0.028) and PFS (p = 0.0016) analysis (Figs. 4D, E). The molecular subtypes were not significantly associated with outcomes in the context of high-grade lesions, perhaps owing to the small sample size. Next, we assessed the prognostic value of the subtypes in relation to other known prognostic variables, including surgical score (R), Eastern Cooperative Oncology Group status, grade, and patient age, using multivariate Cox regression models with stepwise selection. By analysis for association with DSS (Table 1), the subtypes remained statistically significant (p = 0.007) in the presence of both grade (p < 0.001) and Eastern Cooperative Oncology Group status (p = 0.007). When assessed for association with PFS (Table 2), the subtypes again remained significant in the model (p = 0.001), but together with grade (p = 0.002) and surgical score (R; p = 0.03). These findings indicate that the molecular subtypes contribute additive prognostic information not conveyed by conventional prognostic variables.
Table 1.
Cox Proportional Hazards Regression Analyses for Associations with Disease-Specific Survival
| Covariate* | Univariable hazard ratio (95% CI) |
p Value† | Multivariable‡ hazard ratio (95% CI) |
p Value |
|---|---|---|---|---|
| Subtypes | 3.64 (1.53–8.70) | 0.004 | 3.53 (1.41–8.85) | 0.007 |
| Grade | 6.24 (2.56–15.23) | <0.001 | 6.84 (2.57–18.17) | <0.001 |
| R | 2.51 (1.54–4.08) | <0.001 | — | — |
| ECOG | 2.91 (1.71–4.95) | <0.001 | 2.07 (1.22–3.52) | 0.007 |
| Age | 0.99 (0.96–1.03) | 0.63 | — | — |
Subtypes and grade were entered as binary variables; R (Surgical Score) and ECOG status were entered as semi-continuous variables (R = 1, 2, 3; ECOG = 0, 1, 2, 3); age was entered as a continuous variable.
Wald p value.
Stepwise selection procedure with entry significance level 0.05.
ECOG, Eastern Cooperative Oncology Group.
Table 2.
Cox Proportional Hazards Regression Analyses for Associations with Progression-Free Survival
| Covariate* | Univariable hazard ratio (95% CI) |
p Value† | Multivariable‡ hazard ratio (95% CI) |
p Value |
|---|---|---|---|---|
| Subtypes | 3.16 (1.64–6.10) | <0.001 | 3.27 (1.58–6.75) | 0.001 |
| Grade | 3.52 (1.71–7.23) | <0.001 | 3.32 (1.57–7.01) | 0.002 |
| R | 2.03 (1.38–2.98) | <0.001 | 1.56 (1.04–2.35) | 0.03 |
| ECOG | 1.77 (1.13–2.80) | 0.01 | — | — |
| Age | 1.00 (0.97–1.02) | 0.8 | — | — |
Subtypes and grade were entered as binary variables; R (Surgical Score) and ECOG status were entered as semi-continuous variables (R = 1, 2, 3; ECOG = 0, 1, 2, 3); age was entered as a continuous variable.
Wald p value.
Stepwise selection procedure with entry significance level 0.05.
ECOG, Eastern Cooperative Oncology Group.
Inferred pathologic states differentiate the prognostic subtypes
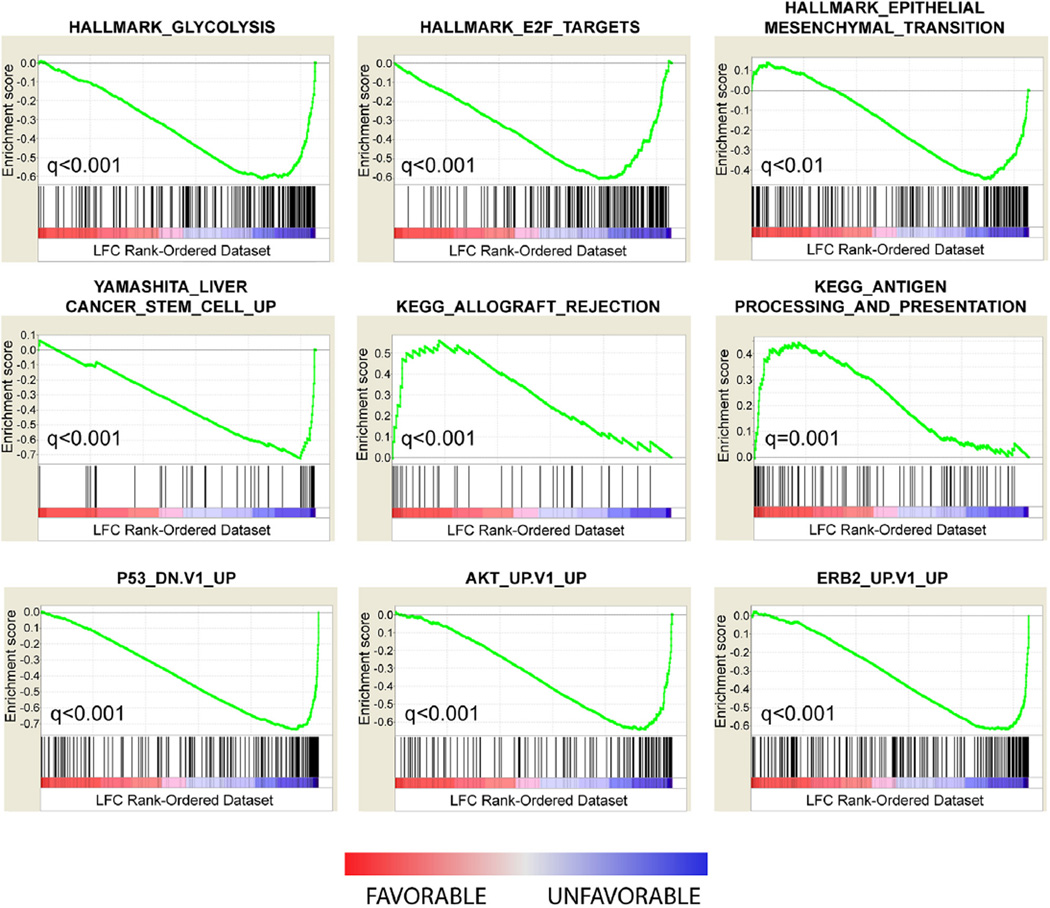
To gain insight into the pathologic properties that distinguish the good and poor outcomes subtypes, we examined their opposing gene expression patterns by GSEA.19 Gene set enrichment analysis is a statistical method that seeks to identify gene sets or signatures defined by biologic states, molecular processes, or signaling networks that are enriched in one condition (ie, subtype) relative to another. To perform the analysis, appendiceal tumors were assigned to molecular subtypes according to Figure 3A, and the genes represented on the microarray were ranked in descending order according to their mean LFC (difference) between the 2 subtypes. In this way, genes overexpressed in the favorable subtype were differentiated from those overexpressed in the poor prognosis subtype according to the magnitude of gene differential expression. A number of highly significant associations were observed with the top most significant results shown in Figure 5. Biologic hallmark gene sets representing glycolysis, epithelial to mesenchymal transition, and E2F target genes were found to be highly significantly enriched in the poor prognosis subtype. Among the curated gene sets, Kyoto Encyclopedia of Genes and Genomes. pathway signatures20 reflecting allograft rejection and antigen processing and presentation were significantly over-represented in the favorable prognostic subtype. Finally, oncogenic signatures reflective of genes overexpressed in the context of p53 mutation, AKT activation, HER2 (erb-B2) overexpression, and cancer stem cells isolated from hepatocellular carcinomas were also found to be significantly enriched in the poor prognosis subtype. Together, these findings suggest that a number of known oncogenic process and pathways can contribute to the aggressive behavior of the poor prognosis subtype, and biology reminiscent of anti-tumor immunity might underlie the favorable subtype.
Figure 5.
Biologic processes and pathways delineate the prognostic subtypes. Gene set enrichment analysis was performed on the genes differentially expression between subtypes. Gene set enrichment analysis gene set titles are listed above each panel. Enrichment scores (green line) and their corresponding false discovery rate-adjusted significance (q-values) are shown. Black vertical bars indicate the position of genes belonging to the gene set along the rank-ordered dataset. Red indicates overexpression (greater mean log fold change [LFC]) in the favorable subtype; blue indicates overexpression in the poor prognosis subtype. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Finally, we cross-referenced the genes of the 139-gene cassette with the Drug Gene Interaction Database.21 This analysis revealed a number of cancer-associated genes, the products of which are targets of existing or emerging anti-neoplastic drugs, including erb-B3 (HER-3), c-MET, FGFR3, CDH1, GPRC5A, DDR1, CA2, CA9, CEACAM5, MST1R, MUC1, and SLC2A1.
DISCUSSION
Appendiceal cancer is an uncommon tumor that all too frequently presents with peritoneal dissemination. There is considerable interest in aggressive CRS and HIPEC for the treatment of peritoneal dissemination from AC (as well as from a variety of other primary tumors). The utility of CRS and HIPEC has been most strongly touted for LGA, as the majority of patients are 10-year survivors if cytoreduction is complete. However, approximately 15% to 30% of patients with low-grade disease and complete cytoreduction, the “best of the best” cases will recur, with most of these dying of peritoneal disease.11,12,22,23 This observation led us to ask whether or not a genetic signature could account for the failures of such an aggressive treatment. In this work, we report the novel discovery of a reproducible prognostic gene signature in AC that discerns differentially aggressive molecular subtypes of low-grade disease. Compared with conventional prognostic markers, such as surgical score, Eastern Cooperative Oncology Group status, and patient age,11,12 the molecular subtypes identified by the signature provided additive prognostic value. This suggests a potential diagnostic role for the molecular subtypes in predicting patient outcomes.
Long-term survival in the context of LGA must be considered in the 10-year range and beyond for most patients. However, there is a minority who will fare poorly despite optimal cytoreduction and intraperitoneal chemotherapy. Therefore, an improved ability to predict better survival outcomes will provide valuable reassurance for patients. However, such a gene-based stratification begs the question of the potential of adjuvant therapy. Systemic chemotherapy is of very limited efficacy for LGA patients.24 The low failure rates associated with the favorable subtype make any adjuvant therapy highly unlikely to be efficacious. By contrast, with the unfavorable subtype and its higher failure rate, consideration must be given to an adjuvant, likely systemic, therapy. Such an adjuvant might be best determined by analysis of genetic targets in lesions via a “targeted” approach. We observed that a number of genes overexpressed in the poor prognosis subtype are known targets of conventional and emerging cancer therapies that could conceivably benefit AC patients. For example, the MET proto-oncogene is known to be amplified and overexpressed in a number of cancer types, including lung, liver, kidney, and gastric cancers.25 MET is a master activator of oncogenic signaling pathways (PI3K/AKT, MAPK/ERK, WNT/beta-catenin, NOTCH) and drives many oncogenic processes, including cellular transformation, invasion, angiogenesis, and metastasis.26 Blocking antibodies and small molecule inhibitors that target MET activation are showing clinical utility. In clinical trials of both non-small cell lung cancer and gastric adenocarcinoma, patients whose tumors overexpressed MET showed significantly better overall survival with novel inhibitors of MET activation as compared with those whose tumors did not overexpress MET.27,28 The genes comprising the prognostic gene cassette might themselves represent new therapeutic targets for AC patients diagnosed with the aggressive molecular subtype.
Overexpression of the 139-gene cassette is the defining characteristic of the poor prognosis subtype. Significantly, among these genes are prominent markers of cancer stem cells, including CD24, PROM1 (or CD133), and EPCAM, all of which are associated with cancer “stemness” in gastrointestinal tumors.29,30 This suggests that the unfavorable AC subtype might be driven, in part, by a stem cell-like phenotype. Indeed, this hypothesis is supported by the GSEA finding (Fig. 5) indicating that a gene signature of EPCAM-positive stem cells in hepatocellular carcinoma31 is significantly enriched in the poor prognosis subtype. Similarly, the GSEA finding of an epithelial to mesenchymal gene signature, enriched in the poor prognosis subtype, might reflect the observation that molecular programs underlying epithelial to mesenchymal transition are characteristic of both normal and neoplastic stem cell populations.32,33
We recognize that the data represent a modest number of patients. However, the rarity of the lesion makes gathering large numbers of cases treated in the same way difficult to obtain. In addition, although prognostic, the 139-gene signature might be better defined by a smaller number of genes. A smaller number of genes in the signature would certainly simplify future studies and will require additional analysis of this and other datasets.
CONCLUSIONS
We have identified a prognostic gene signature for low-grade AC metastatic to the peritoneum. Such genetic signatures suggest that the subset of more aggressive low-grade AC has significantly different clinical outcomes even after such aggressive therapy as CRS and HIPEC. Although the value of pathologic analysis defining this subset remains important, this signature improves considerably our ability to prognosticate beyond light microscopy. Genetic testing is already known to be of substantial value, supplementing microscopic evaluation for many common malignancies. Although adding genetic analysis to the already available descriptors of tumor will complicate staging, it will also improve our ability to prognosticate and define therapy tailored to the patient’s tumor.
Supplementary Material
Acknowledgments
Support: The authors acknowledge the support of the Orin Smith Family Foundation, the National Organization of Rare Diseases and the core laboratories of the Wake Forest Baptist Comprehensive Cancer Center: Genomics, Biostatistics and Bioinformatics, and the Tumor/Tissue and Pathology shared resources supported by NCI CCSG P30CA012197.
Abbreviations and Acronyms
- AC
appendiceal cancer
- CRS
cytoreductive surgery
- DSS
disease-specific survival
- GSEA
Gene Set Enrichment Analysis
- HIPEC
hyperthermic intraperitoneal chemotherapy
- LFC
log fold change
- LGA
low-grade appendiceal
- PFS
progression-free survival
- PSD
peritoneal surface disease
- RNA
ribonucleic acid
Footnotes
Disclosure Information: Nothing to disclose.
Presented at the Southern Surgical Association 127th Annual Meeting, Hot Springs, VA, December 2015.
Author Contributions
Study conception and design: Levine, Miller
Acquisition of data: Levine, Votanopoulos, Qasem, Philip, Cummins, Ruiz, Shen, Miller
Analysis and interpretation of data: Levine, Votanopoulos, Qasem, Philip, Cummins, Chou, D’Agostino, Shen, Miller
Drafting of manuscript: Levine, Miller
Critical revision: Levine, Votanopoulos, Qasem, Philip, Cummins, Chou, Ruiz, D’Agostino, Shen, Miller
REFERENCES
- 1.Collins DC. 71,000 Human appendix specimens. A final report, summarizing forty years’ study. Am J Proctol. 1963;14:265–281. [PubMed] [Google Scholar]
- 2.Connor SJ, Hanna GB, Frizelle FA. Appendiceal tumors: retrospective clinicopathologic analysis of appendiceal tumors from 7,970 appendectomies. Dis Colon Rectum. 1998;41:75–80. doi: 10.1007/BF02236899. [DOI] [PubMed] [Google Scholar]
- 3.McCusker ME, Cote TR, Clegg LX, Sobin LH. Primary malignant neoplasms of the appendix: a population-based study from the surveillance, epidemiology and end-results program, 1973–1998. Cancer. 2002;94:3307–3312. doi: 10.1002/cncr.10589. [DOI] [PubMed] [Google Scholar]
- 4.Marmor S, Portschy PR, Tuttle TM, Virnig BA. The rise in appendiceal cancer incidence: 2000–2009. J Gastrointest Surg. 2015;19:743–750. doi: 10.1007/s11605-014-2726-7. [DOI] [PubMed] [Google Scholar]
- 5.Trivedi AN, Levine EA, Mishra G. Adenocarcinoma of the appendix is rarely detected by colonoscopy. J Gastrointest Surg. 2009;13:668–675. doi: 10.1007/s11605-008-0774-6. [DOI] [PubMed] [Google Scholar]
- 6.Youssef H, Newman C, Chandrakumaran K, et al. Operative findings, early complications, and long-term survival in 456 patients with pseudomyxoma peritonei syndrome of appendiceal origin. Dis Colon Rectum. 2011;54:293–299. doi: 10.1007/DCR.0b013e318202f026. [DOI] [PubMed] [Google Scholar]
- 7.Baratti D, Kusamura S, Nonaka D, et al. Pseudomyxoma peritonei:clinical pathological and biological prognostic factors in patients treated with cytoreductive surgery and hyperthermic intraperitoneal chemotherapy (HIPEC) Ann Surg Oncol. 2008;15:526–534. doi: 10.1245/s10434-007-9691-2. [DOI] [PubMed] [Google Scholar]
- 8.Bradley RF, Stewart JH, Russell GB, et al. Pseudomyxoma peritonei of appendiceal origin: a clinicopathologic analysis of 101 patients uniformly treated at a single institution, with literature review. Am J Surg Pathol. 2006;30:551–559. doi: 10.1097/01.pas.0000202039.74837.7d. [DOI] [PubMed] [Google Scholar]
- 9.Chua TC, Moran BJ, Sugarbaker PH, et al. Early- and long-term outcome data of patients with pseudomyxoma peritonei from appendiceal origin treated by a strategy of cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. J Clin Oncol. 2012;30:2449–2456. doi: 10.1200/JCO.2011.39.7166. [DOI] [PubMed] [Google Scholar]
- 10.Elias D, Gilly F, Quenet F, et al. Pseudomyxoma peritonei: a French multicentric study of 301 patients treated with cytoreductive surgery and intraperitoneal chemotherapy. Eur J Surg Oncol. 2010;36:456–462. doi: 10.1016/j.ejso.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Levine EA, Stewart JH, Shen P, et al. Intraperitoneal chemotherapy for peritoneal surface malignancy: experience with 1,000 patients. J Am Coll Surg. 2014;218:573–585. doi: 10.1016/j.jamcollsurg.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Votanopoulos KI, Russell G, Randle RW, et al. Peritoneal surface disease (PSD) from appendiceal cancer treated with cytoreductive surgery (CRS) and hyperthermic intraperitoneal chemotherapy (HIPEC): overview of 481 cases. Ann Surg Oncol. 2015;22:1274–1279. doi: 10.1245/s10434-014-4147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronnett BM, Zahn CM, Kurman RJ, et al. Disseminated peritoneal adenomucinosis and peritoneal mucinous carcinomatosis. A clinicopathologic analysis of 109 cases with emphasison distinguishing pathologic features, site of origin, prognosis, and relationship to “pseudomyxoma peritonei”. Am J Surg Pathol. 1995;19:1390–1408. doi: 10.1097/00000478-199512000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Levine EA, Blazer DG, 3rd, Kim MK, et al. Gene expression profiling of peritoneal metastases from appendiceal and colon cancer demonstrates unique biologic signatures and predicts patient outcomes. J Am Coll Surg. 2012;214:599–606. doi: 10.1016/j.jamcollsurg.2011.12.028. discussion 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R Development Core Team. R. A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 17.Ihaka R, Gentleman R. A language for data analysis and graphics. J Computat Graph Stat. 1996;5:299–314. [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffith M, Griffith OL, Coffman AC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10:1209–1210. doi: 10.1038/nmeth.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine EA. Appendiceal carcinoma with peritoneal dissemination: outcomes for the best of the best. Ann Surg Oncol. 2007;14:2440–2442. doi: 10.1245/s10434-007-9534-1. [DOI] [PubMed] [Google Scholar]
- 23.Yan TD, Bijelic L, Sugarbaker PH. Critical analysis of treatment failure after complete cytoreductive surgery and perioperative intraperitoneal chemotherapy for peritoneal dissemination from appendiceal mucinous neoplasms. Ann Surg Oncol. 2007;14:2289–2299. doi: 10.1245/s10434-007-9462-0. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro JF, Chase JL, Wolff RA, et al. Modern systemic chemotherapy in surgically unresectable neoplasms of appendiceal origin: a single-institution experience. Cancer. 2010;116:316–322. doi: 10.1002/cncr.24715. [DOI] [PubMed] [Google Scholar]
- 25.Kawakami H, Okamoto I, Okamoto W, et al. Targeting MET amplification as a new oncogenic driver. Cancers (Basel) 2014;6:1540–1552. doi: 10.3390/cancers6031540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 27.Iveson T, Donehower RC, Davidenko I, et al. Rilotumumabin combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: an open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. Lancet Oncol. 2014;15:1007–1018. doi: 10.1016/S1470-2045(14)70023-3. [DOI] [PubMed] [Google Scholar]
- 28.Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2013;31:4105–4114. doi: 10.1200/JCO.2012.47.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherciu I, Barbalan A, Pirici D, et al. Stem cells, colorectal cancer and cancer stem cell markers correlations. Curr Health Sci J. 2014;40:153–161. doi: 10.12865/CHSJ.40.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rassouli FB, Matin MM, Saeinasab M. Cancer stem cells in human digestive tract malignancies. Tumour Biol. 2015 doi: 10.1007/s13277-015-4155-y. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.Yamashita T, Ji J, Budhu A, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morel AP, Lievre M, Thomas C, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.