Summary
Binding of the transcription factor Ets-1 to DNA is allosterically regulated by a serine rich region (SRR) that modulates the dynamic character of the adjacent structured DNA-binding ETS domain and its flanking autoinhibitory elements. Multi-site phosphorylation of the flexible SRR in response to Ca+2 signaling mediates variable regulation of Ets-1 DNA-binding affinity. In this study, we further investigated the mechanism of this regulation. First, thermal and urea denaturation experiments demonstrated that phosphorylation of the predominantly unstructured SRR imparts enhanced thermodynamic stability on the well-folded ETS domain and its inhibitory module. We next identified a minimal fragment (residues 279–440) that exhibits both enhanced autoinhibition of Ets-1 DNA-binding and allosteric reinforcement by phosphorylation. To test for intramolecular interactions between the SRR and the rest of the fragment that were not detectable by 1H-1H NOE measurements, paramagnetic relaxation enhancements were performed using a Cu2+ bound to the N-terminal ATCUN motif. Increased relaxation detected for specific amides and methyls revealed a preferential interaction surface for the flexible SRR extending from the inhibitory module to the DNA binding interface. Phosphorylation enhanced the localization of the SRR to this surface. We therefore hypothesize that the positioning of the SRR at the DNA binding interface and its role in shifting Ets-1 to an inhibited conformation are linked. In particular, transient interactions dampen the conformational flexibility of the ETS domain and inhibitory module required for high affinity binding, as well as possibly occlude the DNA interaction site. Surprisingly, the phosphorylation-dependent effects were relatively insensitive to changes in ionic strength, suggesting that electrostatic forces are not the dominant mechanism for mediating these interactions. The results of this study highlight the role of flexibility and transient binding in the variable regulation of Ets-1 activity.
Keywords: Transcription, allosteric regulation, protein dynamics, NMR spectroscopy, paramagnetic relaxation, Ets-1
Introduction
Regions of intrinsic disorder are being identified with increasing frequency as targets of protein regulation1, 2, 3. Two general features of these regions have been described: first, they adopt an ordered conformation in order to perform their function; and second, they are often sites of post-translational modifications, which either induce or disrupt the active conformation. For example, the tail of stathmin, which is disordered in its apo form, yet adopts a stable β-hairpin when bound to tubulin, leading to the disruption of microtubule formation. Phosphorylation of this region abolishes the hairpin, thus disrupting binding and allowing microtubule formation to proceed4. Conversely, phosphorylation of the intrinsically disordered KID domain of the DNA-binding transcription factor CREB enhances recognition by the KIX domain of co-activator p300. The phosphorylated KID domain undergoes a disorder-to-order transition upon interaction with the KIX domain, forming two α-helices that provide a stable interface for association with p300 and allowing activation of transcription5. In both cases, the change in the physicochemical properties of the modified protein conferred by addition of phosphates plays a role in either inducing or disrupting the active structure. In contrast, our recent studies of the transcription factor Ets-1 show that a predominantly unstructured, multiply phosphorylated region acts as an intramolecular allosteric regulator of the well-folded DNA-binding domain6. The interplay between the unstructured and structured regions of Ets-1 is the focus of this investigation.
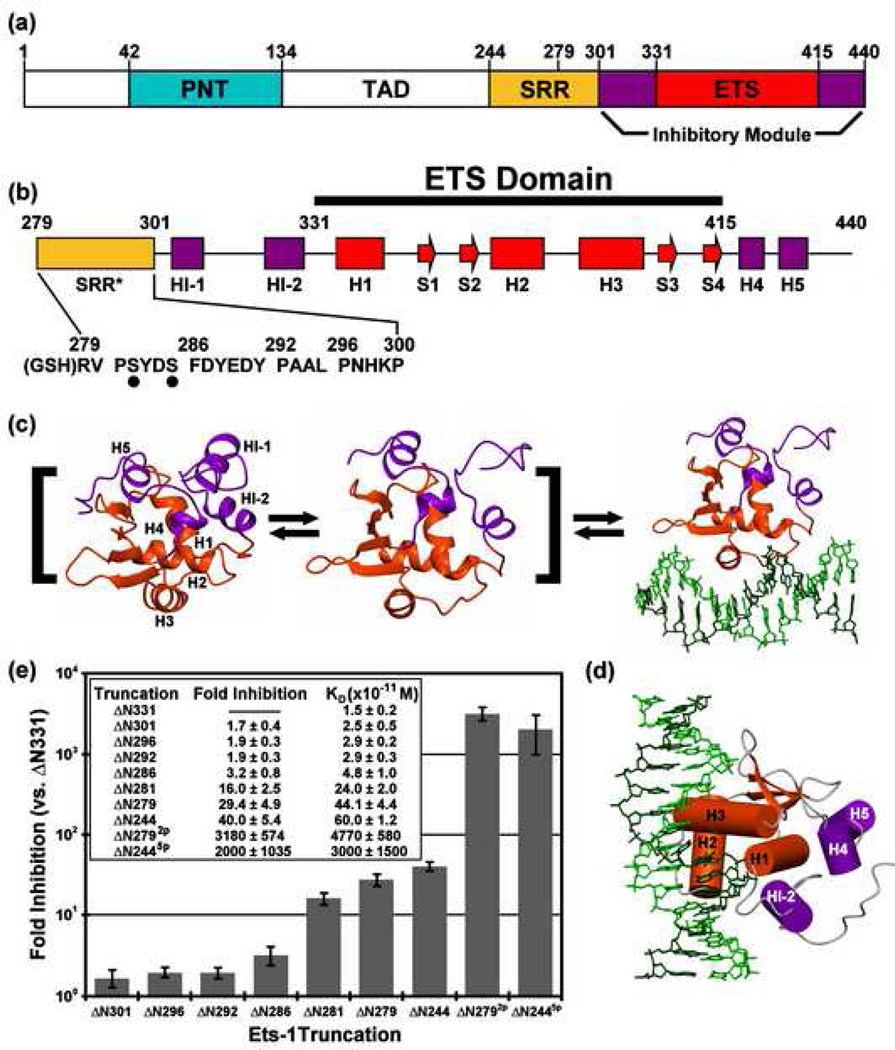
The binding of Ets-1 to DNA is mediated by the ETS domain, which contains three α-helices and a four-stranded β-sheet, and is modulated by flanking regions that fold into an inhibitory module composed of four α-helices (Fig. 1)7, 8. The inhibitory module packs along a surface of the ETS domain at helix H1, opposite the helix (H2)-turn-helix(H3) DNA recognition interface. DNA binding is accompanied by a structural rearrangement that disrupts the inhibitory module, as most dramatically highlighted by the unfolding of the marginally stable N-terminal inhibitory helix HI-18, 9, 10. Proper packing of the inhibitory module against the ETS domain is required for autoinhibition, as demonstrated by the higher affinity of variants containing mutations that disrupt the hydrophobic core of the module11. The presence of an adjacent disordered serine rich region (residues 244–300, termed the SRR) accentuates repression of the ETS domain and inhibitory modules (residues 301–440, also termed ΔN301 or the "regulatable unit") and recapitulates binding of the full-length protein6. Multi-site phosphorylation of the SRR in response to Ca+2 signaling further attenuates DNA binding12, with each added phosphate causing progressively increased repression6. Protein partnerships, both heterotypic and homotypic, can also counteract autoinhibition, thereby enhancing DNA binding13, 14, 15. Thus, Ets-1 displays a 1000-fold range in DNA binding affinity, from a fully activated state by deletion or biological partners to a fully phosphorylated, repressed state.
Fig. 1.
(a) The structural organization of Ets-1, showing the protein interaction PNT domain (cyan), transactivation domain (TAD, white), and serine rich region (SRR, yellow), along with the regulatable unit, composed of the DNA binding ETS domain (red), and inhibitory module (purple). (b) The secondary structural elements of ΔN279 (α-helix, H; β-strand, S) and sequence of residues 279–300 of the truncated SRR*, including sites of Ca2+-dependent kinase phosphorylation (S282 and S285; dots) and a Gly-Ser-His motif resulting from thrombin cleavage of an N-terminal His6-tag. (c) Allosteric model of autoinhibition in which free Ets-1 exists in equilibrium between rigid-inactive and flexible-active states, the latter characterized most conspicuously by the unfolding of HI-1. Structures shown are for isolated ΔN301 (left; 1R36.pdb), DNA-bound ΔN280 (right; 1MDM.pdb), and a proposed DNA-free state with HI-1 unfolded (middle). (d) A view of DNA-bound ΔN280, oriented as in cylinder model of Figure 7d. (e) A summary of the electrophoretic mobility shift assays of DNA binding by Ets-1 truncation constructs, showing equilibrium dissociation constants, KD, and the fold inhibition relative to ΔN331, the completely de-repressed fragment of Ets-1. The shorter fragments (ΔN301, ΔN296, ΔN292, and ΔN286) have modest effects, whereas the longer constructs have a higher level of inhibition. Phosphorylation of S282 and S285 (ΔN2792p) significantly represses DNA binding relative to unmodified state ΔN279. This recapitulates the behavior of ΔN2446. ΔN281 results from deletion of V280 from ΔN280 during E. coli expression, and ΔN279 and ΔN2792p have an N-terminal Gly-Ser-His motif.
The mechanism of autoinhibition and its reinforcement by phosphorylation was partially elucidated by the discovery of the dynamic nature of the ETS domain and its inhibitory elements, even in the absence of DNA6, 8. The degree of this internal motion correlates with the DNA binding affinity of various deletion fragments and phosphorylated forms of Ets-1. Specifically, the presence of the SRR and its phosphorylation reduces the msec-µsec timescale motions of the inhibitory module and ETS domain, as well as imparting its overall stabilization as evidenced by increased protection from amide hydrogen exchange (HX)6. Along with chemical shift perturbation mapping, these NMR-based observations indicated that Ets-1 exists in a conformational equilibrium between at least two states. The active state, which can bind DNA, exhibits conformational mobility within HI-1 and HI-2 of the inhibitory module and, importantly, the DNA recognition helix H36. This led to the model that the dynamic nature of Ets-1 reflects the sampling of conformations necessary to bind to DNA recognition sites with high affinity, and that a reduction in these dynamics shifts the equilibrium to a more rigid, low affinity state.
The shift between dynamic-active and rigid-inactive states is reminiscent of the classical allosteric transition between the relaxed and tense forms of a protein16. However, distinct from this model, the phosphorylation-dependent exchange between the two states of Ets-1 is not all-or-none. Progressive mutation of phosphoacceptor sites within the SRR yielded species with progressively decreasing inhibition, indicating that the affinity of Ets-1 for DNA can be fine-tuned in vitro6 and in vivo17 by multi-site phosphorylation. Importantly, each added phosphate also shifts the equilibrium toward the inhibited conformation, further bolstering the idea that decreasing the active state population is the mode for reducing DNA affinity. Surprisingly, the phosphorylated SRR directs this conformational transition despite being predominantly unstructured and flexible on the sub-nsec timescale6.
To further our understanding of the mechanism of Ets-1 autoinhibition and to determine how structured and unstructured protein segments interact, we investigated several unresolved features of the allosteric model. Does the SRR directly interact with the ETS domain and inhibitory helices to mediate its stabilizing effect? Does this change upon phosphorylation? What is the physicochemical nature of this interaction between an unstructured, flexible region and a dynamic structured region? In particular, are electrostatic forces involved due to the addition of negative charge upon phosphorylation? To address these questions, we demonstrate that SRR phosphorylation increases the global thermodynamic stability of an Ets-1 fragment. Using NMR spectroscopy, we also show that the SRR is predominantly unstructured both before and after phosphorylation. However, addition of phosphates to the SRR subtly dampens its fast timescale motions detected by 15N relaxation, as well as restricting the conformational dynamics of the regulatable unit as evidenced by reduced amide HX. Importantly, paramagnetic relaxation enhancement (PRE) measurements revealed that the flexible SRR localizes to a surface that extends from the inhibitory module to the DNA binding interface, and that this localization is more persistent upon phosphorylation. These observations suggest that transient intramolecular interactions can affect the dynamic nature of the ETS domain and its inhibitory elements, thus supporting an allosteric mechanism of autoinhibition. The location of the SRR near the DNA binding surface might also augment autoinhibition through a steric mechanism.
Results
SRR phosphorylation thermodynamically stabilizes the regulatable unit
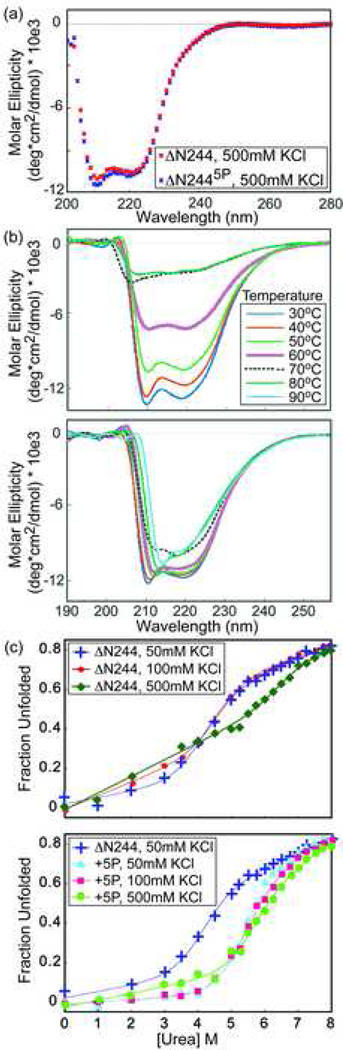
We focused our investigation on the interaction between the flexible, unstructured SRR (residues 244–300) and the structured, but dynamic, regulatable unit (residues 301–440). The motions of this unit, composed of the DNA-binding ETS domain and its flanking inhibitory module, are dampened in the presence of the SRR and more so in response to multi-site phosphorylation6. To test whether this effect is detectable as a change in global thermodynamic stability, CD spectroscopy was used to monitor thermal- and urea-induced denaturation of ΔN244 and ΔN2445p (the ΔN244 variant phosphorylated at S251, S270, S273, S282, and S285). The secondary structure of ΔN244 and ΔN2445p remained essentially identical as indicated by their equivalent molar ellipticities (Fig. 2a). However phosphorylation dramatically shifted the midpoint temperature of denaturation (Tm) from ~ 55 °C to over 90 °C (Fig. 2b). Similarly, phosphorylation increased the midpoint urea denaturation concentration ([urea]1/2) from ~ 4.8 M for ΔN244 to ~ 5.6 M for ΔN2445p (Fig. 2c). Thus, without any change in helical content, SRR phosphorylation substantially stabilizes the regulatable unit. Unfortunately, calculation of the free energy of this stabilization was unreliable due to poorly defined baselines for the urea unfolded proteins.
Fig. 2.
Phosphorylation stabilizes Ets-1. (a) The CD spectra of ΔN244 and ΔN2445p are essentially superimposable, indicating that phosphorylation does not alter the secondary structure of the Ets-1 fragment. (b) Signal averaged CD spectra of ΔN244 and ΔN2445p recorded with increasing temperature in 10°C intervals. ΔN244 had a midpoint unfolding temperature of ~ 55 °C, whereas ΔN2445p remained well folded at 90 °C. (c) Urea denaturation of ΔN244 versus ΔN2445p in the presence of 50 mM, 100 mM, and 500 mM KCl. Lines show the data fit (see Methods). Phosphorylation elevated the [urea]1/2 from ~4.8 M to ~5.6 M in 50 mM KCl. Increasing ionic strength reduced the co-operativity of ΔN244 unfolding, but did not markedly change the stability of ΔN2445p. Samples were in 25 mM potassium phosphate, pH 7.9, and 100 mM KCl at 25 °C unless stated otherwise.
ΔN279 is a minimally sized fragment that exhibits phosphorylation-enhanced autoinhibition
We previously used fragments ΔN301, ΔN280, and ΔN2445p to investigate autoinhibition and the effect of phosphorylation on the regulatable unit of Ets-1 by NMR spectroscopic analyses6, 8, 18. This experimental design was necessary due to limited solubility of the non-phosphorylated ΔN244 (~100 µM) under NMR sample conditions. We sought to define a smaller fragment of Ets-1 that recapitulated both the autoinhibition and phosphorylation effects and was more amenable to NMR spectroscopic analysis in the unmodified state. ΔN280, previously characterized by NMR methods, displays autoinhibition levels comparable to that of native, unmodified full length Ets-19, 18, 19. Importantly, two phosphoacceptor sites, S282 and S285, which act additively with the more N-terminal sites in ΔN244, are retained in ΔN280. In addition, although all residues of the ΔN2445p SRR are conformationally dynamic6, a Lipari-Szabo model-free analysis20 of their amide 15N T1, T2, and heteronuclear 1H{15N}-NOE parameters showed non-uniform mobility. Specifically, residues 280–300 have restricted fast timescale motions relative to the remaining N-terminal portion of the SRR (Supplemental Figs. S1 and S2). Accordingly, we chose to focus on the former region, and developed an Ets-1 species spanning residues 279–440. This fragment is preceded by a short tripeptide (Gly-Ser-His) resulting from the proteolytic cleavage of a His6-tag. For simplicity, we denote this as ΔN279 (Fig. 1b).
To ensure that ΔN279 with the minimally-sized SRR* (residues 279–300) retained both the autoinhibition and phosphorylation effects, we measured the DNA binding activity of a series of N-terminal truncations by quantitative EMSA and compared these data to the affinity of unrepressed ΔN331 to assess the level of autoinhibition (Fig. 1e). All deletions exhibited reduced autoinhibition, indicating that residues 279–300 are necessary for full repression. Importantly, mass spectrometry and gel electrophoresis confirmed the efficient phosphorylation of ΔN279 at S282 and S285 (data not shown). The resulting ΔN2792p exhibited a KD of ~5 × 10−8 M, representing a ~100 fold reduction relative to unmodified ΔN279. Thus, ΔN279 recapitulated both autoinhibition and phosphorylation effects. The signals from backbone amide and sidechain nuclei of ΔN279 and ΔN2792p were assigned using a suite of heteronuclear NMR experiments (Supplemental Figs. S3 and S4)8. We confirmed by chemical shift analyses that the secondary structure elements of the inhibitory module and ETS domain were the same as observed in previous X-ray crystallographic and NMR spectroscopic studies of various Ets-1 fragments8, 10 (Fig. 3 and Supplemental Fig. S2). Therefore, we were poised to study the effects of phosphorylation of the SRR using a single well-characterized species, ΔN279. To distinguish the full length SRR of ΔN244 from the truncated one in ΔN279, we designate the latter as SRR*.
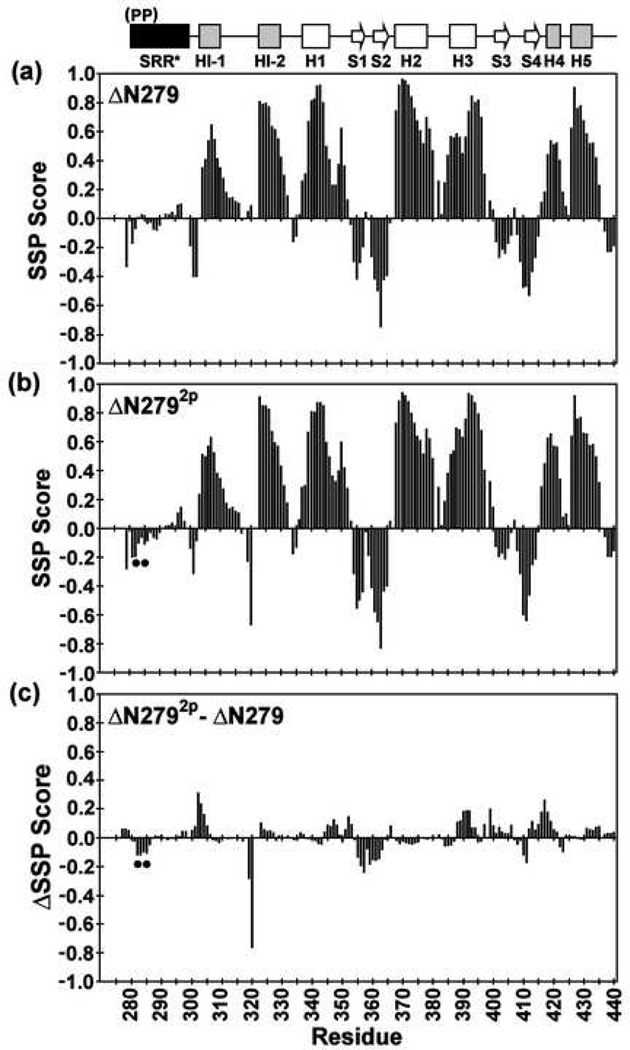
Fig. 3.
The SRR* of ΔN279 and ΔN2792p lack any predominant secondary structure, and phosphorylation does not alter the secondary structure of the inhibitory module or ETS domain. Shown are the secondary structural propensity (SSP) scores for (a) ΔN279 and (b) ΔN2792p, along with the (c) change ΔSSP upon phosphorylation, derived from available main chain 1H, 13C, and 15N chemical shifts (and using random coil phosphoserine chemical shifts25). Values approaching +1 and −1 are indicative of well defined α-helices and β-strands, respectively24. The phosphoserines are indicated by dots, and missing data points correspond to residues with unassigned NMR signals.
SRR* is unstructured and dynamic
Previous biophysical comparison of ΔN2445p and ΔN280 indicated that the SRR was unstructured and suggested that there were no losses or gains of structural elements upon phosphorylation6, 18 (Supplemental Fig. S2). These findings were re-investigated with the better-behaved ΔN279 species, thus enabling more complete assignment and a detailed assessment of any changes within the same species upon SRR* phosphorylation. Secondary structure elements were identified by analyzing the 1Hα, 13Cα, 13Cβ, and 13CO chemical shifts with the CSI21, RCI22, TALOS23, and SSP24 algorithms. By all these criteria, and in contrast to the well-structured regulatable unit, the SRR* residues exhibited random coil chemical shifts in ΔN279 and ΔN2792p (Fig. 3). Thus, the SRR* lacks any predominant secondary structure in both its unmodified and phosphorylated states. The possible presence of structural elements was also tested by amide HX using a combination of 1H-1H magnetization transfer and 1H-2H exchange experiments. In both ΔN279 and ΔN2792p, the SRR* exhibited amide HX rates within a factor of ~5 of those predicted for a random coil polypeptide of the same sequence (Fig. 4a,c). These data sets confirmed that the truncated SRR* does not adopt any predominant conformation in either the unmodified or phosphorylated states of ΔN279. Finally, 15N- and 13C-edited NOESY-HSQC spectra were recorded to determine if any persistent inter-residue contacts were detectable either within the SRR* or between the SRR* and the regulatable unit. Only intra- and nearest neighbor inter-residue 1H-1H NOE interactions were observed within the SRR*, and no long-range NOEs were detected between the SRR* and the regulatable unit in ΔN279 or ΔN2792p (data not shown). Together, these assays yielded no evidence of any persistent structural elements within either form of the SRR*, nor any long-lived interactions between the SRR* and the regulatable unit.
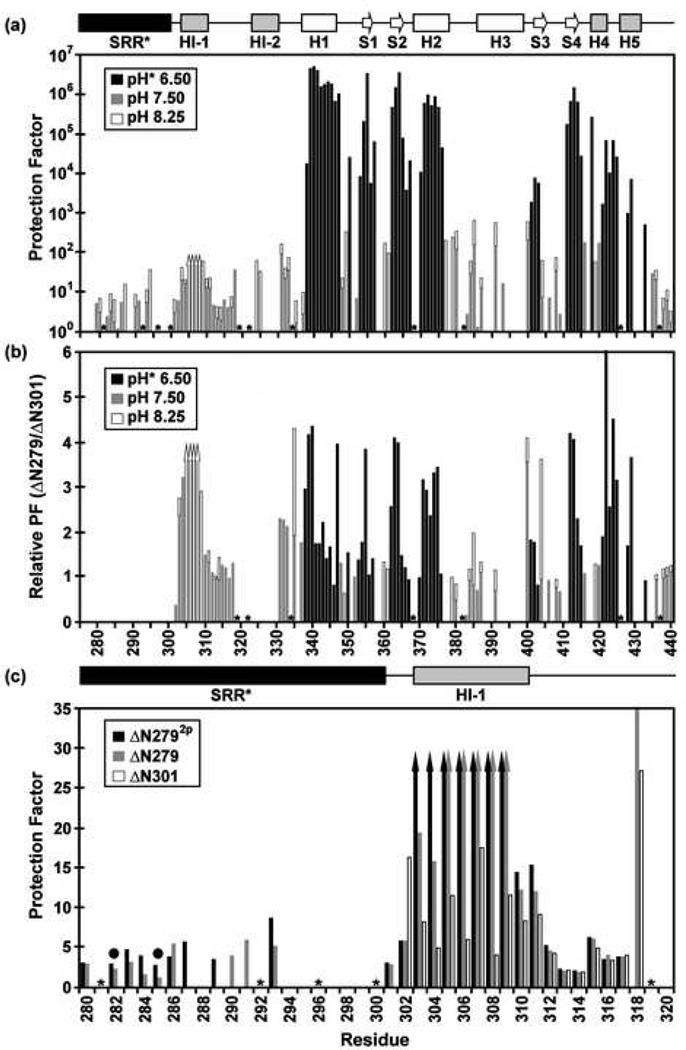
Fig. 4.
Amide HX measurements provide insights into the structure, stability, and dynamics of the Ets-1 deletion fragments (500 mM NaCl, 28 °C). (a) The comprehensive backbone amide HX protection factors of ΔN279, determined from slow H2O-D2O exchange (pH* 6.50, black) and fast H2O-H2O CLEANEX (pH 7.50, gray; pH 8.25, white) measurements. Residues 305–308 exchanged too slowly to be characterized by CLEANEX, yet too fast to be detected after transfer into D2O buffer, and thus their protection factors fall within the range of ~ 30 to 200 (up arrows). The SRR* is predominantly unstructured, showing little protection against HX. The inhibitory helices HI-1 and H1-2, as well as the DNA recognition helix H3, undergo relatively facile HX. (b) However, the presence of the SRR* stabilizes the inhibitory module and ETS domain against HX by up to ~ 4 fold. Shown are relative protection factors for ΔN279 versus ΔN301, obtained from H2O-D2O exchange (pH* 6.5, black) and/or CLEANEX measurements (pH 7.5, gray; pH 8.25, white). (c) Phosphorylation of S282 and S285 further stabilizes the N-terminal inhibitory helix of ΔN279 against amide HX. Shown are the amide protection factors of residues 280–320 in ΔN2792p (black), ΔN279 (gray), and ΔN301 (white), acquired from CLEANEX exchange experiments at pH 7.5. Up arrows indicate minimal estimated values. Dark circles indicate the phosphoacceptor serines, and missing data correspond to prolines (*) or amides with weak or overlapping NMR signals. Slow amide HX kinetics, which require measurements over many months, were not carried out with ΔN2792p.
Phosphorylation partially dampens the flexibility of the unstructured SRR*
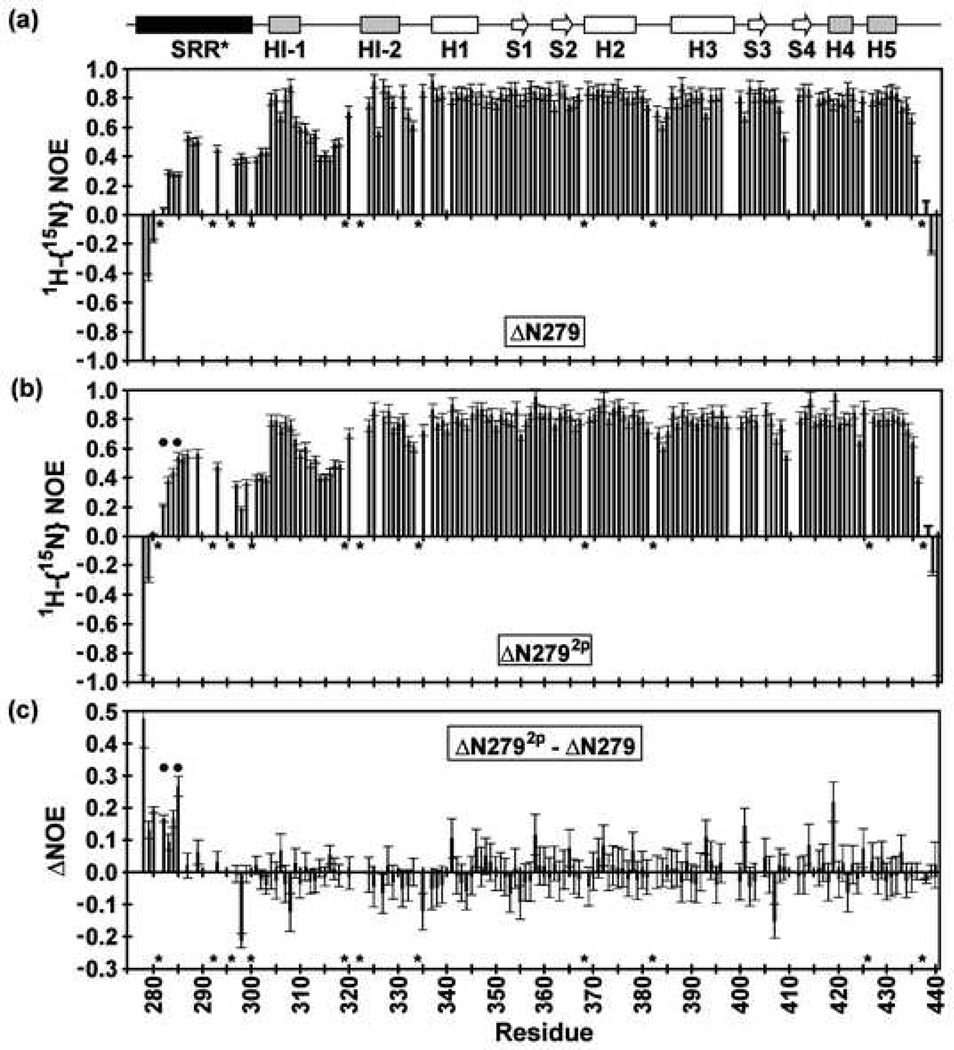
Previous analysis of ΔN2445p indicated the SRR was conformationally dynamic6, but no direct test of phosphorylation effects on the same species was possible. Thus, 15N relaxation experiments were performed with ΔN279 in its unmodified and phosphorylated states. Amides within the regulatable unit of ΔN279 exhibited average T1, T2, and 1H{15N}-NOE values of 883 ± 74 msec, 54 ± 8 msec, and 0.82 ± 0.08, respectively (500 mM NaCl, 28 °C). In contrast, amides within the SRR* displayed markedly different relaxation properties with average T1, T2, and 1H{15N}-NOE values of 645 ± 66 msec, 113 ± 39 msec, and 0.34 ± 0.20, respectively (Fig. 5a). These parameters corresponded to isotropic Lipari-Szabo model-free S2 values of 0.97 ± 0.06 for the regulatable unit) and 0.49 ± 0.16 for the SRR*. In particular, the longer T2 lifetimes and lower 1H{15N}-NOE values indicated substantially greater, but importantly not unrestricted, conformational mobility on a sub-nsec timescale for the truncated SRR* than for the structured ETS domain and inhibitory module. Upon phosphorylation, the SRR* residues of ΔN2792p remained conformationally flexible relative to the regulatable unit (Fig. 5b,c). However, the 1H{15N}-NOE values of these residues increased slightly to 0.39 ± 0.17, and the T2 values decreased to 82 ± 27 msec, whereas, the rest of the protein was not measurably perturbed. Thus, phosphorylation of S282 and S285 partially dampens the fast timescale mobility of the truncated SRR*.
Fig. 5.
The heteronuclear 1H{15N}-NOE values of (a) ΔN279, (b) ΔN2792p and (c) the difference, ΔNOE = NOE (ΔN2792p) – NOE (ΔN279), recorded in 500 mM NaCl and at 28 °C. The uniformly high NOE ratios of residues 301–440 in both ΔN279 and ΔN2792p are indicative of well-ordered backbone segments, except in regions such as the HI-1/HI-2 and H2/H3 loops, as discussed in detail for ΔN301 [Lee, 2005 #1]. Conversely, as shown by NOE ratios < 0.6, the SRR* in both ΔN279 and ΔN2792p exhibits flexibility on the sub-nsec timescale. This motion is not totally unrestricted as the NOE values are mostly > 0.2. Furthermore, positive ΔNOE values for residues 279–285 indicate that phosphorylation marginally dampens this fast timescale motion. Dark circles indicate the phosphoacceptor serines, and missing data correspond to prolines (*) or amides with weak or overlapping NMR signals.
SRR* and phosphorylation-dependent changes in the regulatable unit
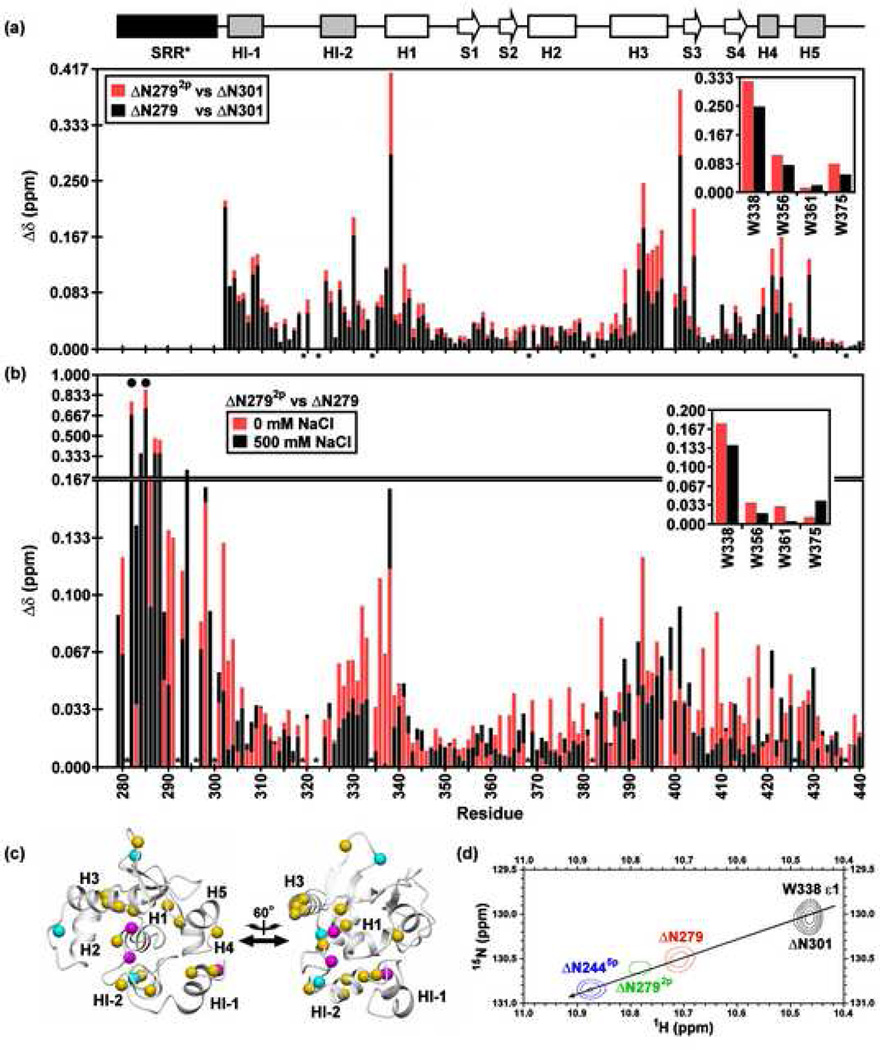
The SRR* is dynamic and lacks any predominant structure, yet this region stabilizes the regulatable unit against global unfolding in a phosphorylation-dependent manner. Insights into the mechanism of this stabilization were provided by a comparison of the amide 1HN and 15N chemical shifts of ΔN301, ΔN279, and ΔN2792p (Fig. 6a) Although not inducing any persistent, regular structure within the SRR*, phosphorylation altered the amide chemical shifts of S282 and S285, as well as those of adjacent residues (Fig 6b). Such local changes are expected at least in part from inductive and electric field effects25. More interestingly, whereas the majority of the peaks displayed in the 1H-15N HSQC spectra of ΔN301 and ΔN279 were superimposable, clear perturbations due to the presence of the SRR* occurred for corresponding residues within the inhibitory module, H1, and the DNA-recognition helix H3 of the ETS domain (Fig. 6a). These perturbations increased upon phosphorylation to form ΔN2792p (Fig. 6a,b and Supplemental Fig. S5). Amide chemical shifts are highly sensitive to subtle local environmental changes, and thus these results indicated that structural alterations occur in a region of the regulatable unit by the unmodified and, even more so, the phosphorylated SRR*. This region includes residues in the inhibitory module and the DNA binding interface, bridged via a hydrophobic network involving helix H1 (Fig. 6c)6. Importantly, as observed previously in the longer ΔN244, mutants with varying phosphorylation levels exhibited chemical shift perturbations that followed a co-linear relationship between chemical shift and DNA binding affinity (Fig. 6d)6. This correlation confirms an allosteric shift in the conformational equilibrium of the regulatable unit with increasing autoinhibition.
Fig. 6.
Amide 1HN and 15N chemical shift perturbations of the inhibitory module and DNA binding interface by the SRR* are enhanced by phosphorylation and partially reduced with increasing ionic strength. Shown are the combined backbone amide and tryptophan indole (insert) chemical shift perturbations (Δδ) for corresponding residues in (a) ΔN279 (black) and ΔN2792p (red) versus ΔN301 at 500 mM NaCl, and (b) ΔN2792p versus ΔN279 in 0 mM (red) and 500 mM (black) NaCl. A difference plot of the data in (b) is provided as Supplemental Fig. S5. The histogram bar for the smaller change is shown in front of that for the larger change. The absence of a bar indicates that the Δδ could not be unambiguously measured for a given residue due to spectral overlap or weak signals in at least one species or condition. Dark circles and asterisks represent the phosphoacceptor serines and prolines, respectively. (c) As expected, S282 and S285 underwent the largest chemical shift changes upon phosphorylation, with smaller effects occurring for adjacent amides within the SRR*. However, spectral perturbations also occurred in the inhibitory module, H1, and the DNA binding interface, as shown by mapping the amide shift difference between ΔN2792p and ΔN279 at 0 mM NaCl onto the ΔN301 structure (magenta, Δδ > 0.15 ppm; cyan, 0.10 to 0.15 ppm; yellow, 0.05 to 0.10 ppm). The two views are rotated by 60° about the vertical axis. (d) A co-linear relationship is observed between chemical shift and increasing autoinhibition, indicating an allosteric shift in the conformational equilibrium of the regulatable core6. This is illustrated by an overlay of the sidechain indole signals of W338 in four constructs at 500 mM NaCl (ΔN301, black; ΔN279, red; ΔN2792p, green; ΔN2445p, blue). ΔN301 is poorly soluble in low ionic strength buffers, thus precluding comparisons at reduced salt concentrations.
HX measurements were also used to monitor the effects of the SRR* on the local and global stability of the regulatable unit in ΔN279. As summarized in Figure 4a, HI-1 and HI-2 are marginally stable with HX protection factors only ~10 fold greater that those of the unstructured SRR*. Furthermore, amides within H3 of the ETS domain also exhibited modest protection factors of ~1000, indicative of significant local conformational fluctuations of the DNA recognition helix. In contrast, amides in helices H1 and H2 and b-strands S1, S2, and S4 of ΔN279 underwent HX at rates near six orders of magnitude slower than expected for a random coil polypeptide, demonstrating that they form the stable core of the ETS domain and likely exchange via a global unfolding pathway. Qualitatively, the HX profile of ΔN279 is similar to that previously measured for ΔN3018. Quantitatively, however, amides throughout the regulatable unit exchanged by up to ~ 4 fold slower in ΔN279 than in ΔN301, demonstrating that the SRR* stabilizes the regulatable unit against fluctuations leading to HX by ~ −0.8 kcal/mol (Fig. 4b). This parallels the increased autoinhibition of ΔN279 relative to ΔN301. Notably, the largest measurable increase in HX protection (6 fold) occurred for L422, a residue that is important for the integrity of the autoinhibitory module because it is located at the C-terminus of H4, which abuts tail-to-head with HI-18, 10. Phosphorylation of S282 and S285 further reduced the HX rates of the regulatable unit. A clear trend for this is observed at the termini of HI-1 and within HI-1/HI-2 loop, for which protection factors progressively increased by at least 6 fold in the order of ΔN301, ΔN279, and ΔN2792p (Fig. 4c). Thus, as observed in a previous study of ΔN2445p, both amide HX and 15N relaxation measurements (Fig. 5) revealed an increased damping of internal motions of the regulatable unit with increasing autoinhibition.
SRR* transiently interacts with the regulatable unit
Previously, we postulated that the structural and dynamic properties of the regulatable unit are allosterically modulated through transient interactions with the SRR. However, such interactions were not observed through NOESY measurements (not shown). The lack of detectable intramolecular 1H-1H NOEs between the SSR* and the regulatable unit, which in well ordered proteins typically arise from protons within ~ 5 Å of one another, may result from many factors. These include low populations and/or an ensemble of conformations with the SRR* contacting the remainder of the protein, unfavorable correlation times for these interactions, and/or spectral degeneracy.
In contrast to 1H-1H NOE measurements, PRE experiments are sensitive to longer-range, as well as transient short range, interactions26. Therefore, we utilized the ATCUN (Amino Terminal Cu2+ Ni2+) motif engineered into the N-terminus of ΔN279 to help localize the SRR* with respect to the regulatable unit by PRE. This motif, formed by the N-terminal Gly-Ser-His tripeptide remaining after thrombin cleavage of the His6-tag, binds Cu2+ with high affinity (KD ~ 10−15M)27, 28. In the case of an ordered ATCUN, proton-Cu2+ distances of ~10 to 20 Å can be measured from enhanced 1H relaxation due to the unpaired electron in the paramagnetic metal ion. The signals from protons closer than this lower limit are generally broadened beyond detection. However, conformational averaging will attenuate this 1/r6-dependent effect26. Thus, we have used this approach as a qualitative indicator of the proximity of amide and methyl protons in ΔN279 and ΔN2792p to the bound Cu2+ probe.
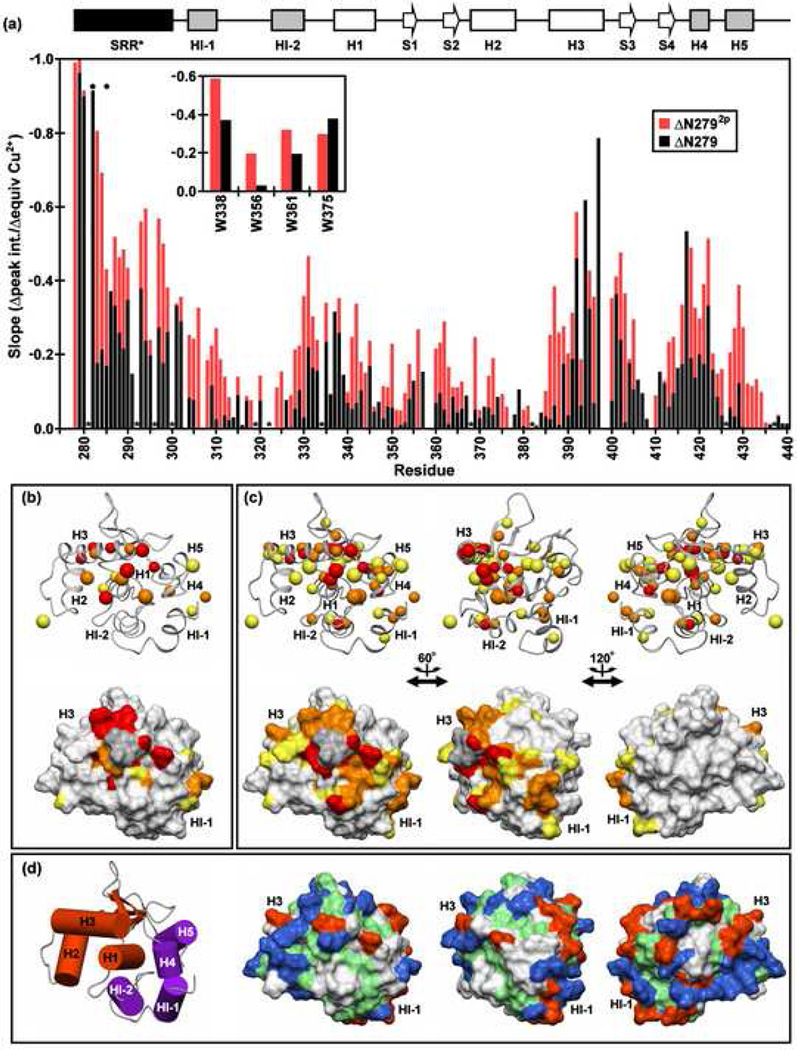
PRE measurements revealed a preferred localization of the ATCUN motif in ΔN279 with respect to the regulatable unit. As expected, 1H-15N and 1H-13C HSQC signals from SRR* residues immediately adjacent to this motif, such as V280 and S282, became undetectable upon formation of a 1:1 Cu+2:protein complex (Fig. 7a). However, significant PRE's were also observed for residues located in the loop preceding H1 (notably, L337 and W338), in H3, in the loop between H3 and S3 (I401 and I402), and within and adjacent to H4 (D417 and L422). When mapped onto the structure of ΔN301, residues most affected by the ATCUN-bound Cu+2 lie along a region on one face extending from HI-1 of the inhibitory module to the recognition helix H3 of the DNA binding interface (Fig. 7b).
Fig. 7.
The SRR* is localized to a region of the regulatable unit spanning from the inhibitory helix HI-1 to the DNA recognition helix H3, and phosphorylation enhances this localization. (a) Paramagnetic relaxation enhancements (PRE) observed for the backbone amides and tryptophan indoles (inset) of ΔN279 (black) and ΔN2792p (red) in 500 mM NaCl, as reflected by a linear regression analysis of the plots of 1H-15N HSQC signal intensities versus equivalents Cu2+ added. Resonances which completely broaden upon addition of ~ 1 equivalent CuSO4 have slopes approaching −1.0. The absence of a bar indicates that data could not be unambiguously measured for a given residue due to spectral overlap or a weak signal. Dark circles and asterisks represent the phosphoacceptor serines and prolines, respectively. The PRE slope values of (b) ΔN279 and (c) ΔN2792p (rotated about the vertical axis as indicated) are mapped onto the ΔN301 structure. The upper and lower views are the ribbon and corresponding surface diagrams, respectively. The smaller balls represent backbone amides, while the larger balls indicate either Asn/Gln sidechain NH2 or aliphatic methyl groups. The color scheme is red, slope < −0.4 (i.e. largest PRE); orange, −0.4 ↔ −0.325; yellow, −0.325 ↔ −0.25. Residues for which data were unavailable are shaded in grey. (d) Insights into enhanced DNA binding inhibition may be derived from the surface diagram of ΔN301, showing negatively-charged (Asp, Glu; red), positively-charged (Arg, Lys, His; blue), hydrophobic (green), and neutral polar/Gly (white) residues. The orientations match those of panel (c). A cylinder model of the regulatable unit, colored and positioned as in Fig. 1e, is provided for orientation of the leftmost structures in panels in (b–d).
Phosphorylation of N279 caused a similar, but stronger PRE effect (Fig 7a,c). Once again, relaxation enhancement was observed within the inhibitory module, H1, and H3. However, in the case of ΔN2792p, a larger number of residues at this interface were affected by paramagnetic relaxation. Together, these data indicated that the SRR* interacts with one face of the regulatable unit and that this effect is enhanced upon the phosphorylation of S282 and S285. Residues within same region also displayed chemical shift perturbations due to the presence of the SRR* (Fig. 6). We postulate that these PRE's and spectral shift changes reflect direct interactions of the SRR* to allosterically modulate the structure and dynamics of the regulatable unit. This correspondence also indicates that phosphorylation contributes to autoinhibition by reinforcing these interactions.
SRR* interactions are not predominantly electrostatic
The SRR* is net negatively-charged due to four asparate/glutamate residues, and this charge is increased upon phosphorylation of S282 and S285 (Fig. 1b). These observations prompted the hypothesis that electrostatic forces might mediate the interactions between the SRR and the regulatable unit. To test this possibility, urea denaturation assays were performed with ΔN2445p under conditions of varying ionic strength. Surprisingly, there was little change in the stability of ΔN2445p upon increasing the concentration of KCl from 50 to 500 mM (Fig. 2c). In contrast, the mid-point of the denaturation profile of ΔN244 remained similar at 50 and 100 mM KCl, yet showed a loss of cooperativity at 500 mM. These data argue against a major role of electrostatic interactions in the global stabilization of ΔN244 by SRR phosphorylation. To further explore this phenomenon, we also compared the amide chemical shifts of ΔN279 and ΔN2792p under native conditions at varying ionic strengths (Fig. 6a,b and Supplemental Fig. S5). For most (~2/3) residues, the magnitude of shift perturbations resulting from phosphorylation increased when the concentration of NaCl was lowered from 500 mM to 0 mM. Thus, the effect of SRR* phosphorylation on the regulatable unit is partially ionic strength dependent. However, the observation of clear spectral differences between ΔN279 and ΔN2792p at even 500 mM NaCl suggests that electrostatic interactions are not the dominant driving force for phosphorylation-dependent autoinhibition.
Discussion
Allosteric model of autoinhibition
In this report we describe new insights into the mechanism of phosphorylation-dependent autoinhibition of Ets-1 DNA binding. Our previous allosteric model for autoinhibition was developed from two major lines of evidence, namely the labile nature of helix HI-1, including its unfolding upon DNA binding, and the impact of both the SRR and its phosphorylation on the dynamic character of the ETS domain and flanking inhibitory module6, 7, 8, 9, 10, 19. The model proposes that a conformational equilibrium exists for the ETS domain and associated inhibitory module in the absence of DNA and that this equilibrium can be the target of regulation by the flanking SRR, as well as by its phosphorylation (Fig. 1c). This report expands the evidence for the allosteric mechanism by showing direct evidence for the phosphorylation-dependent dampening of the fast timescale mobility of the predominantly unstructured SRR* in ΔN279, along with dampening of the internal dynamics (ΔN279) and enhanced stability (ΔN279, Δ244) of the regulatable unit. In addition, new mechanistic insights came from the mapping of intramolecular interactions between SRR* and the regulatable unit, including the recognition helix, via chemical shift and PRE analyses. Importantly, the analyses were performed on ΔN279, a more easily manipulatable deletion fragment containing a truncated SRR* with two phosphoacceptor serines. The presence of residues 279–300 from the SRR increases the autoinhibition of ΔN279 by ~15 fold, from the modest level exhibited by the regulatable unit (ΔN301) to a level similar to that observed with native Ets-1 and ΔN2446, 8, 9. Phosphorylation of S282 and S285 reinforces this effect by an additional ~100 fold (Fig. 1e), again, to levels observed with native Ets-1 and ΔN2446, 12.
Surprisingly, although required for autoinhibition, the truncated SRR* lacks any predominant structure in both ΔN279 and ΔN2792p, as evidenced by random coil chemical shifts, the absence of detectable 1H-1H NOE interactions to the regulatable unit, and minimal protection from amide HX (Figs. 3, 4 and Supplemental Fig. S2). However, the conformational disorder of the SRR* is not completely unrestricted, as 15N relaxation measurements revealed that the SRR* exhibits nsec-psec timescale mobility greater than that of the well-ordered regions of the regulatable unit, yet dampened relative to the highly flexible residues at the termini of a deletion fragment such as ΔN244 (Fig. 5 and Supplemental Figure S1). The availability of ΔN279 in both its unphosphorylated and phosphorylated state also enabled analyses that demonstrated the mobility of the SRR* was further, albeit modestly, dampened upon phosphorylation. These observations support the original allosteric model of autoinhibition by correlating phosphorylation effects on reducing SRR* dynamics with increasing effectiveness of the SRR* in inhibiting DNA binding.
The role of the truncated SRR* in mediating autoinhibition was investigated thermodynamically and spectroscopically by analyzing the effects of its presence and its phosphorylation on the regulatable unit. The SRR* stabilizes the regulatable unit against fluctuations that lead to amide HX (Fig. 4). Phosphorylation of S282 and S285 enhances this effect, as observed most notably for the labile helix HI-1, which was shown previously to be unfolded in the DNA bound state9, 10. Within the context of ΔN244, phosphorylation also dramatically stabilizes the regulatable unit against thermal- and denaturant-induced global unfolding (Fig. 2) and dampens msec-µsec timescale motions detected by relaxation dispersion experiments6. Detailed chemical shift comparisons between ΔN301, ΔN279, and ΔN2792p indicate that the truncated SRR* either directly or indirectly perturbs the structure of a contiguous region of the regulatable unit encompassing the inhibitory module and helices H1 and H3 of the ETS domain, and that the perturbations increase with phosphorylation (Fig. 6). Interestingly, a comparison of SSP scores, which predict secondary structure from chemical shift information, shows small but distinct patterns of increased α-helical and β-strand values for several regions of ΔN2792p versus ΔN279, including the N-terminus of HI-1 and H4 in the inhibitory module and S1, S2, and the DNA-recognition helix H3 in the ETS domain (Fig. 3c). Although it is unclear how to interpret SSP scores for folded regions of proteins24, based on the correlation of secondary chemical shifts and dynamics noted by Wishart and co-workers22, this suggests that the spectral changes occurring upon phosphorylation also reflect the stabilization of the regulatable unit into a more rigid conformation.
In our previous work on Ets-1 DNA binding, we used similar data to propose an allosteric mechanism of regulation. According to this model, the ETS domain and inhibitory module pack together to form a regulatable unit that is conformationally flexible as evidenced by amide HX, 15N and 13C relaxation, and proteolysis measurements6, 8. The presence of the SRR and increasing levels of phosphorylation progressively shift the equilibrium structure and dynamics of the regulatable unit to a more rigid state with reduced DNA affinity. A co-linear change in the amide chemical shifts of residues in the inhibitory module and DNA binding interface that correlates with increasing autoinhibition provided clear evidence for an equilibrium between dynamic-active and rigid-inactive states6. Such a diagnostic co-linear change was also exhibited by ΔN279 and ΔN2792p (Fig. 6d). Note that this mechanism is allosteric from two complementary perspectives: DNA binding via the ETS domain is coupled to a conformational change in the inhibitory module that is highlighted by the unfolding of HI-1, a helix distal to the "DNA effector", and the phosphorylatable SRR serves as an "intramolecular effector", shifting the conformation, dynamic properties, and stability of the regulatable unit to a more rigid inactive state, refractory to DNA binding.
To explore the mechanism by which the SRR, a predominantly unstructured and conformationally flexible element, could be an intramolecular allosteric effector, we sought experimental evidence for its direct interaction with the regulatable unit. PRE effects, measured using Cu+2 bound to the ATCUN motif of ΔN279 (Fig. 7), revealed transient interactions consistent with the flexible, yet not unrestricted, dynamic properties of the SRR. Phosphorylation enhanced the magnitude or lifetime of these interactions. These PRE effects correlate with the partially dampened fast timescale mobility and reduced HX of the SRR* in ΔN2792p versus ΔN279 (Figs. 3, 4, 5, and 6). In addition, PRE mapping demonstrated that the truncated SRR* interacts with both the inhibitory module and the DNA recognition helix H3 (Figs. 6 and 7) along one contiguous surface of the regulatable unit. Many of these same residues displayed chemical shift perturbations upon addition of the SRR* and further upon phosphorylation. As noted previously, these elements are linked by a dynamic, hydrophobic network that includes the intervening helix H1 of the ETS domain6. Thus, we speculate that transient surface contacts made by the SRR allosterically transduce effects broadly through the regulatable unit.
Electrostatic forces are not dominant for allosteric regulation
We next considered the possible physicochemical nature of the transient interactions leading to allosteric control of Ets-1 DNA binding. The SRR* has four negatively-charged residues (D284, D287, E289, and D290) in segments required for autoinhibition, as demonstrated through deletion analysis (Fig. 1e). Phosphorylation of S282 and S285 increases further the net negative charge of the SRR*. Also, many of the residues most affected by the presence of the SRR*, as shown by PRE measurements, lie along the positively-charged DNA binding interface of the ETS domain. Accordingly, we asked whether electrostatic interactions might play a role in mediating the transient interactions of the SRR* with the regulatable unit. Previously, we observed that the msec-µsec timescale motions of ΔN279 and ΔN2792p detected by NMR relaxation-dispersion measurements were dampened in buffers of reduced salt concentration [6 and not shown]. Also, the phosphorylation-dependent chemical shift perturbations of the regulatable unit by the SRR* generally decreased with increasing ionic strength (Fig. 6a,b and Supplemental Fig. S5), suggesting some electrostatic contributions to autoinhibition. However, even in 500 mM NaCl, clear differences existed between the 1H-15N HSQC spectra of ΔN2792p and ΔN279. More dramatically, the stability of ΔN2445p against urea-induced denaturation showed no change between measurements made in 50 and 500 mM KCl solutions (Fig. 2). It is difficult to gauge the extent of screening expected for the intramolecular association of the phosphorylated SRR* with the regulatory unit. Nevertheless, persistent NMR spectral perturbations and the lack of a significant difference in the [urea]1/2 of ΔN2445p over this large range of ionic strength argues strongly against a dominant role of electrostatic interactions in mediating the phosphorylation-dependent stabilization of the regulatable unit, and by inference, in DNA binding autoinhibition.
Alternatively, we note that the truncated SRR* contains several hydrophobic amino acids (V280, Y283, F286, Y288, Y291, L295) immediately adjacent to negatively-charged Asp or Glu residues and the phosphoacceptor serines. Perhaps the association of the SRR* with the regulatable unit involves hydrophobic contacts to that neutral sidechains on a surface patch between HI-1 and H3 that also show PRE effects upon addition of Cu+2 to ΔN279 (Fig. 7d). A mechanistic model for this could involve phosphorylation-dependent changes in some fluctuating aromatic/hydrophobic clustering within the SRR to enable greater aromatic/hydrophobic interaction with the regulatable unit. All known Ets-1 vertebrate orthologs display overall >95% identity at the amino acid level, and thus the importance of these hydrophobic residues is not predictable from homology arguments. However, we note that the above residues lie in a region of Ets-1 that shares >80% amino acid sequence conservation relative to highly-related Ets-2, which also displays autoinhibitory properties. Five of the six hydrophobic residues are conserved in 16 Ets-1 and Ets-2 vertebrate orthologs, with the sixth being a variant only in mouse Ets-1. Site-directed mutational studies of these individual residues within the SRR* should help define the relative roles of electrostatic and hydrophobic interactions in autoinhibition.
A possible steric component to regulation of DNA binding
One feature of our findings suggests additional possible mechanistic contributions to the inhibition of DNA binding. PRE measurements demonstrated that the SRR* transiently interacts with the DNA binding interface of the ETS domain. Such localization of the negatively-charged phosphorylated SRR near the recognition helix H3 could reinforce autoinhibition by sterically blocking and/or electrostatically repelling DNA. Previous kinetic studies revealed that autoinhibition results mainly from a reduction in the lifetime of the bound Ets-1/DNA complex19. However, a small reduction in association rates, as would be expected for steric blockage mechanism, was also observed. Note that a global electrostatic repulsion mechanism is unlikely as disruption of the inhibitory module through mutation of its hydrophobic core leads to loss of autoinhibition and insensitivity to phosphorylation effects without changing the net charge of the Ets-1 fragments studied10, 11, 12. That is, the simple presence of the SRR and phosphorylated serine residues does not attenuate DNA binding affinity. Together, these data indicate that the allosteric and any possible steric contributions to autoinhibition are likely intimately linked. That is, the integrity of the inhibitory module is thermodynamically and structurally coupled to the proper juxtaposition of the SRR with respect to regulatory unit, as required for both modulating the equilibrium populations of dynamic-active and rigid-inactive states and for possible blockage of the DNA binding interface. Although the allosteric component of this mechanism has been well established, dissecting the nature and magnitude of the steric contribution requires more detailed mutational and kinetic analysis.
Adoption of structure is not required for SRR regulation of Ets-1 autoinhibition
Most studies of intrinsically disordered protein segments report a transition to a stable conformation in order to function1 (see however29). In contrast, our previous work and the new results in this report show that the multiply phosphorylated SRR of Ets-1 is able to inhibit binding by over two orders of magnitude in KD despite its predominantly unstructured and flexible character. A similar example of a functionally important regulator that retains disorder both before and after multiple phosphorylation events is found with the yeast cyclin-dependent kinase inhibitor Sic130. However, Sic1 displays a switch-like behavior in that it associates with the WD40 domain of the SCF ubiquitin ligase subunit Cdc4 only after any 6 of its 9 possible sites have been phosphorylated31. Each of the phosphorylation sites is a sub-optimal binding motif for the WD40 domain, and the switch has been proposed to result from a threshold level of cumulative "polyelectrostatic" interactions necessary for net binding of Sic1 to Cdc430. In the case of Ets-1, however, electrostatic forces do not appear to be crucial to the autoinhibitory mechanism and variable or "rheostat-type" regulation, rather than an all-or-none switch, is accomplished by progressive addition/removal of phosphates to the flexible SRR segment6. As part of this rheostatic mechanism, a correlation between dampened nsec-psec timescale dynamics of the SRR, but not adoption of a predominant structure, and reduced msec-µsec conformational flexibility of the regulatable ETS domain and inhibitory module are observed. A link between changes in the fast motions of flexible segments to regulation of intermediate timescale dynamics has been recently documented for enzymatic catalysis32, 33. Further characterization such dynamic mechanisms in biological processes is of great importance due to the preponderance of unstructured protein segments encoded by the genome.
Flexible SRR allows for rapid response to cell signals
The properties of Ets-1 are ideal for rapid and sensitive integration of signals. Intrinsically disordered regions, such as the SRR, are frequent targets of post-translational modification. In their unstructured forms, they allow unfettered access to the chemical moieties of the sidechains, and have low energy barriers to adopting the conformation required for rapid binding and modification via signal transduction machinery. For example, in response to Ca+2 release, Ca+2 dependent kinases, such as CamKII, are activated and, in turn, phosphorylate multiple serines within the flexible SRR. However, as noted above, phosphorylation does not induce and persistent structure within the SRR, nor does it cause stable binding of this region to the regulatable unit of Ets-1. Instead, the phosphorylated SRR region stays available for modification by other kinases or, importantly, phosphatases such as calcineurin, which can also become activated depending on the nature and length of Ca+2 stimulation34. Furthermore, we have previously shown that increasing the phosphorylation level of the SRR progressively attenuates the affinity of Ets-1 for DNA. Thus, by maintaining flexibility, regardless of modification state, the SRR can be rapidly modified and the DNA binding affinity of Ets-1 fine-tuned by the kinase/phosphatase balance in the cell. Based on these structural and biochemical properties, the SRR of Ets-1 can serve as an integrator for Ca+2 signaling, thereby modulating gene expression at the level of DNA binding with exquisite sensitivity.
Methods
Sample expression, purification, and phosphorylation
Expression and purification of Ets-1 fragments were performed according to previous methods6. Genes encoding ΔN244, ΔN280, ΔN286, ΔN292, ΔN296, and ΔN301 were sub-cloned into the pET22b vector (Invitrogen), expressed in E. coli BL29(λDE3) cells, and purified by conventional ion exchange and gel filtration chromatography. The gene encoding ΔN279 with an N-terminal His6-tag was cloned into the pET28a vector, followed by mutation of the codon from the vector encoding a methionine to the native R279 by the QuikChange method (Stratagene). ΔN279 was expressed in E. coli BL29(λDE3) cells. After lysis by sonication into 50 mM Tris, pH 7.5, 500 mM NaCl, 1 mM PMSF, 1 mM DTT, and 20 mM imidazole, and centrifugation at 40,000 rpm for 40 min., the supernatant was loaded on a 5 ml Nickel Sepharose column (GE Biosciences), and eluted in the same buffer with a gradient of 20 to 500 mM imidazole. Peak fractions were combined with thrombin (5 units/mg ΔN279; Sigma) and 2.5 mM CaCl2, dialyzed overnight against 20 mM citrate, pH 5.3, 100 mM NaCl, 1 mM DTT, and purified as described above for non-tagged constructs. After thrombin cleavage, the resulting ΔN279 contains three additional, non-native N-terminal residues (Gly-Ser-His). Uniformly 15N-, uniformly 15N/13C-, and uniformly 15N-/selectively 13C-methyl Ile/Val/Leu labeled samples of ΔN244 and ΔN279 were prepared according to published methods6, 8. Sample concentrations were quantified via UV absorbance using a predicted ε280 of 38120 M−1cm−1 (ExPASy Protparam, http://ca.expasy.org/tools/protparam.html).
All Ets-1 fragments were phosphorylated using the same published protocol6. Samples at a final concentration of ~25 µM were incubated for 1 hr at 30°C in 50 mM HEPES, pH 7.5, 10 mM magnesium acetate, 0.5 mM CaCl2, 2 mM DTT, 1 µM calmodulin, 200 nM calmodulin kinase II, and 1 mM ATP. The reaction mixtures were then diluted 1:10 with 25 mM Tris, pH 7.9, 10% glycerol, and 1 mM EDTA, loaded onto a strong anion exchange column (MonoQ, Pharmacia), and eluted with a KCl gradient from 50 to 400 mM. Peak fractions were pooled, analyzed by ESI-MS, and stored at 4°C.
Electrophoretic mobility shift assays
EMSAs were performed on Ets-1 fragments as previously described6, 12 using a high affinity ETS binding site composed of a 9 bp consensus site (underlined) embedded in the following complimentary 27-mers that include 4 bp overhangs: 5'-TCGACGGCCAAGCCGGAAGTGAGTGCC-3' (top strand); 5’-TCGAGGCACTCACTTCCGGCTTGGCCG-3’ (bottom strand).
Circular dichroism and denaturation measurements
Circular dichroism (CD) spectra of 30 µM protein samples, dialyzed into 25 mM potassium phosphate, pH 7.9, 100 mM KCl, 1 mM β-mercaptoethanol, and 0.1 mM EDTA, were recorded using a Jasco J-720 spectropolarimeter with a 0.1 cm pathlength cuvette. The final spectra represent the average of 6 scans at 25 °C. Thermal denaturation measurements were performed using a water-circulating temperature-controlled 0.5 cm pathlength cuvette. The water temperature was regulated by a Neslab RET-110. Samples were allowed to equilibrate for 10 minutes between temperature changes, and the final spectra represent the average of 3 scans at each temperature. Urea denaturation measurements were carried out with a series of protein samples in 0 to 8 M urea. A 10 M urea stock was prepared with 0.2 µM filtered dialysis buffer, and then aliquots were added to fixed volume of protein to generate equimolar samples at the desired urea concentrations. The CD signal θobs of each sample was measured at 222 nm and 25 °C with an AVIV 62DS spectropolarimeter. The value reported represents the average of readings acquired at a rate of 1 s−1 for 100 seconds. Data were analyzed by normalizing the blank-corrected CD signal versus the most negative signal recorded (θmin) followed by fitting to a standard 2-state equation for urea-induced protein unfolding35:
with yF = yF° + bF[urea] and yU = yU° + bU[urea]. In this equation, yF° and yU° are the folded and unfolded baseline intercepts, bf and bu are the folded and unfolded baseline slopes, m is the linear dependence of free energy of unfolding on [urea], and [urea]1/2 is the midpoint denaturant concentration for unfolding.
Spectral assignments
All NMR spectra were recorded on a cryoprobe-equipped Varian Inova 600 MHz spectrometer at 28 °C. A typical NMR sample consisted of 0.1 to 0.3 mM protein in 0.35 to 0.50 mL of 20 mM phosphate buffer (pH 6.5) containing 0.02% NaN3, 5 mM DTT, 10% D2O, and either 0, 50, or 500 mM NaCl. The sample conditions were adjusted by dialysis or by concentration and buffer exchange using MicroSep 3K (Pall Life Sciences) or Amicon Ultra 5K Centrifugal Filter Devices (Millipore Corp.). Spectral processing and analysis were performed with NMRpipe36 and Sparky37.
Assignments for the backbone and selected sidechain 1H, 15N, and 13C resonances of ΔN279 and ΔN2792p were obtained using multidimensional NMR experiments acquired on a series of uniformly 15N-, uniformly 15N/13C-, or uniformly 15N-/selectively 13C-methyl Ile/Val/Leu labeled proteins, as previously described8. Simultaneous 3D 15N-13C-NOESY-HSQC, centered on the methyl (τm = 140 msec), aliphatic (τm = 150 msec), and aromatic (τm = 150 msec) 13C regions, as well as 3D-15N NOESY-HSQC (τm = 150 msec) spectra were acquired for both ΔN279 and ΔN2792p. The first three residues (Gly-Ser-His) were not assignable due to rapid amide HX. Ionic strength-dependent chemical shifts were obtained from 1H-15N HSQC spectra recorded with ~ 0.5 mL samples of ~ 0.1 to 0.3 mM 15N-labeled protein, initially in 0 mM NaCl, to which µL aliquots of sample buffer containing 5 M NaCl were added. Chemical shift perturbations were calculated from the combined 1H and 15N shift differences as and mapped onto the NMR-derived structure of ΔN301 (pdb accession code 1R36.pdb) using Molmol38.
Backbone amide hydrogen exchange
Slow amide proton-deuterium HX rates, kex, for ΔN279 were measured from a series of 1H-15N HSQC spectra recorded after rapid transfer of the 15N-labeled protein into D2O buffer (28 °C, pH* 6.5, 500 mM NaCl), as previously described for ΔN3018. Rapid amide proton-proton HX rates were acquired for both ΔN279 and ΔN2792p (28 °C, 500 mM NaCl, pH 7.5 and pH 8.25) using the CLEANEX-PM method39. Predicted exchange rates, kpred, for an unstructured protein containing the ΔN279 sequence were calculated with the program SPHERE40 using poly-dl-alanine reference data corrected for amino acid type, pH, temperature, and isotope effects41,42. In the case of ΔN2792p, reference kpred values for phosphorylated serines were estimated by replacing S282 and S285 in the sequence with aspartates. Amide-specific protection factors were calculated as the ratio kpred/kex. As such, a protection factor can be interpreted as the inverse of an equilibrium constant describing fluctuations between a closed, non-exchangeable state and a transiently-exposed, exchange-competent state, and thus provides a measure of the residue-specific free energy changes, ΔG°HX = RTln(kpred/kex), governing local or global conformational equilibria detected by HX.
Backbone amide relaxation
Backbone amide 15N relaxation parameters of the 15N-labeled ΔN279 and ΔN2792p samples in both 50 mM and 500 mM NaCl were acquired at 28 °C, as previously described43. Curve fitting and relaxation rate calculations were performed with either Sparky37 or a Matlab (The MathWorks, Inc.) macro supplied by W.-Y. Choy (Univ. of Toronto). Error analysis was facilitated by a 200-step Monte Carlo routine. Due to the dynamic nature of residues 280–300, only per-residue Lipari-Szabo model-free fits20 were obtained using a Matlab macro provided by L.E. Kay (Univ. of Toronto).
ATCUN paramagnetic relaxation enhancement measurements
Immediately prior to Cu2+ titrations, ΔN279 and ΔN2792p samples were exchanged into 20 mM Bis-Tris (pH 6.5), 0.02% NaN3, and 500 mM NaCl with no reducing agents. These conditions were necessary to avoid the formation of insoluble Cu+2 complexes and the reduction of paramagnetic Cu2+ to diamagnetic Cu1+. Cu2+ titrations were performed by adding µL aliquots of a stock solution of 5 mM CuSO4 in this buffer to a ~ 0.5 mL solution containing 0.1 – 0.2 mM protein. Relatively low protein concentrations were used to limit the possible effects of nonspecific Cu2+ binding or intermolecular paramagnetic relaxation. The titrations were monitored with 1H-15N and/or 1H-13C HSQC spectra. An approximate 1:1 ratio of Cu+2:protein was ascertained when the normalized intensities of the methyl and amide peaks of V280 approached a baseline value, typically 0–5 % of the original value.
1H-15N HSQC-detected amide 1HN T2 (=1/R2) relaxation measurements28 of ΔN279 and ΔN2792p with 0 and ~ 1 equivalents CuSO4 were recorded at 28 °C. Residue-specific amide paramagnetic relaxation enhancement (PRE) values were calculated as ΔR2 = R2 (Cu2+-bound) – R2 (Cu2+-free). Alternatively, PRE values were qualitatively evaluated during the Cu2+ titration by plotting the normalized 1H-15N and/or 1H-13C-HSQC peak intensities versus the equivalents Cu2+ added. Since a linear relationship between the reduction of peak intensities and added equivalents of Cu2+ was observed due to tight binding, linear regression analyses were performed using data points ranging from 0:1 to ~ 1:1 Cu2+ to protein ratios. Slopes decreasing from 0 to −1.0, therefore, represent HSQC peaks that are increasingly affected by high affinity Cu+2 binding to the limit of being broadened beyond detection upon complete saturation of the ATCUN motif.
Supplementary Material
Acknowledgments
We are grateful to Lewis Kay, Algirdas Velyvis, Wing-Yiu (James) Choy, Oscar Millet for providing data analysis programs, and to Mark Okon for assistance with NMR spectroscopy. This research was supported by grants from National Cancer Institute of Canada with funds from the Canadian Cancer Society (to L.P.M.) and the National Institutes of Health grants GM38663 (to B.J.G.) and CA42014-I (to the Huntsman Cancer Institute for support of core facilities). B.J.G. also acknowledges funding from the Huntsman Cancer Institute/Huntsman Cancer Foundation. Instrument support was provided by the Canadian Institutes for Health Research, the Protein Engineering Network of Centres of Excellence, the Canadian Foundation for Innovation, the British Columbia Knowledge Development Fund, the UBC Blusson Fund, and the Michael Smith Foundation for Health Research. The work by M.A.P. was performed under NIH grants T32-CA93247 and GM08537, and he is currently a fellow of the Leukemia and Lymphoma Society (5416-07).
Abbreviations used
- ATCUN
amino terminal copper and nickel binding motif
- CD
circular dichroism
- EMSA
electrophoretic mobility shift assay
- ESI-MS
electrospray ionization mass spectrometry
- HSQC
heteronuclear single quantum correlation
- HTH
helix-turn-helix
- HX
hydrogen exchange
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- PF
protection factor
- pH*
the pH meter reading uncorrected for isotope effects
- PRE
paramagnetic relaxation enhancement
- SRR
serine rich region (residues 244–301)
- SRR*
truncated serine rich region (residues 279–301)
- Ets-1
fragments are indicated by the boundaries of truncations from the N-terminus of the protein e.g. ΔN301 is a deletion of residues 1–300 and corresponds to Ets-1(301–440)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 2.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr. Opin. in Struct. Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nuc. Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinmetz MO. Structure and thermodynamics of the tubulin-stathmin interaction. J. Struct. Biol. 2007;158:137–147. doi: 10.1016/j.jsb.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 6.Pufall MA, Lee GM, Nelson ML, Kang H-S, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 7.Pufall MA, Graves BJ. Autoinhibitory domains: Modular effectors of cellular regulation. Ann. Rev. Cell Devel. Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 8.Lee GM, Donaldson LW, Pufall MA, Kang H-S, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 9.Petersen JM, Skalicky JJ, Donaldson LW, McIntosh LP, Alber T, Graves BJ. Modulation of transcription factor Ets-1 DNA-binding - DNA-induced unfolding of an alpha-helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 10.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J. Biol. Chem. 2002;277:45529–45536. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 11.Wang H, McIntosh LP, Graves BJ. Inhibitory module of Ets-1 allosterically regulates DNA binding through a dipole-facilitated phosphate contact. J. Biol. Chem. 2002;277:2225–2233. doi: 10.1074/jbc.M109430200. [DOI] [PubMed] [Google Scholar]
- 12.Cowley DO, Graves BJ. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes & Devel. 2000;14:366–376. [PMC free article] [PubMed] [Google Scholar]
- 13.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha 2. Mol. Cell. Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baillat D, Bègue A, Stéhelin D, Aumercier M. Ets-1 transcription factor binds cooperatively to the palindromic head to head Ets-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J. Biol. Chem. 2002;277:29386–29398. doi: 10.1074/jbc.M200088200. [DOI] [PubMed] [Google Scholar]
- 15.Lamber EP, Vanhille L, Textor LC, Kachalova GS, Sieweke MH, Wilmanns M. Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J. 2008 doi: 10.1038/emboj.2008.117. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kern D, Zuiderweg ERP. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Grundstrom T. Calcium regulation of GM-SCF by calmodulin-dependent kinase II phosphorylation of Ets1. Mol. Biol. Cell. 2002;13:4497–4507. doi: 10.1091/mbc.E02-03-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skalicky JJ, Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Structural coupling of the inhibitory regions flanking the Ets domain of murine Ets-1. Prot. Sci. 1996;5:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonsen MD, Petersen JM, Xu Q, Graves BJ. Characterization of the cooperative function of inhibitory sequences of Ets-1. Mol. Cell. Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic-resonance relaxation in macromolecules.1. Theory and range of validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 21.Wishart DS, Sykes BD. The C-13 chemical-shift index - a simple method for the identification of protein secondary structure using C-13 chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 22.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 23.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 24.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: Implications for fibrillation. Prot. Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bienkiewicz EA, Lumb KJ. Random-coil chemical shifts of phosphorylated amino acids. J. Biomol. NMR. 1999;15:203–206. doi: 10.1023/a:1008375029746. [DOI] [PubMed] [Google Scholar]
- 26.Clore GM, Tang C, Iwahara J. Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 2007;17:603–616. doi: 10.1016/j.sbi.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harford C, Sarkar B. Amino terminal Cu(ii)- and Ni(ii)-binding (ATCUN) motif of proteins and peptides: Metal binding, DNA cleavage, and other properties. Acc. Chem. Res. 1997;30:123–130. [Google Scholar]
- 28.Donaldson LW, Skrynnikov NR, Choy WY, Muhandiram DR, Sarkar B, Forman-Kay JD, Kay LE. Structural characterization of proteins with an attached ATCUN motif by paramagnetic relaxation enhancement NMR spectroscopy. J. Am. Chem. Soc. 2001;123:9843–9847. doi: 10.1021/ja011241p. [DOI] [PubMed] [Google Scholar]
- 29.Tompa P, Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends. Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc. Nat. Acad. Sci. (USA) 2007;104:9650–9655. doi: 10.1073/pnas.0702580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nash P, Tang XJ, Orlicky S, Chen QH, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 32.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:U913–U927. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 33.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hubner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:U838–U913. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 34.Salazar C, Politi AZ, Hofer T. Decoding of calcium oscillations by phosphorylation cycles: Analytic results. Biophys. J. 2008;94:1203–1215. doi: 10.1529/biophysj.107.113084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pace CN, Shirley BA, Thomson JA. Measuring the conformational stability of a protein. In: Creighton TE, editor. Protein structure - a practical approach. Oxford: IRL Press; 1989. pp. 311–330. [Google Scholar]
- 36.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Goddard TD, Kneller DG. Sparky 3. San Francisco: University of California; 1999. [Google Scholar]
- 38.Koradi R, Billeter M, Wuthrich K. MolMol: A program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 39.Hwang TL, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated clean chemical exchange (CLEANX-PM) approach with a fast-hsqc (fhsqc) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y-Z. Protein and peptide structure and interactions studied by hydrogen exchange and NMR. Ph.D. University of Pennsylvania; 1995. [Google Scholar]
- 41.Bai YW, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen-exchange. Prot. Struct. Funct. Gen. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connelly GP, Bai YW, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen-exchange. Prot. Struct. Funct. Gen. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 43.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Formankay JD, Kay LE. Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studied by N-15 NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.