Abstract
In order to determine if the expression of the activation marker CD38 can correlate with HIV disease progression independently of cycling, we performed a cluster-based multivariate correlation analysis of total circulating CD4+ T cell counts and viral loads with frequencies of CD38 and Ki67 expression on CD4+ lymphocytes from patients with untreated HIV infection, stratified in maturation subpopulations, and subpopulation subsets defined by the expression of CXCR5, CXCR3, and CCR4. The frequencies of the activated phenotypes %CD38+ Ki67− and %CD38+ Ki67+ of the CXCR5− CXCR3− CCR4+ (“pre-Th2”) central memory (TCM) cell subset clustered together, comprising a significant negative correlate of total circulating CD4+ T cell counts and a positive correlate of viral load in multivariate analysis. Frequency of cycling-uncoupled CD38 expression in “pre-Th2” TCM cells was a negative correlate of total circulating CD4+ T cell counts in univariate analysis, which was not the case of their %CD38+ Ki67+. CXCR5+ CXCR3− CCR4− TCM cells were underrepresented in patients, and their absolute counts correlated negatively with their %CD38+ Ki67− but not with their % CD38+ Ki67+. Our results may imply that CD38 expression either reflects or participates in pathogenic mechanisms of HIV disease independently of cell cycling.
1. Introduction
T cell activation is a strong predictor of CD4+ T cell loss in HIV infection [1], particularly when assessed by the expression of CD38, which shows a remarkable value as a predictor of HIV disease progression in diverse settings [1–3]. T cell activation has accordingly been deemed a possible indirect mechanism of CD4+ T cell depletion in HIV disease [4, 5].
A number of studies on activation have also measured the expression of the nuclear and perinuclear protein Ki67 initially considered to indicate proliferation [6, 7] and later delimited as an indicator that cells are in cycle [8] and undergoing turnover [9]. However, the expression of Ki67 does not always correlate with that of CD38, and these molecules show different predictive value depending on the T cell subset on which they are analyzed [5, 10–12]. In several studies Ki67+ CD4+ T cells comprise only a fraction of CD38+ cells [13–16], and these molecules show different expression dynamics during antiretroviral treatment and in other settings [17–19]. Therefore, it is important to investigate their relative contributions to the association of T cell activation and overall CD4 T cell loss.
The differences between CD38 and Ki67 as predictors may also reflect that their relationship with CD4+ T cell loss depends on the cell population that is studied which makes it potentially relevant to detect activation in different maturation subpopulations and additionally in relevant subsets within maturation subpopulations. Among subpopulations, central memory CD4 T cells (TCM cells) have important self-renewal and differentiation capacities [20–22] and are crucial to the relative homeostasis of memory cells during the chronic phase of HIV infection [23–26].
Different subsets of TCM cells have been identified by their expression of CXCR5, CXCR3, and CCR4 chemokine receptors. These subsets display specialized responses in vitro to TCR engagement or homeostatic cytokines, either proliferating and self-renewing (CXCR5+ CXCR3− CCR4− TCM cells) or proliferating and differentiating to Th1 cells (CXCR5− CXCR3+ CCR4− “pre-Th1” cells) or to Th2 cells (CXCR5− CXCR3− CCR4+ “pre-Th2” cells) [20, 27]. This further subdivision of TCM cells may be useful to investigate differential associations of CD38 and Ki67 with HIV disease progression, since their specialized functions correspond to those required by TCM cells for their regenerative capacity in untreated HIV infection. Additionally, these chemokine receptors are by themselves important in T cell function and in HIV disease pathogenesis. CXCR5 is expressed by TCM cells with B cell-help capacity [28] and by follicular helper cells, which are important in HIV control [29]. T cells expressing CXCR3 and CCR5, HIV's coreceptor, home to inflammatory sites [30], where CD4 T cell turnover is high [31]. CCR4 confers T cells the capacity to home to lung mucosal tissues [32], also critical in HIV disease [33]. Thus, we considered the subdivision of TCM (and TEM) cells according to the expression of these receptors as potentially informative.
Our objective was to study both the joint or independent participation of CD38 expression and cell cycling (assessed by Ki67 expression), measured in different subsets within the different maturation subpopulations of circulating CD4+ and CD8+ T cells, as correlates of HIV disease progression, and to determine if they are mutually dependent.
2. Methods
This study was approved by the Institutional Boards of Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas (INER) and Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (INCMN), Mexico. Blood samples were collected from 11 HIV+ antiretroviral-naive patients from the Department of Infectious Diseases of INCMN and from 11 healthy HIV− controls. Both groups had 9 men and 2 women. Patients signed informed consent according to the Helsinki Protocol. Patients did not have any active opportunistic infection or malignancy, and none was receiving immunomodulatory drugs. CD4+ T cell counts were not available for one patient.
2.1. Phenotyping of Subsets of CD4+ and CD8+ Lymphocytes and Activation Phenotypes
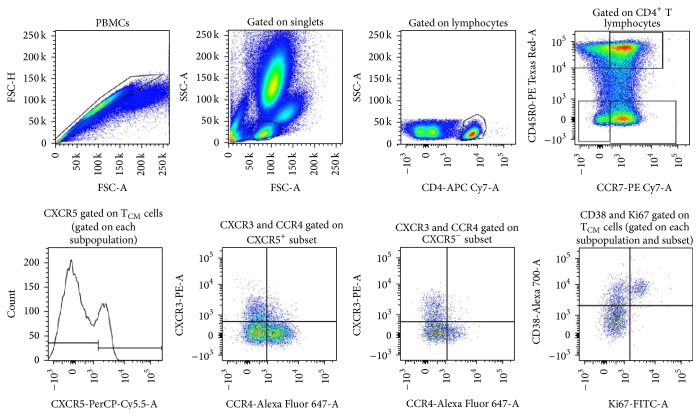
To determine frequency, activation phenotype, and functionality of maturation subsets of CD4+ T cells [20, 27], peripheral blood mononuclear cells (PBMCs) from HIV+ patients and healthy controls were obtained and processed completely immediately after sampling. Cells were incubated for 30 minutes at 4–8°C away from light with titrated biotin-conjugated monoclonal antibody specific for CD45RO (BioLegend, San Diego, CA, USA, Supplemental Material, Table A available online at http://dx.doi.org/10.1155/2016/9510756), washed with phosphate-buffered solution containing 10% bovine serum albumin, and stained with Streptavidin conjugated with PE-Texas Red (BD Biosciences, San Jose, CA, USA, Supplemental Material, Table A). This was followed by incubation with fluorochrome-conjugated monoclonal antibodies specific for surface molecules in the same conditions (Supplemental Material, Table A). Cells were then washed with PBS (Lonza, Walkersville, MD, USA), fixed for 30 minutes with 500 μL 4% p-formaldehyde (JK Baker, Mexico City, Mexico), and washed twice with 1 mL of a 1/2 dilution of Permeabilization Wash Buffer (10x BioLegend). Cells were then stained with anti Ki67-FITC (BD Biosciences, San Jose, CA, USA) in Perm/Wash buffer for 40 minutes on ice and away from light. Appropriate negative controls for each marker were used, consisting of cells stained with isotype controls plus the necessary fluorochrome-conjugated specific antibodies to eliminated spillover (Supplemental Material, Table B). Cells were analyzed in a FACSCanto II cytometer (Becton Dickinson, San Jose, CA, USA) and analyzed with FlowJo software (Tree Star, San Carlos, CA, USA). Lymphocytes were identified by their side scatter and forward scatter properties, and, among them, we selected CD4high cells (excluding all CD4dim events) as well as CD8high cells. Central memory cells (TCM) were delineated as CD45ROhigh CCR7+ CD4high or CD8high lymphocytes. Effector memory cells (TEM) were CD45ROhigh CCR7− CD4+ CD4high CD8high lymphocytes, naive cells (TN) were CD45RO− CCR7+ CD4high CD8high lymphocytes, and terminally differentiated cells were identified by the phenotype CD45RO− CCR7−. This broadly used strategy excludes most of the possible contaminating cells other than CD4+ and CD8+ T cells (Figure 1).
Figure 1.
Successive analysis of CD4+ and CD8+ lymphocyte subsets and expression of CD38 and/or Ki67. The CD38 versus Ki67 plot, yielding three possible activation phenotypes, was analyzed in each gating stratum (layer); third plot, whole CD4 high lymphocytes (or CD8+ lymphocytes); each quadrant in the fourth plot, maturation subpopulations; each of eight possible patterns of expression of CXCR5 CCR4 and CXCR3 within each maturation subpopulation.
2.2. Data Analysis
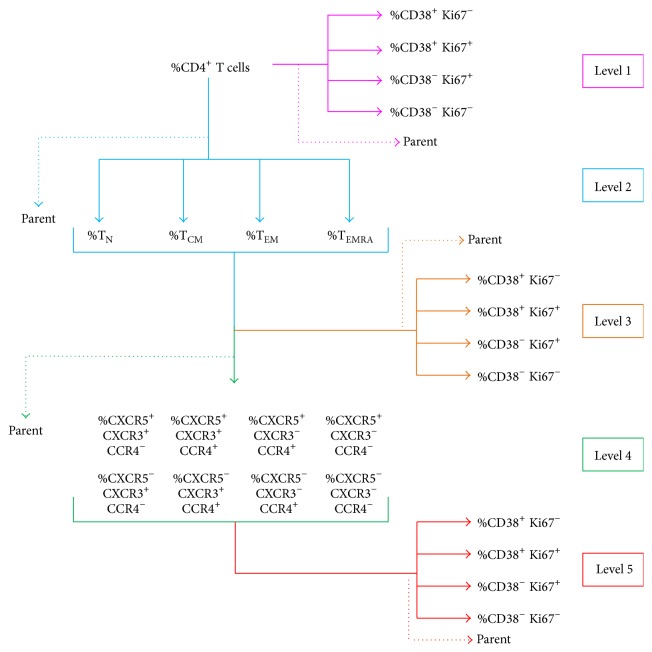
We obtained groups of variables for analysis by subsequent subgating, depicted as levels in Figure 2. We determined the frequency (%) of cells in each classification level of CD4+ T cells as follows (Figure 2):
-
Level 1, activation phenotypes (CD38 and Ki67 expression patterns) on total CD4+ and CD8+ T cells.
-
Level 2, maturation subpopulations on total CD4+ and CD8+ T cells.
-
Level 3, activation phenotypes on subpopulations of level 2.
-
Level 4, subsets within subpopulations, discriminated by the expression of chemokine receptors CXCR5, CXCR3, and CCR4 [20, 27].
-
Level 5, activation phenotypes of each subset.
Figure 2.
Groups of variables used in each cluster-based multivariate analysis. Subsequent gating generated groups of variables depicted as levels: level 1, CD4+ T cells; level 2, maturation subpopulations (naive, central memory, effector memory, and terminally differentiated); level 4, the eight possible combinations of expressions of CXCR5, CCR4, and CXCR3 within each maturation subset; and level 5, frequency of activation phenotypes within each subset. The frequencies (%) of activation phenotypes were analyzed at all higher levels. In each analysis level, the parent population was included to correct for differences in the percentage of the parent population as a confounder.
Gating was performed only on subpopulations comprising at least 300 events, which would assure meaningful percentages in the daughter population under the assumption of independent and identically distributed (iid) samples under a binomial distribution and considering only the last gate's variance. In this case, percentages derived from ≥300 events will have an 80% power for effect sizes of h ≥ 0.16.
Mann-Whitney test was used to analyze univariate differences between HIV+ patients and controls. Correlations were carried out by Spearman's ranked correlation. These tests were performed with StatView (Brain Power Inc., Calabasas, CA, USA) and Prism (GraphPad Software Inc., La Jolla, CA, USA) software.
2.3. Multivariate Analysis
Our analysis strategy started with a principal component analysis (PCA) to account for correlations between variables. PCA reduced variables to clusters of variables and generated a new variable consisting of the mean of their standardized values. Variables resulting from PCA were then utilized in multivariate analysis. We selected the PCA dimensions that cumulatively explained 80% of the dataset variance. As centroids for a k-means clustering algorithm we used for each variable the value of the individual that most contributed to the dimension to which the variable in question belonged. The final clusters had a 95% confidence measured by bootstrapping 1000 PCA clustering algorithms. This analysis was performed using R 2.1.1 with FactoMineR package [34]. Each cluster of variables had within it variables that had positive and negative estimates, and these are reported only as positive or negative, but the whole score for the dimension comprising all the variables was analyzed through normal multivariate logistic (in the case of HIV versus control) or linear (in the case of T CD4+ cell/mL of blood count or viral load) regressions, with associated P values.
2.4. Correction for Confounders
Statistical analysis required assuring that effects at a given subset were not influenced by differences in the parental population from which the subset was subgated. To control for these confounders, we included in the analysis of each cell group the frequency of its parent population (indicated in Figure 2 by a dotted arrow). For example, in the analysis using percentage of each activation phenotype in TN cells (level 3), the frequency of TN cells within CD4 T cells was also included.
3. Results
Patients had a median age of 33.1 years (range: 19 to 50), not differing significantly from controls (median: 27.3 years; range: 20 to 43). Patients had median 116 565 HIV RNA copies/mL blood (range: 1527–421 290), and median 323 CD4+ T cells/mm3 blood (range: 96–561).
3.1. Multivariate Correlates of Disease Progression
Cluster-based multivariate analysis showed a significant correlation of CD4+ T cell counts and viral load with one cluster composed of the %CD38+ Ki67− of CXCR5− CXCR3− CCR4+ TCM cells and the %Ki67+ CD38+ of the same subset as positive coefficients and the other phenotypes (Ki67+ CD38− and Ki67− CD38−) as negative coefficients (P = 0.036 for CD4+ T cell count, P = 0.030 for viral load, Table 1). This was the only multivariate correlate of CD4+ T cell counts that we found within all levels. Multivariate logistic regression showed a correlation of infection with a cluster containing %CD38+ Ki67− of CXCR5− CXCR3− CCR4+ TEM cells as the only positive coefficient with CD38 expression thus determining the significance (Table 1).
Table 1.
Clusters of variables with significant correlation with HIV disease. P values correspond to a multivariate linear regression using main clusters as independent variables (see Section 2) and CD4+ T cell counts and viral load as dependent variable or status (HIV+ or HIV−) as dependent variable.
| Correlate | Coefficient sign in cluster model | CD4+ T cells/mm3 blood∗ | HIV RNA copies/mL blood | ||
|---|---|---|---|---|---|
| Variables in cluster | Sign of regression coefficient | P | Sign of regression coefficient | P | |
|
| |||||
| TCM CXCR5− CCR4+ CXCR3−/Ki67− CD38+ | + | − | 0.036 | + | 0.030 |
| TCM CXCR5− CCR4+ CXCR3−/Ki67+ CD38+ | + | ||||
| TCM CXCR5− CCR4+ CXCR3−/Ki67+ CD38− | − | ||||
| TCM CXCR5− CCR4+ CXCR3−/Ki67− CD38− | − | ||||
|
| |||||
| HIV+ versus control | |||||
|
| |||||
| TEM CXCR5− CCR4+ CXCR3−/Ki67− CD38+ | + | + | 0.043 | ||
| TEM CXCR5− CCR4+ CXCR3−/Ki67+ CD38+ | − | ||||
| TEM CXCR5− CCR4+ CXCR3−/Ki67+ CD38− | − | ||||
| TEM CXCR5− CCR4+ CXCR3−/Ki67− CD38− | − | ||||
3.2. Linear Correlation of Relevant Activation Phenotypes with CD4+ T Cell Count
Since %CD38+ Ki67− and %CD38+ Ki67+ of CXCR5− CXCR3− CCR4+ TCM cells clustered as multivariate correlates of CD4+ T cell counts, we asked if both activation phenotypes were overrepresented among CXCR5− CXCR3− CCR4+ TCM and TEM cells from patients and if each one separately correlated with CD4+ T cell counts.
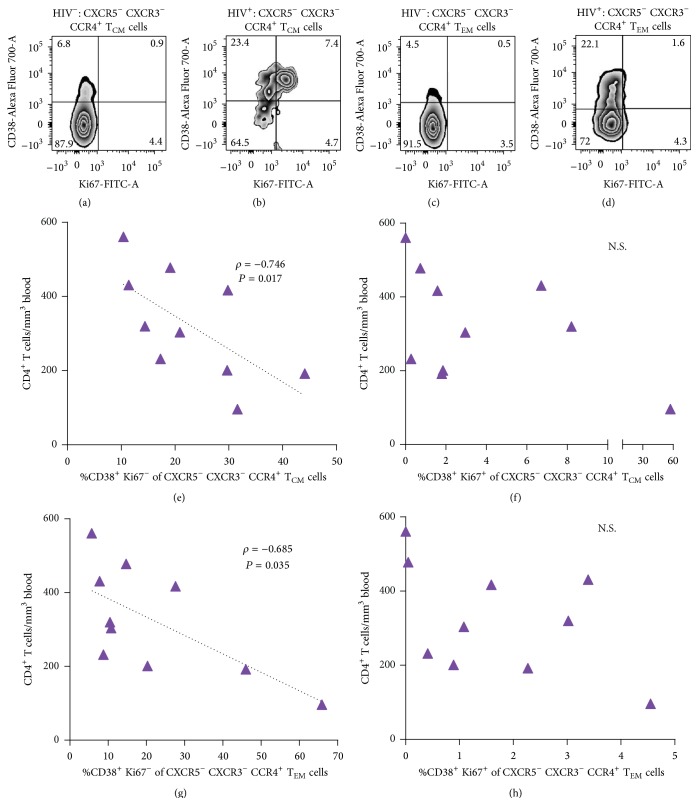
Among the three possible activation phenotypes (CD38+ Ki67−, CD38+ Ki67+, and CD38− Ki67+), the CD38+ Ki67+ phenotype had an increased frequency in patients' CXCR5− CXCR3− CCR4+ TCM cells, compared with their counterparts from controls (P = 0.009, Figures 3(a) and 3(b), upper right quadrants), and comprised the majority of Ki67+ cells, as previously described [35]. Contrastingly, a mean 23.4% of patients' CXCR5− CXCR3− CCR4+ TCM cells showed Ki67-uncoupled CD38 expression (CD38+ Ki67−, Figure 3(b)), which comprised a majority of all CD38+ cells, and were also significantly more frequent, compared with controls (P = 0.0003).
Figure 3.
Relevant activation phenotypes and univariate correlation with CD4+ T cell counts. (a) Contour plot of CD38 and/or Ki67 expression in the CXCR5− CCR4+ CXCR3− subset of TCM cells from a representative HIV− control and a representative HIV+ patient. (b) CD38 and/or Ki67 expression on the CXCR5− CCR4+ CXCR3− subset of TEM cells from a representative HIV− control and a representative HIV+ patient. Numbers in each quadrant correspond to the group's mean frequency, as a percentage of the CXCR5− CXCR3− CCR4+ subset. (c) Correlation of total circulating CD4+ T cell counts in HIV+ patients with the percentage of CXCR5− CXCR3− CCR4+ TCM cells with the CD38+ Ki67− phenotype or (d) with the CD38+ Ki67+ phenotype. (e) Correlation of total circulating CD4+ T cell counts in HIV+ patients with the percentage of CXCR5− CXCR3− CCR4+ TEM cells with the CD38+ Ki67− phenotype or (f) with the CD38+ Ki67+ phenotype. Analysis was made with Spearman's correlation.
Patients' TEM cell subset with the same pattern of expression of chemokine receptors (CXCR5− CXCR3− CCR4+) showed an increased percent of Ki67-uncoupled CD38 expression (mean 22.1%, Figures 3(c) and 3(d), upper left quadrants), also significantly greater than its percentage among the controls' counterparts (P = 0.0001). Notably, the frequency of cells coexpressing CD38 and Ki67 did not differ between patients and controls and was negligible (Figures 3(c) and 3(d), lower right quadrant).
The frequency of the CD38+ Ki67− phenotype on CXCR5− CXCR3− CCR4+ TCM cells (Figure 3(e)) and on CXCR5− CXCR3− CCR4+ TEM (Figure 3(g)) showed a significant negative correlation with CD4+ T cell counts (ρ = −0.746, P = 0.017, and ρ = −0.685, P = 0.035, resp.), while the frequencies of the CD38+ Ki67+ phenotype on these subsets showed no significant correlation with CD4+ T cell counts (Figures 3(f) and 3(h)).
Cycling of other cell subsets, like CXCR5− CXCR3− CCR4+ TEM cells, as well as CXCR5+ CXCR3+ CCR4− TCM and TEM cells, was also negatively correlated with CD4+ T cell counts; however, these associations were lost in multivariate analysis.
3.3. The CD38+ Ki67− Phenotype Is the Only Negative Correlate of an Underrepresented TCM Cell Subset
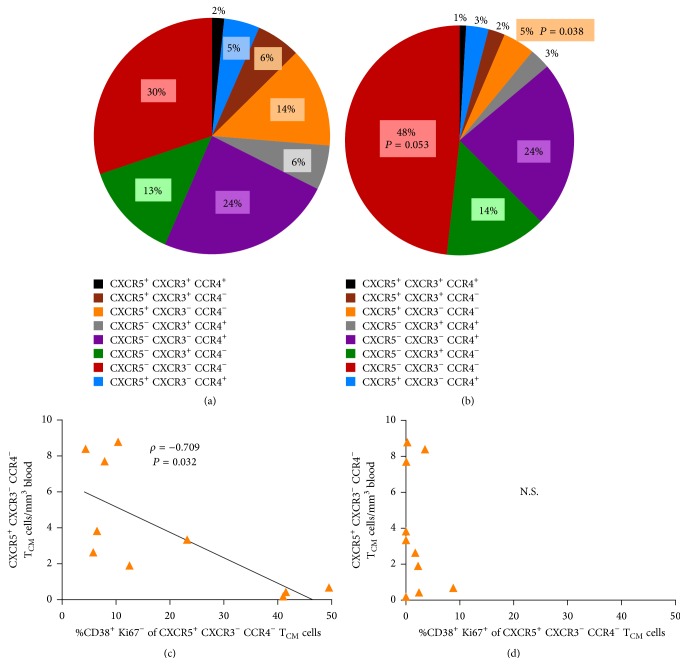
The proportion of CXCR5+ CXCR3− CCR4− cells among total TCM cells (14 ± 4.165%, mean ± 1 SEM, Figure 4(a)) was significantly reduced in HIV+ patients (5% ± 1.138%) compared with controls (P = 0.038, Figure 4(b)). The absolute counts of cells from this TCM subset correlated negatively with their own %CD38+ Ki67− (ρ = −0.709, P = 0.032) but not with their %CD38+ Ki67+ (Figures 4(c) and 4(d)), which constitutes another instance of the Ki67-uncoupled CD38 expression as a negative correlate of CD4 T cell counts.
Figure 4.
Effect of HIV on the relative subset composition of TCM cells and its association with activation. Each slice of the pie chart corresponds to the mean percentage of each subset from a group, color-coded according to the list below (a) and (b). Pie charts correspond to controls' TCM cells (a) and patients' TCM cells (b). P values correspond to the comparison of patients and controls with Mann-Whitney's test. (c, d) Correlation of absolute counts of CXCR5+ CXCR3− CCR4− TCM cells from patients with the percentage of this subset with the CD38+ Ki67− activated phenotype (c) or with the CD38+ Ki67+ phenotype (d).
There were analogous changes in the subset composition of TEM cells, where both the underrepresentation of CXCR5+ CXCR3− CCR4− cells and the overrepresentation of CXCR5− CXCR3− CCR4− cells were significant (P = 0.008, P = 0.014, correspondingly, not shown). TN and TEMRA cells were almost entirely CXCR5− CXCR3− and had only a small percentage of CCR4+ cells within CXCR5− cells (not shown).
4. Discussion
In the present cross-sectional study of patients and controls, we found evidence that CD38 expression determines the correlation of activation and HIV disease progression independently of cell cycling, adjusted to a multivariate model that accounts for T cell maturation subpopulations and subsets within them. According to our analysis, cycling and noncycling cells from a CXCR5− CXCR3− CCR4+ subset of central memory CD4+ T cells (previously reported as “pre-Th2” cells [20]) clustered together on the basis of CD38 expression. In turn, this cluster was a negative correlate of circulating CD4+ T cell counts and a positive correlate of viral load. The small sample size of this study may have hampered the recognition of additional multivariate correlates of HIV disease progression. Also, since we did not determine additional indicators of activation, like, for instance, metabolic changes [36], the present study does not address the actual activation state of noncycling CD38+ cells, a sizable proportion of CD38+ cells. Despite these limitations, the thoroughness of our analysis evidences the fact that CD38 expression in a particular subset of CD4+ T cells has an inherent relevance as a correlate of CD4+ T cell counts, independent of whether CD38+ cells are in cycle.
Chronic T cell activation in HIV disease has been determined phenotypically by the detection of surface molecules (activation markers) like CD69, CD25 [37, 38], and notably CD38 and HLADR [2], which have different functions and are thus associated with different cellular processes [39]. The expression of Ki67, indicating cycling and turnover of T cells, has been used as a surrogate of activation markers [6]. It may correlate with the expression of phenotypic activation markers [35], and more importantly, it correlates with decreased CD4+ T cell numbers [5, 40]. Cycling may have a causal role in CD4+ T cell loss in HIV infection, since cycling CD4+ T cells from HIV+ patients show an increased turnover [9], are under cycle arrest [41], or die after entering S phase [42]. In contrast, whether activation markers, especially CD38 [2], reflect or participate in other processes leading to HIV disease progression remains to be elucidated.
The independence of CD38 in the present study as a correlate of disease progression has important implications. Either CD38 expression in “pre-Th2” TCM cells is reflecting pathogenic processes leading to CD4 T cell loss or CD38 itself is participating in pathogenic mechanisms. In this regard, even though CD38 expression is highly correlated with T cell activation, its function in this process remains unknown. CD38 is a well-known ectoenzyme that catalyzes the transformation of NAD into ADPR, cADPR, and NAADP, and these “second messengers” can regulate the functionality of T cells [43–46]. Also regarding a possible role of CD38, we have previously reported that CD38+ CD4+ TCM cells from HIV-infected patients show a response to TCR engagement dominated by IFN-γ, instead of IL-2, and disconnected of CD40L induction, as well as lack of response to CD28 costimulation [47, 48]. Such functionality could be less propitious to proliferation, a requirement for self-renewal and differentiation [21, 49], and might thus underlie the association of CD38 expression on “pre-Th2 TCM” (CXCR5− CXCR3− CCR4+) cells and overall CD4+ T cell loss in patients with HIV infection. These previous studies, along with our present findings, make a strong case for CD38 as an actor in pathogenesis of HIV disease. Mechanistic studies of this molecule in cells from patients with HIV are warranted.
There are additional ways in which activation of pre-Th2 TCM cells could lead to depletion. Among human helper T cells, CCR4 is distinctly expressed by cells that express GATA3 and produce IL-4 when stimulated [50]. CCR4 expression in Th2-polarized central memory and effector memory cells directs homing to lungs [51–53] or other tissues with inflammation [54, 55], which are sites of increased CD4+ T cell turnover during chronic HIV infection [23, 31]. Importantly, CD38 expression may make them permissive to HIV infection [56, 57]. Additionally, IL-4 has been found to enable productive infection of CD38+ T cells by X4-tropic HIV-1 [58].
We observed a significant underrepresentation of CXCR5+ CXCR3− CCR4− cells among TCM and TEM cells from HIV+ patients. CXCR5+ CXCR3− CCR4− TCM cells have been shown in vitro to self-renew by proliferating without differentiating after TCR-mediated or homeostatic cytokines [20]. Although the frequency of this TCM subset was not a multivariate correlate of CD4+ T cell counts, it could be expected that TCM cells from HIV-infected patients would be less able to maintain the memory pool, given their reduced percentage of CXCR5+ CXCR3− CCR4− cells. Additionally, the use of surface CXCR5 in our subset delineation brings forth the possibility that CXCR5+ CXCR3− CCR4− TCM and TEM cells could contain circulating cellular subsets with a follicular helper functionality [28, 59, 60]. Their relative underrepresentation contrasts with the previously reported expansion of dysfunctional follicular helper cells in lymph nodes from HIV+ patients [61]. The knowledge of the relationship between these two B cell-helping compartments could help understand this contrast.
5. Conclusions
Our study addresses the call for studying the biology of the diverse activation markers in the context of HIV infection [62]. Our findings indicate that focusing research on CD38's has informative potential, by possibly showing pathogenic mechanisms reflected by CD38 expression or mechanisms in which CD38 participates. Studying the subset level within the CD4+ maturation subpopulations is meaningful in HIV pathogenesis research.
Supplementary Material
Table A shows the panel of fluorochrome-conjugated antibodies used for cell phenotyping by flow cytometry, including clone and origin. Table B shows the mixture of antibodies used to delineate positive and negative cells in the cytometry plots used to detect the expression of each marker.
Acknowledgments
The authors gratefully thank Audelia Alanís, from INCMNS, for assuring the availability of samples, and thank patients and controls that donated the blood samples. This study was funded with research Grant no. FOSISS-2009-114484 from Consejo Nacional de Ciencia y Tecnología, Mexico, and National Institutes of Health R01 Grant no. AI91526.
Competing Interests
The authors declare that there are no competing interests.
Authors' Contributions
Daniela Würsch took and processed the samples, performed the experiments, and analyzed flow cytometry data. Christopher E. Ormsby performed the statistical analysis, Dámaris P. Romero-Rodríguez took samples, did flow cytometry analysis, and reviewed the paper critically, Gustavo Olvera-García made a critical review of the experiments and the paper, Joaquín Zúñiga critically reviewed the project and the paper, Wei Jiang interpreted results and critically reviewed the paper, and Santiago Pérez-Patrigeon designed the study, recruited patients, and wrote the paper. Enrique Espinosa conceived and designed the study, analyzed and interpreted results, and wrote the paper. Daniela Würsch and Christopher E. Ormsby contributed equally to this work.
References
- 1.Deeks S. G., Kitchen C. M. R., Liu L., et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104(4):942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 2.Karim R., Mack W. J., Stiller T., et al. Association of HIV clinical disease progression with profiles of early immune activation: results from a cluster analysis approach. AIDS. 2013;27(9):1473–1481. doi: 10.1097/qad.0b013e3283601bad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carbone J., Gil J., Benito J. M., et al. Increased levels of activated subsets of CD4 T cells add to the prognostic value of low CD4 T cell counts in a cohort of HIV-infected drug users. AIDS. 2000;14(18):2823–2829. doi: 10.1097/00002030-200012220-00003. [DOI] [PubMed] [Google Scholar]
- 4.Paiardini M., Müller-Trutwin M. HIV-associated chronic immune activation. Immunological Reviews. 2013;254(1):78–101. doi: 10.1111/imr.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sousa A. E., Carneiro J., Meier-Schellersheim M., Grossman Z., Victorino R. M. M. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. The Journal of Immunology. 2002;169(6):3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- 6.Orenstein J. M., Bhat N., Yoder C., et al. Rapid activation of lymph nodes and mononuclear cell HIV expression upon interrupting highly active antiretroviral therapy in patients after prolonged viral suppression. AIDS. 2000;14(12):1709–1715. doi: 10.1097/00002030-200008180-00004. [DOI] [PubMed] [Google Scholar]
- 7.Hazenberg M. D., Cohen Stuart J. W. T., Otto S. A., et al. T-cell division in human immunodeficiency virus (HIV)-1 infection is mainly due to immune activation: a longitudinal analysis in patients before and during highly active antiretroviral therapy (HAART) Blood. 2000;95(1):249–255. [PubMed] [Google Scholar]
- 8.Yamamoto S., Ibusuki M., Yamamoto Y., et al. Clinical relevance of Ki67 gene expression analysis using formalin-fixed paraffin-embedded breast cancer specimens. Breast Cancer. 2013;20(3):262–270. doi: 10.1007/s12282-012-0332-7. [DOI] [PubMed] [Google Scholar]
- 9.Ribeiro R. M., Mohri H., Ho D. D., Perelson A. S. In vivo dynamics of t cell activation, proliferation, and death in HIV-1 infection: why are CD4+ but not CD8+ T cells depleted? Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15572–15577. doi: 10.1073/pnas.242358099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazenberg M. D., Otto S. A., Hamann D., et al. Depletion of naive CD4 T cells by CXCR4-using HIV-1 variants occurs mainly through increased T-cell death and activation. AIDS. 2003;17(10):1419–1424. doi: 10.1097/00002030-200307040-00001. [DOI] [PubMed] [Google Scholar]
- 11.Orendi J. M., Bloem A. C., Borleffs J. C. C., et al. Activation and cell cycle antigens in CD4+ and CD8+ T cells correlate with plasma human immunodeficiency virus (HIV-1) RNA level in HIV-1 infection. Journal of Infectious Diseases. 1998;178(5):1279–1287. doi: 10.1086/314451. [DOI] [PubMed] [Google Scholar]
- 12.Papagno L., Spina C. A., Marchant A., et al. Immune activation and CD8+ T-cell differentiation towards senescence in HIV-1 infection. PLoS Biology. 2004;2(2) doi: 10.1371/journal.pbio.0020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Presicce P., Orsborn K., King E., Pratt J., Fichtenbaum C. J., Chougnet C. A. Frequency of circulating regulatory T cells increases during chronic HIV infection and is largely controlled by highly active antiretroviral therapy. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028118.e28118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eller M. A., Blom K. G., Gonzalez V. D., et al. Innate and adaptive immune responses both contribute to pathological cd4 t cell activation in hiv-1 infected ugandans. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0018779.e18779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaspan H. B., Liebenberg L., Hanekom W., et al. Immune activation in the female genital tract during HIV infection predicts mucosal CD4 depletion and HIV shedding. Journal of Infectious Diseases. 2011;204(10):1550–1556. doi: 10.1093/infdis/jir591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Read S. W., DeGrezia M., Ciccone E. J., et al. The effect of leflunomide on cycling and activation of T-cells in HIV-1-infected participants. PLoS ONE. 2010;5(8) doi: 10.1371/journal.pone.0011937.e11937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carsenti-Dellamonica H., Saïdi H., Ticchioni M., et al. The suppression of immune activation during enfuvirtide-based salvage therapy is associated with reduced CCR5 expression and decreased concentrations of circulating interleukin-12 and IP-10 during 48 weeks of longitudinal follow-up. HIV Medicine. 2011;12(2):65–77. doi: 10.1111/j.1468-1293.2010.00848.x. [DOI] [PubMed] [Google Scholar]
- 18.Antonelli L. R. V., Mahnke Y., Hodge J. N., et al. Elevated frequencies of highly activated CD4+ T cells in HIV+ patients developing immune reconstitution inflammatory syndrome. Blood. 2010;116(19):3818–3827. doi: 10.1182/blood-2010-05-285080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez S., Price P., McKinnon E. J., Nolan R. C., French M. A. Low CD4+ T-cell counts in HIV patients receiving effective antiretroviral therapy are associated with CD4+ T-cell activation and senescence but not with lower effector memory T-cell function. Clinical Immunology. 2006;120(2):163–170. doi: 10.1016/j.clim.2006.04.570. [DOI] [PubMed] [Google Scholar]
- 20.Rivino L., Messi M., Jarrossay D., Lanzavecchia A., Sallusto F., Geginat J. Chemokine receptor expression identifies pre-T helper (Th)1, pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. Journal of Experimental Medicine. 2004;200(6):725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang A., Chandran S., Shah S. A., et al. The stoichiometric production of IL-2 and IFN-γmRNA defines memory T cells that can self-renew after adoptive transfer in humans. Science Translational Medicine. 2012;4(149) doi: 10.1126/scitranslmed.3004306.149ra120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Chang Li X., Xiao X., Sun R., Tian Z., Wei H. CD4+CD62L+ central memory T cells can be converted to Foxp3+ T Cells. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0077322.e77322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okoye A. A., Picker L. J. CD4+ T-cell depletion in hiv infection: mechanisms of immunological failure. Immunological Reviews. 2013;254(1):54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Grevenynghe J., Procopio F. A., He Z., et al. Transcription factor FOXO3a controls the persistence of memory CD4+ T cells during HIV infection. Nature Medicine. 2008;14(3):266–274. doi: 10.1038/nm1728. [DOI] [PubMed] [Google Scholar]
- 25.Bai F., Tincati C., Merlini E., et al. Reduced central memory CD4+ T cells and increased T-cell activation characterise treatment-naive patients newly diagnosed at late stage of hiv infection. AIDS Research and Treatment. 2012;2012:10. doi: 10.1155/2012/314849.314849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter S. J., Lacabaratz C., Lambotte O., et al. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. Journal of Virology. 2007;81(24):13904–13915. doi: 10.1128/jvi.01401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim C. H., Rott L., Kunkel E. J., et al. Rules of chemokine receptor association with T cell polarization in vivo. The Journal of Clinical Investigation. 2001;108(9):1331–1339. doi: 10.1172/jci200113543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chevalier N., Jarrossay D., Ho E., et al. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. The Journal of Immunology. 2011;186(10):5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 29.Cohen K., Altfeld M., Alter G., Stamatatos L. Early preservation of CXCR5+ PD-1+ helper T cells and B cell activation predict the breadth of neutralizing antibody responses in chronic HIV-1 infection. Journal of Virology. 2014;88(22):13310–13321. doi: 10.1128/jvi.02186-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner J. E., Steinmetz O. M., Stahl R. A., Panzer U. Targeting of Th1-associated chemokine receptors CXCR3 and CCR5 as therapeutic strategy for inflammatory diseases. Mini-Reviews in Medicinal Chemistry. 2007;7(11):1089–1096. doi: 10.2174/138955707782331768. [DOI] [PubMed] [Google Scholar]
- 31.Grossman Z., Meier-Schellersheim M., Paul W. E., Picker L. J. Pathogenesis of HIV infection: what the virus spares is as important as what it destroys. Nature Medicine. 2006;12(3):289–295. doi: 10.1038/nm1380. [DOI] [PubMed] [Google Scholar]
- 32.Al-Banna N. A., Vaci M., Slauenwhite D., Johnston B., Issekutz T. B. CCR4 and CXCR3 play different roles in the migration of T cells to inflammation in skin, arthritic joints, and lymph nodes. European Journal of Immunology. 2014;44(6):1633–1643. doi: 10.1002/eji.201343995. [DOI] [PubMed] [Google Scholar]
- 33.Brenchley J. M., Knox K. S., Asher A. I., et al. High frequencies of polyfunctional HIV-specific T cells are associated with preservation of mucosal CD4 T cells in bronchoalveolar lavage. Mucosal Immunology. 2008;1(1):49–58. doi: 10.1038/mi.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. http://www.R-project.org/ [Google Scholar]
- 35.Jiang W., Souheil-Antoine Y., Funderburg N. T., et al. Cycling memory CD4+ T cells in HIV disease have a diverse T cell receptor repertoire and a phenotype consistent with bystander activation. Journal of Virology. 2014;88(10):5369–5380. doi: 10.1128/jvi.00017-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer C. S., Ostrowski M., Gouillou M., et al. Increased glucose metabolic activity is associated with CD4+ T-cell activation and depletion during chronic HIV infection. AIDS. 2014;28(3):297–309. doi: 10.1097/qad.0000000000000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sieg S. F., Rodriguez B., Asaad R., Jiang W., Bazdar D. A., Lederman M. M. Peripheral S-phase T cells in HIV disease have a central memory phenotype and rarely have evidence of recent T cell receptor engagement. Journal of Infectious Diseases. 2005;192(1):62–70. doi: 10.1086/430620. [DOI] [PubMed] [Google Scholar]
- 38.Bordoni V., Casetti R., Capuano G., et al. A novel 8-color flow cytometry panel to study activation, maturation and senescence of CD4 and CD8 T lymphocytes in HIV-infected individuals at different stages of disease. International Journal of Immunopathology and Pharmacology. 2012;25(2):415–424. doi: 10.1177/039463201202500211. [DOI] [PubMed] [Google Scholar]
- 39.Chattopadhyay P. K., Roederer M. Good cell, bad cell: flow cytometry reveals T-cell subsets important in HIV disease. Cytometry Part A. 2010;77(7):614–622. doi: 10.1002/cyto.a.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen Stuart J. W. T., Hazebergh M. D., Hamann D., et al. The dominant source of CD4+ and CD8+ T-cell activation in HIV infection is antigenic stimulation. Journal of Acquired Immune Deficiency Syndromes. 2000;25(3):203–211. doi: 10.1097/00126334-200011010-00001. [DOI] [PubMed] [Google Scholar]
- 41.Combadère B., Blanc C., Li T., et al. CD4+Ki67+ lymphocytes in HIV-infected patients are effector T cells accumulated in the G1 phase of the cell cycle. European Journal of Immunology. 2000;30(12):3598–3603. doi: 10.1002/1521-4141(200012)30:12<3598::aid-immu3598>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 42.Sieg S. F., Bazdar D. A., Lederman M. M. S-phase entry leads to cell death in circulating T cells from HIV-infected persons. Journal of Leukocyte Biology. 2008;83(6):1382–1387. doi: 10.1189/jlb.0907643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ernst I. M. A., Fliegert R., Guse A. H. Adenine dinucleotide second messengers and T-lymphocyte calcium signaling. Frontiers in Immunology. 2013;4, article 259 doi: 10.3389/fimmu.2013.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melzer N., Hicking G., Göbel K., Wiendl H. TRPM2 cation channels modulate T cell effector functions and contribute to autoimmune CNS inflammation. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0047617.e47617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis L. C., Morgan A. J., Chen J.-L., et al. NAADP Activates two-pore channels on T cell cytolytic granules to stimulate exocytosis and killing. Current Biology. 2012;22(24):2331–2337. doi: 10.1016/j.cub.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolf I. M. A., Diercks B.-P., Gattkowski E., et al. Frontrunners of T cell activation: Initial, localized Ca2+ signals mediated by NAADP and the type 1 ryanodine receptor. Science Signaling. 2015;8(398) doi: 10.1126/scisignal.aab0863.ra102 [DOI] [PubMed] [Google Scholar]
- 47.Espinosa E., Ormsby C. E., Reyes-Terán G., Asaad R., Sieg S. F., Lederman M. M. Dissociation of CD154 and cytokine expression patterns in CD38+ CD4+ memory T cells in chronic HIV-1 infection. Journal of Acquired Immune Deficiency Syndromes. 2010;55(4):439–445. doi: 10.1097/qai.0b013e3181ef991d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olvera-García G., Espinosa E., Sieg S. F., Lederman M. M. Cytomegalovirus-specific responses of CD38+ memory T cells are skewed towards IFN-γ and dissociated from CD154 in HIV-1 infection. AIDS. 2014;28(3):311–316. doi: 10.1097/QAD.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Appay V., Van Lier R. A. W., Sallusto F., Roederer M. Phenotype and function of human T lymphocyte subsets: consensus and issues. Cytometry Part A. 2008;73(11):975–983. doi: 10.1002/cyto.a.20643. [DOI] [PubMed] [Google Scholar]
- 50.Becattini S., Latorre D., Mele F., et al. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 2015;347(6220):400–406. doi: 10.1126/science.1260668. [DOI] [PubMed] [Google Scholar]
- 51.Mikhak Z., Strassner J. P., Luster A. D. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. Journal of Experimental Medicine. 2013;210(9):1855–1869. doi: 10.1084/jem.20130091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nouri-Aria K. T., Wilson D., Francis J. N., et al. CCR4 in human allergen-induced late responses in the skin and lung. European Journal of Immunology. 2002;32(7):1933–1938. doi: 10.1002/1521-4141(200207)32:7<1933::AID-IMMU1933>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 53.Sato T., Komai M., Iwase M., et al. Inhibitory effect of the new orally active CCR4 antagonist K327 on CCR4+CD4+ T cell migration into the lung of mice with ovalbumin-induced lung allergic inflammation. Pharmacology. 2009;84(3):171–182. doi: 10.1159/000235748. [DOI] [PubMed] [Google Scholar]
- 54.Yang X., Jiao Y.-M., Wang R., et al. High CCR5 density on central memory CD4+ T cells in acute HIV-1 infection is mostly associated with rapid disease progression. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049526.e49526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim S. H., Cleary M. M., Fox H. S., Chantry D., Sarvetnick N. CCR4-bearing T cells participate in autoimmune diabetes. Journal of Clinical Investigation. 2002;110(11):1675–1686. doi: 10.1172/JCI200215547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gosselin A., Monteiro P., Chomont N., et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+ CD4+ T cells are highly permissive to HIV-1 infection. Journal of Immunology. 2010;184(3):1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agrawal L., Vanhorn-Ali Z., Alkhatib G. Multiple determinants are involved in HIV coreceptor use as demonstrated by CCR4/CCL22 interaction in peripheral blood mononuclear cells (PBMCs) Journal of Leukocyte Biology. 2002;72(5):1063–1074. [PubMed] [Google Scholar]
- 58.Li Y.-G., Iwabu Y., Warachit J., et al. Interleukin-4 up-regulates T-tropic human immunodeficiency virus type 1 transcription in primary CD4+ CD38+ T-lymphocyte subset. Microbiology and Immunology. 2005;49(2):155–165. doi: 10.1111/j.1348-0421.2005.tb03715.x. [DOI] [PubMed] [Google Scholar]
- 59.Locci M., Havenar-Daughton C., Landais E., et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39(4):758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morita R., Schmitt N., Bentebibel S.-E., et al. Human blood CXCR5+CD4+ T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34(1):108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lindqvist M., van Lunzen J., Soghoian D. Z., et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. The Journal of Clinical Investigation. 2012;122(9):3271–3280. doi: 10.1172/jci64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cockerham L. R., Siliciano J. D., Sinclair E., et al. CD4+ and CD8+ T cell activation are associated with HIV DNA in resting CD4+ T cells. PLoS ONE. 2014;9(10) doi: 10.1371/journal.pone.0110731.e110731 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A shows the panel of fluorochrome-conjugated antibodies used for cell phenotyping by flow cytometry, including clone and origin. Table B shows the mixture of antibodies used to delineate positive and negative cells in the cytometry plots used to detect the expression of each marker.