Abstract
Prediction of gene regulatory sequences using phylogenetic footprinting has advanced considerably but lacks experimental validation. Here, we report whether transcription factor binding sites predicted by dot plotting or web-based Trafac analysis could be validated by chromatin immunoprecipitation assays. MYC overexpression enhances glycolysis without hypoxia and hence may contribute to altered tumor metabolism. Because the full spectrum of glycolytic genes directly regulated by Myc is not known, we chose Myc as a model transcription factor to determine whether it binds target glycolytic genes that have conserved canonical Myc binding sites or E boxes (5′-CACGTG-3′). Conserved canonical E boxes in ENO1, HK2, and LDHA occur in 31- to 111-bp islands with high interspecies sequence identity (>65%). Trafac analysis revealed another region in ENO1 that corresponds to a murine region with a noncanonical E box. Myc bound all these conserved regions well in the human P493-6 B lymphocytes. We also determined whether Myc could bind nonconserved canonical E boxes found in the remaining human glycolytic genes. Myc bound PFKM, but it did not significantly bind GPI, PGK1, and PKM2. Binding to BPGM, PGAM2, and PKLR was not detected. Both GAPD and TPI1 do not have conserved E boxes but are induced and bound by Myc through regions with noncanonical E boxes. Our results indicate that Myc binds well to conserved canonical E boxes, but not nonconserved E boxes. However, the binding of Myc to unpredicted genomic regions with noncanonical E boxes reveals a limitation of phylogenetic footprinting. In aggregate, these observations indicate that Myc is an important regulator of glycolytic genes, suggesting that MYC plays a key role in a switch to glycolytic metabolism during cell proliferation or tumorigenesis.
Defining transcriptional regulatory networks is essential for our understanding of embryonic development, cell growth, and tumorigenesis. Throughout evolution, biologically important genes and their regulatory elements have been selectively conserved (34). Completing sequencing the genomes of a variety of species provides a unique opportunity for the identification of transcriptional regulatory regions through interspecies sequence comparison, also known as phylogenetic footprinting. In particular, noncoding sequences with interspecies sequence identity approaching that of exonic sequences are enriched with putative transcription factor binding sites.
A number of approaches that allow the prediction of transcriptional regulatory regions in any genomic region through comparisons of mouse and human sequences have been reported (4, 19, 21, 24). These approaches identify conserved regions that contain putative transcription factor binding sites. However, a purely computational approach tends to be replete with the uncertainty as to whether a predicted cis-regulatory module is biologically functional.
Several algorithms have been developed and experimentally evaluated for the discovery of candidate regulatory regions, such as those in the Drosophila genome (2, 26). In mammalian systems, we and other groups have evaluated the functionality of the transcriptional regulation of conserved cis elements. These studies, however, addressed individual genes or a small set of genes rather than a series of functionally related genes, such as those encoding a specific biochemical pathway. In our previous studies, putative direct Myc target genes were randomly selected and subjected to chromatin immunoprecipitation (ChIP) assays to identify Myc binding sites (43). In addition to class I genes in which Myc binding regions are highly conserved among species, we identified another group of genes termed class II genes, in which Myc binding regions do not contain conserved sequences (18). Thus, experimental validation of these computational approaches is particularly important in a well-defined model system involving a set of coordinately regulated genes, such as those encoding components of a metabolic pathway.
Although Myc and its target genes have been studied at a broader genome-wide level (6, 7, 14, 16, 17, 23, 25, 27, 29-31, 35, 41), the coupling of comparative interspecies sequence analysis and experimental validation of Myc target genes involved in a single metabolic pathway has not been thoroughly studied. It is particularly intriguing to note that ODC is not only the first identified bona fide Myc target gene, but it also contains a phylogenetically conserved intronic region bearing tandem canonical E boxes (1). MYC overexpression has been suggested to aberrantly enhance tumor glycolysis even in the presence of oxygen, a phenomenon termed the Warburg effect (8, 39). Thus, we chose Myc as a model transcription factor and the glycolytic genes as model target genes to predict Myc binding regulatory regions, which can then be experimentally tested. We have previously found that Myc specifically transactivates LDHA and increases the expression of other glycolytic enzyme genes (32, 37). Numerous studies using global gene expression profiling methods, such as serial analysis of gene expression (SAGE) and DNA microarrays, have found that Myc increases the expression of specific glycolytic enzyme genes, though these increases may be direct or indirect effects of Myc (27-29, 35).
To identify the direct target genes of Myc and its binding sites, we and other groups have applied various assays, including an in vitro reporter assay, electrophoresis mobility shift assay (EMSA), and the Myc-estrogen receptor (MYC-ER) system (11). Through the use of the MYC-ER system, several glycolytic genes, such as ENO1, GPI, HK2, LDHA, and PFKM, have been identified as direct targets of Myc (7, 29, 32, 35). However, these experimental approaches did not provide physical evidence that Myc directly activates the transcription of these genes through its association with specific genomic regions. The MYC-ER fusion protein system has been particularly used as a standard for the study of direct Myc target genes, as it allows the identification of MYC-ER-induced targets upon estrogenic ligand stimulation in the presence of cycloheximide, which prevents secondary transcriptional events (11). However, estrogenic ligands and cycloheximide used in these experiments may confound the effects of Myc, and the MYC-ER system is unable to reveal Myc target genes that are in feed-forward loops. In these loops, the expression of terminal target genes are dependent on both Myc and an intermediate transcription factor that is also a direct target of Myc.
We reasoned that functionally important genomic regions for Myc binding have been preferentially conserved in the direct target genes. Given that exonic sequence identity in the human and mouse genomes is estimated at about 85% (40), we have performed manual sequence alignments using dot plotting to identify nonexonic regions with at least 65% sequence identity in 30-bp segments. These cutoff criteria are adequately stringent to predict a particular class of Myc target genes, as previously reported (18, 43). We also used the Trafac server to identify potential Myc binding sites in glycolytic genes (19). A number of programs, including RepeatMasker (masking out repeat elements), PipMaker-BLASTZ (sequence alignment algorithm), and MatInspector Professional (transcription factor binding sequence scan), are integrated into the Trafac system to perform phylogenetic footprinting analysis (19). Trafac analysis predicts both canonical and noncanonical E boxes that reside in regions that have at least 50% sequence identity in the human and mouse genomes. Within these conserved segments, we sought to identify canonical Myc binding sites or E boxes with the consensus sequence 5′-CACGTG-3′ and determine whether Myc could bind these regions by ChIP assays (10, 14, 23). By performing a ChIP assay, we can identify immunoprecipitated regions of the genome that are cross-linked to the bound Myc protein by amplifying the Myc-associated DNA fragments by PCR.
Our approach using Myc and 14 glycolytic genes as a model provides a unique opportunity not only to evaluate phylogenetic footprinting and determine the architecture of the Myc target glycolytic gene network but also to dissect the molecular basis of Myc-induced altered glucose metabolism. Our results provide evidence that MYC enhances aerobic glycolysis by directly up-regulating the expression of ENO1, GAPD, HK2, LDHA, PFKM, and TPI1 genes, whereas Myc binding to GPI, PGK1, and PKM is diminished or absent in the cases of BPGM, PGAM2 (muscle specific), and PKLR (liver specific). This study indicates that conserved, canonical E boxes are predictive of significant Myc binding to glycolytic target genes, but the absence of canonical E boxes does not exclude the possibility of significant Myc association.
MATERIALS AND METHODS
Phylogenetic footprinting analysis.
Genomic or cDNA sequences were downloaded from the University of California at Santa Cruz Genome Bioinformatics website (http://genome.ucsc.edu), Ensembl Genome Browser (http://www.ensembl.org), or National Center for Biotechnology Information (NCBI) Reference Sequence database (http://www.ncbi.nlm.nih.gov/RefSeq/). Canonical (5′-CACGTG-3′) or noncanonical (5′-CATGTG-3′, 5′-CACGCG-3′, 5′-CATGCG-3′, 5′-CACGAG-3′, 5′-CTCGCG-3′, and 5′-CACGTTG-3′) Myc binding sites (E boxes) (3) were identified within the 5 kb upstream of the transcriptional start site through intron 1 using the user-defined nucleic acid motifs feature of OMIGA software (Oxford Molecular Limited, Oxford, United Kingdom). Computational comparison of phylogenetically conserved canonical E boxes and extended flanking sequences was performed using the dot plot feature of OMIGA software with the parameters of a 30-bp window and more than 65% sequence identity. These parameters are less stringent than the criterion of 70% sequence identity in 50-bp windows, which has a sensitivity of only 65% (21). Identifying and visualizing shared transcription factor binding sites in the phylogenetically conserved regulatory regions were performed using a web-based phylogenetic footprinting analysis called Trafac (http://trafac.cchmc.org) as described previously (19).
Cell lines.
The human Burkitt's lymphoma cell line P493-6 carrying an inducible MYC repression system (35) and human breast epithelial A1N4 cells (22) were used for ChIP analysis. P493-6 cells were maintained in RPMI 1640 medium with 10% fetal bovine serum (GIBCO/BRL) and 1% streptomycin and penicillin (Invitrogen). A1N4 cells were maintained in Improved MEM Zinc Option medium (Invitrogen) with 0.5% fetal bovine serum (GIBCO/BRL), 10 ng of epidermal growth factor (EGF) (Invitrogen) per ml, 0.5 ng of hydrocortisone (Clonetics) per ml, and 5 ng of insulin (Clonetics) per ml. Incubating P493-6 cells with 0.1 μg of tetracycline per ml for 72 h led to significant repression of MYC.
ChIP assay.
P493-6 cells left untreated or treated with tetracycline for 72 h and human breast epithelial A1N4 cells stimulated by 20 ng of EGF (Invitrogen) per ml for 24 h were used for all ChIP assays. Cells were cross-linked by formaldehyde, and chromatin was immunoprecipitated as described previously (5). The rabbit polyclonal Myc antibody (sc-764; Santa Cruz Biotechnology) and human hepatocyte growth factor (HGF) antibody (sc-7949; Santa Cruz Biotechnology) were used to precipitate chromatin. The total input was the supernatant from the no-antibody control. For a control, some samples were treated the same as the other samples, but these samples had no chromatin (mock control samples). Real-time PCR quantitation of precipitated chromatin fragments was performed using a SYBR green core reagent kit (PE Applied Biosystems) on an ABI 7700 sequence detection system (PE Applied Biosystems) according to the manufacturer's instructions. Primers were designed using Primer Express software (PE Applied Biosystems) (see Table S1 in the supplemental material for the primer sequences for the ChIP assay). The data were analyzed by SDS 1.91 software (PE Applied Biosystems). Four serial 10-fold dilutions of total input DNA were used to generate a standard curve for each primer pair. Relative amounts of each chromatin fragment were then extrapolated on the basis of their threshold cycle values and determined by the percentage of the total input DNA. For each amplification, melting curves and gel electrophoresis of the PCR product were used to verify the identities of the PCR products. All real-time PCRs were performed in triplicate.
RNA analysis.
MYC and glycolytic mRNA levels were determined by Northern blot analysis or quantitative real-time reverse transcription-PCR (RT-PCR). Total RNA was isolated from P493-6 cells using Trizol (Invitrogen). Five micrograms of RNA was used in Northern blot analysis. RNA was subjected to 1.2% agarose electrophoresis and transferred to a nylon membrane (Nytran). The membrane was probed with a human MYC gene probe or a glycolytic gene probe which had been labeled with 32P using a random primer labeling kit (Stratagene). An ethidium bromide-stained agarose gel of 18S rRNA was used as a loading control.
Quantitative real-time RT-PCR was performed using TaqMan one-step RT-PCR master mix kit (PE Applied Biosystems) with probes and primers. Alternatively, cDNA was reverse transcribed from the total RNA (2 μg) using TaqMan reverse transcription reagents (PE Applied Biosystems), and subsequent real-time PCR of cDNA was performed using the SYBR green core reagent kit on an ABI 7700 sequence detection system. Primers and probes were designed by the Primer Express software (see Table S2 in the supplemental material for the primers and probes). Amplified fragments span intron/exon boundaries in the cDNA sequences. The expression level of human 18S RNA was determined by a predeveloped mixture of TaqMan probe and primers (PE Applied Biosystems) and used for normalization. All PCRs were performed in triplicate.
Western blotting.
Equal amounts of protein extracted from P493-6 cells were subjected to electrophoresis on sodium dodecyl sulfate-10% polyacrylamide gels. Monoclonal anti-Myc antibody (9E10; Oncogene Research Products) and anti-α-tubulin antibody (CP06; Oncogene Research Products) were used for immunoblotting.
RESULTS
Identification of canonical E boxes in glycolytic genes.
The genomic sequences spanning 5 kb upstream of the transcriptional start site through the first exon and including the entire intron 1 of 14 human glycolytic genes (except LDHB) and their murine orthologs were analyzed to identify canonical E boxes (5′-CACGTG-3′). Although it is possible that Myc binding sites may occur outside the regions we studied, this coverage was determined from the precedence that bona fide Myc binding sites appear to cluster within 2 kb of the start site, and there is a prevalence of Myc binding sites in many first introns (14). The analysis shows the locations of canonical E boxes in human and mouse glycolytic genes (Fig. 1 and 2). As summarized in Table 1, canonical E boxes occur in 12 human glycolytic genes but not in GAPD and TPI1. Among the mouse glycolytic genes, Gpi, Pfkm, and Pklr have no canonical E boxes. Intronic canonical E boxes occur in several glycolytic genes. Glycolytic genes in which canonical E boxes were found in both human and mouse genomic sequences were subjected to phylogenetic footprinting to determine whether these human E boxes are phylogenetically conserved.
FIG. 1.
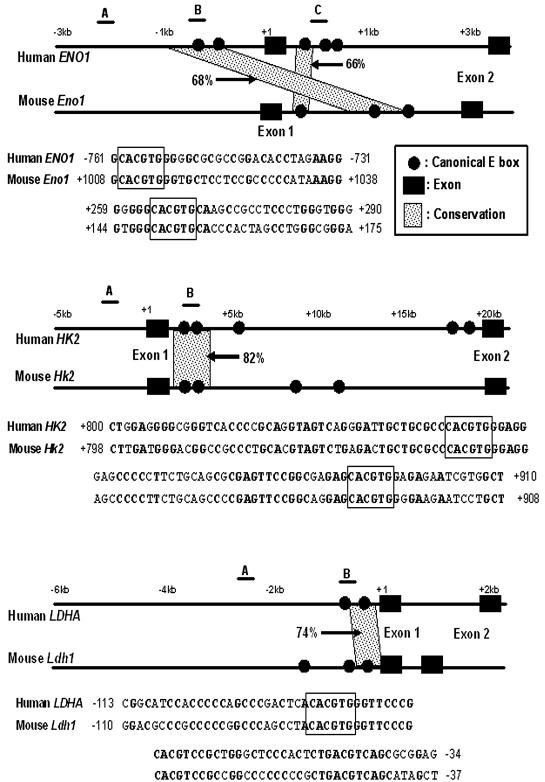
Locations of canonical E boxes in the human and mouse genomic sequences and phylogenetic footprinting analysis. Genomic sequences containing 5 kb upstream of the transcriptional start site through intron 1 were screened for the presence of the canonical E box. The positions of canonical E boxes and exons are mapped in the human (top of each pair) and mouse (bottom of each pair) genomic sequences. The maps show phylogenetically conserved canonical E boxes in ENO1, HK2, and LDHA genes. Conserved canonical E boxes were identified using the dot plot features of OMIGA software. Conservation of the canonical E box and its extended flanking region with more than 65% sequence identity for longer than 30 bp is indicated. The percentages of sequence identity between the two sequences are also indicated in the maps. Sequence alignments of the conserved E box and its extended flanking regions are shown below the maps. Canonical E boxes are boxed in each sequence alignment. Conserved nucleotides are shown in bold type. The regions that are amplified for the ChIP assay are indicated by the lines above the human gene and labeled A, B, or C in the maps.
FIG. 2.
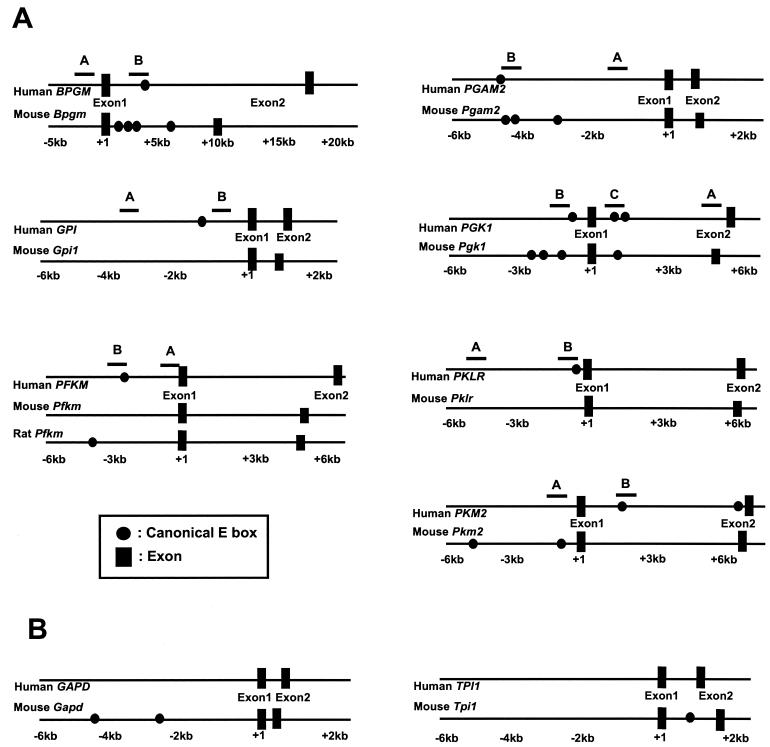
(A) Organization of human and mouse glycolytic genes, which do not contain conserved canonical E-box regions. Mouse Gpi, Pfkm, and Pklr genes do not contain a canonical E box. The sequences in or around the canonical E boxes of BPGM, PFKM, PGAM2, PGK1, and PKM2 were not conserved. Note that most sequences for the mouse Pfkm promoter region are not available. (B) Organization of GAPD and TPI1 genes. Human GAPD and TPI1 genes do not contain a canonical E box. The regions that are amplified for the ChIP assay are indicated by the lines above the human gene and labeled A, B, or C.
TABLE 1.
Summary of data on conservation of canonical E boxes, in vivo Myc binding, and expression profiles of glycolytic gene expression
| Gene | This study
|
Data from previous studiesa
|
||||||
|---|---|---|---|---|---|---|---|---|
| No. of canonical E boxesb
|
Conservation of E-box region | In vivo Myc bindingc | Induction by Mycd | Induction by Myce | Induction by MYC-ER | Myc DNA binding | ||
| Human | Mouse | |||||||
| ENO1 | 5 | 3 | Yes | +++ (0.80) | 8.05 (±0.11) | Up (M, S) | Yes | Yes (ChIP) |
| HK2 | 5 | 4 | Yes | +++ (1.47) | 3.66 (±1.28) | Up (M) | Yes | |
| LDHA | 2 | 3 | Yes | +++ (0.75) | 14.98 (±0.88) | Up (M, S) | Yes | Yes (ChIP, EMSA) |
| GAPD | 0 | 2 | No | +++ (0.76) | 3.72 (±0.36) | Up (G, S) | No | Yes (ChIP) |
| PFKM | 1 | 1 (rat)f | No | ++ (0.29) | 8.68 (±0.03) | Up (M, S) | Yes | |
| TPI1 | 0 | 1 | Yesg | ++ (0.34) | 5.19 (±2.29) | No | ||
| GPI | 1 | 0 | No | +/− (0.11) | 9.84 (±3.23) | Up (M) | Yes/Noh | |
| PGK1 | 3 | 4 | No | +/− (0.17) | 2.85 (±1.24) | Up | NDi | |
| PKM2 | 1 | 2 | No | +/− (0.13) | 3.20 (±0.94) | ND | Yes (ChIP) | |
| BPGM | 1 | 4 | No | − (0.03) | 0.74 (±0.12) | ND | ||
| PGAM2 | 1 | 3 | No | − (0.02) | NA | ND | ||
| PKLR | 1 | 0 | No | − (0.02) | NA | No | ||
| ALDOA | 2 | 2 | No | NA | 6.73 (±0.08) | No | ||
| HK1 | 4 | 8 | No | NA | 1.93 (±0.03) | ND | ||
Data adapted from the MYC target gene database (http://www.MYCcancergene.org) and previous publications (7, 29, 32).
Number of canonical E boxes within the 5 kb upstream of the transcriptional start site through intron 1.
In vivo Myc binding is indicated as follows: +++, strong binding; ++, moderate binding; +/−, weak binding; −, no binding; NA, not available. The percentage of total input is shown in parentheses.
Fold induction in P493-6 cells not treated with tetracycline.
The method is given in parentheses as follows: M, microarray; S, SAGE; G, guess.
Most sequences for the mouse Pfkm reporter region are not available.
Conservation of noncanonical E boxes.
Yes/No, discrepant reports.
ND, not determined.
Phylogenetic footprinting analysis.
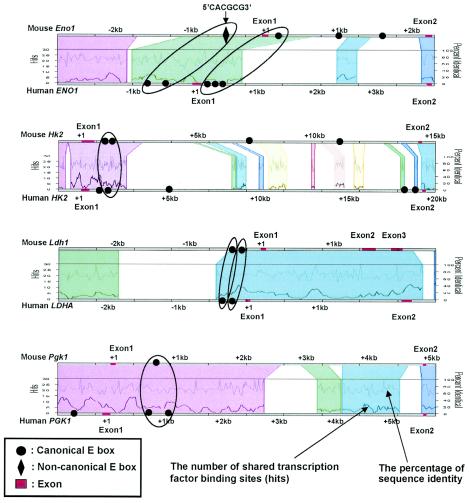
We chose manual sequence alignment using the dot plot function of OMIGA software and the Trafac server for phylogenetic footprinting to predict Myc binding sites. The ALDOA, BPGM, ENO1, HK1, HK2, LDHA, PFKM, PGAM, PGK1, and PKM2 genes were selected for the phylogenetic footprinting analysis, as canonical E boxes for these genes occur in both human and mouse genomic sequences. Using the parameters of a 30-bp window and a minimum of 65% sequence identity, we identified three genes, ENO1, HK2, and LDHA, whose E boxes and their extended flanking sequences showed 66 to 82% sequence identity between the human and mouse genomic sequences for longer than 30 bp (31 to 111 bp) (Fig. 1). Moreover, as shown in a regulogram of a web-based Trafac analysis, a visual representation of common binding sites (hits), conserved canonical E boxes identified with these parameters were consistently detected in the conserved sequence blocks (Fig. 3).
FIG. 3.
Trafac analysis (regulogram) of glycolytic genes. The phylogenetically conserved regions aligned with more than 50% identity are represented as different colored blocks. In each conserved region (indicated by a different color), the number of shared transcription factor binding sites (hits) and the percentage of sequence identity between human and mouse genomic sequences are indicated as two separate line graphs. For HK2, LDHA, and intronic E boxes of ENO1, conserved canonical E boxes identified by dot plot analysis (OMIGA software) as described in the legend to Fig. 1 consistently aligned in the Trafac analysis. Two human canonical E boxes in the promoter region of ENO1 correspond to the mouse noncanonical E box (5′-CACGCG-3′) in the same region. With PGK1, Trafac analysis detected a conserved intronic canonical E box.
Through manual alignment, the promoter and intron 1 regions of HK2 and LDHA display very high sequence conservation. These regions extend over 70 bp, with more than 70% sequence identity (Fig. 1). In ENO1, however, the conserved sequences are not parallel; one human canonical E box in the promoter region aligns with an intronic mouse canonical E box that lies 1 kb downstream of the transcriptional start site (Fig. 1). Both conserved regions in ENO1 are about 30 bp, which is shorter than those in HK2 or LDHA. Trafac analysis further reveals two human ENO1 canonical E boxes that correspond to a mouse noncanonical E box (Fig. 3). Despite manually lowering the stringency to 50% sequence identity with dot plotting, we are unable to align either of the two human ENO1 promoter E boxes with the mouse noncanonical E box (data not shown). The increased sensitivity of Trafac analysis is due to the less stringent criterion of including noncanonical E boxes that occur in larger regions with at least 50% interspecies sequence identity. Trafac analysis does not require local sequence alignment of the E boxes for inclusion as predicted transcription factor binding sites.
Predicted conserved canonical E-box regions bind to Myc in vivo.
To determine whether Myc binds conserved canonical E boxes, we performed ChIP assays. In addition to ENO1, HK2, and LDHA, which display significant conservation (Fig. 1), we selected additional human glycolytic genes to determine whether nonconserved canonical E boxes might be bound by Myc. Human GPI, PFKM, and PKLR genes contain canonical E boxes that are not found in the mouse orthologs. BPGM, PGAM, PGK1, and PKM2 do not have conserved E boxes (Fig. 2A). It should be noted, however, that intronic PGK1 conserved canonical E boxes were detected by Trafac analysis (Fig. 3). By lowering the stringency of our manual alignment to 50% identity, we were able to align these E boxes (data not shown). The P493-6 B-cell line was chosen for the ChIP assay (35, 36). These cells carry a tetracycline-responsive MYC expression system. Tetracycline significantly represses ectopic Myc protein expression, resulting only in residual endogenous Myc expression (Fig. 4A). MYC mRNA level was also significantly repressed, as previously reported (43). Removal of tetracycline resulted in a significant induction of MYC and enhancement of glycolysis (36).
FIG. 4.
(A) Western blot analysis of Myc expression in P493-6 cells left untreated (−) or treated (+) with tetracycline (Tet) for 72 h. (B) Western blot analysis of Myc expression in A1N4, human breast epithelial cells treated with 20 ng of EGF per ml. The number of hours after incubation with EGF is indicated. Tubulin is shown as a loading control in both panels.
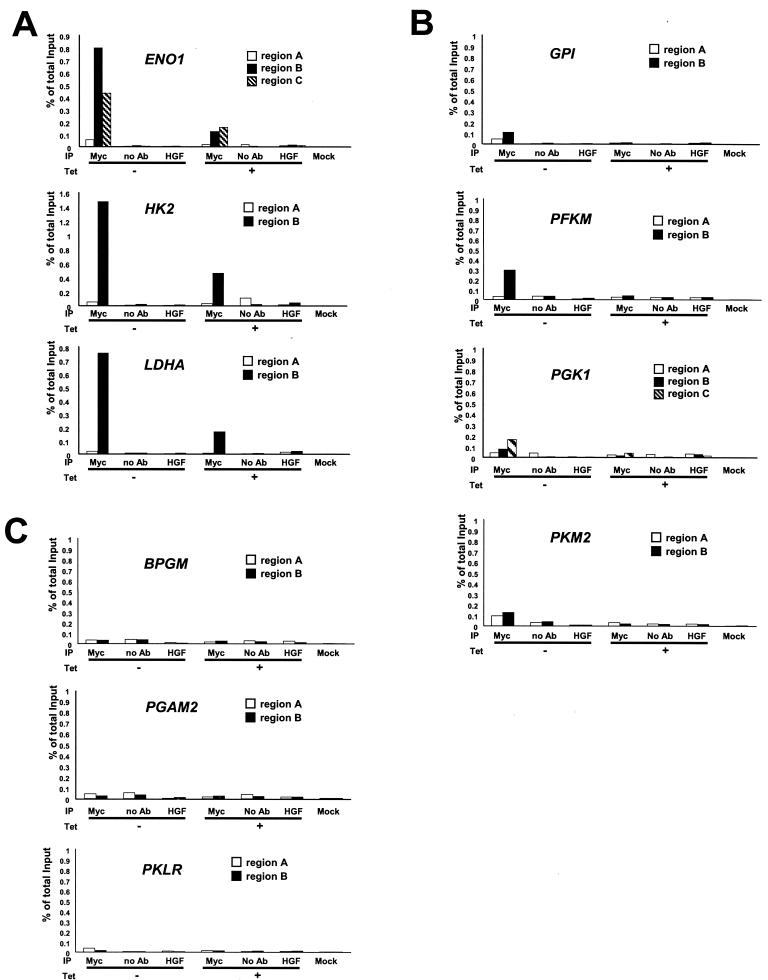
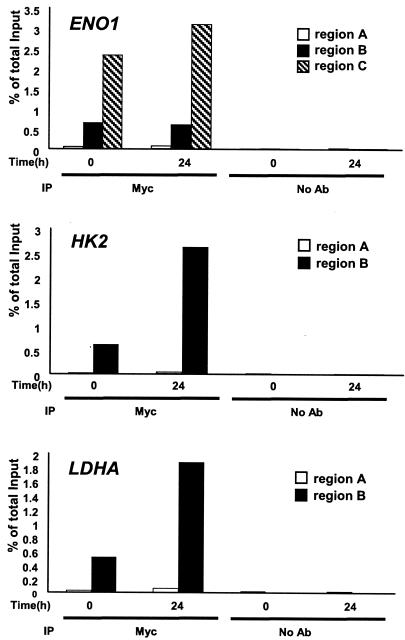
Chromatin fractions from untreated and tetracycline-treated P493-6 cells were immunoprecipitated with polyclonal anti-Myc antibody. For each gene, two or three primer pairs were designed to amplify the DNA regions that contained conserved canonical E boxes (ENO1, HK2, and LDHA), nonconserved E boxes (BPGM, GPI, PFKM, PGAM, PGK1, PKLR, and PKM2), or control regions that are at least 1 kb away from the E boxes (Fig. 1 and 2). Region A represents the control region at least 1 kb away from the E boxes, and regions B and C are the chromatin fragments that contain nonconserved and conserved canonical E boxes, respectively. We observe that all conserved canonical E-box regions in ENO1, HK2, and LDHA displayed strong Myc binding (0.7 to 1.5% of total input DNA), whereas control regions showed only background signal (Fig. 5A). Myc binding to GPI, PGK1, and PKM2 (region B or C) was diminished (Fig. 5B). The canonical E-box region of the human PFKM gene, which is not conserved in the rat Pfkm gene (most sequences for mouse Pfkm promoter region are not available), was bound by Myc but to a lesser extent than the highly conserved E-box regions of ENO1, HK2, and LDHA (Fig. 5B). Myc did not bind the nonconserved E-box regions of BPGM, PGAM2, and PKLR (region B) (Fig. 5C). We confirmed the specificity of ChIP assay by performing immunoprecipitation with anti-HGF antibody, without antibody, and with mock control samples. In all these conditions, we observed only background signals (Fig. 5 and 6).
FIG. 5.
ChIP assay of glycolytic genes in the P493-6 cell system. Glycolytic genes displaying strong Myc binding (A), moderate or weak Myc binding (B), or no Myc binding (C) are shown. Labeled regions (as shown in Fig. 1; region A, B, or C) of each gene were quantitatively amplified by real-time PCR. PCR was performed on the fragmented chromatin precipitated from P493-6 cells left untreated (−) or treated (+) with tetracycline (Tet) for 72 h with anti-Myc or HGF, without antibody (no Ab), or mock control samples as indicated at the bottom of the graph. The white bars represent the percentage of total input of control regions (region A). The black and hatched bars represent the percentage of total input of the chromatin regions that contain conserved canonical E boxes (ENO1, HK2, and LDHA) or nonconserved canonical E boxes (BPGM, GPI, PFKM, PGAM2, PGK1, and PKM2).
FIG. 6.
ChIP assay of glycolytic genes in the A1N4 cell system. Chromatin precipitated from A1N4 cells at the indicated times with anti-Myc antibody or without antibody (no Ab) were subjected to real-time PCR as described in the legend to Fig. 5.
Moderate Myc binding to ENO1, HK2, and LDHA canonical E boxes were detected with endogenous Myc (Fig. 5A) in P493-6 cells treated with tetracycline, which suppresses only ectopic MYC expression. We further confirmed that endogenous Myc could bind chromatin fragments that contain conserved canonical E boxes in another experimental system. We used the A1N4 human breast epithelial cell line (22), which when treated with EGF for 24 h, expresses a significantly higher level of endogenous Myc than EGF-starved A1N4 cells (Fig. 4B). All regions spanning conserved canonical E boxes (regions B and C) displayed strong Myc binding, whereas control regions (region A) showed only background signals (Fig. 6). While we do not know the basis for the relatively high level of Myc binding to target sequences in A1N4 cells, it is intriguing to note that the levels of recovered chromatin from endogenous Myc reproducibly reached approximately 2 to 4%, which is higher than those from exogenous Myc in P493-6 cell system (0.7 to 1.5%). Furthermore, for the ENO1 gene, region C spanning the conserved intronic canonical E box was bound by endogenous Myc at a significantly higher level than the conserved canonical E box in the promoter region (region B). Taken together, these observations suggest that conserved canonical E boxes are bound by ectopic Myc and that they are also significantly bound by endogenous levels of Myc.
The binding of Myc to the promoter or intron 1 regions of ENO1, HK2, and LDHA also correlated with gene expression. Using P493-6 cells, we observed that the mRNA levels of these three genes were also significantly higher in untreated P493-6 cells (high MYC) than in tetracycline-treated P493-6 cells (low MYC) (Table 1). The increased expression of these genes in response to elevated Myc provides further evidence that these genes are functional direct Myc targets.
Identification of Myc binding regions not predicted by phylogenetic footprinting in GAPD and TPI1.
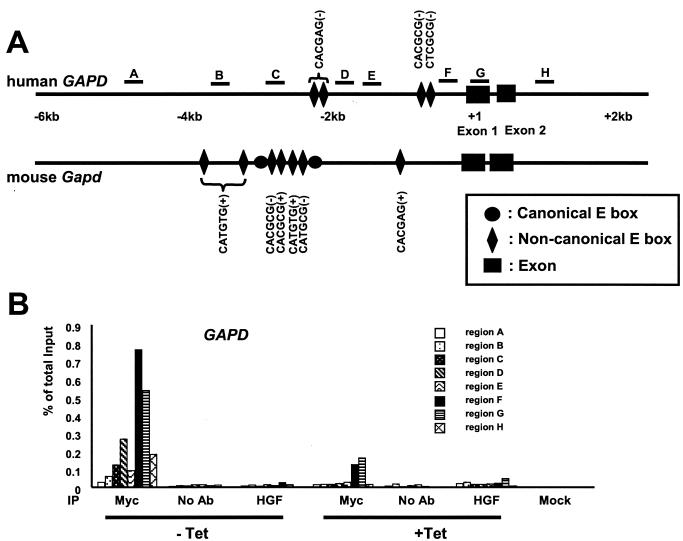
Myc induces the expression of both GAPD and TPI1, although they do not contain canonical E boxes (Fig. 2B and Table 1). To determine whether Myc could directly bind to regions in the promoter or intron 1, we used scanning ChIP assay (42). First, we used five PCR primer pairs to scan the human GAPD locus at approximately 1-kb intervals (regions A, B, C, E, and G) (Fig. 7A). Using the P493-6 cell system, significant Myc binding was found in region G that spans approximately 40 bp upstream of the transcriptional start site through the entire exon 1 (Fig. 7B). We then considered the role of noncanonical E boxes, as we previously demonstrated Myc binding to conserved, noncanonical E boxes in vivo (43). We identify four noncanonical E boxes in the human GAPD gene and seven noncanonical E boxes in the mouse Gapd gene (Fig. 7A) (see Materials and Methods for the sequence variations of noncanonical E boxes). Two pairs of human noncanonical E boxes are located either within 0.4 kb or within 2 kb upstream of the transcriptional start site. However, these noncanonical E boxes were not significantly conserved. We then performed an extended scan with additional PCR primer pairs to pinpoint the fragments that lie within 100 bp of the noncanonical E boxes (regions D and F) (Fig. 7). Region H was used as an additional control. Figure 7B demonstrates Myc binding to region F (0.8% of total input) and region D (0.3% of total input). These results suggest that Myc binds a GAPD promoter region containing noncanonical E boxes that are not conserved in the mouse Gapd gene.
FIG. 7.
Scanning ChIP assay of the human GAPD gene. (A) Locations of canonical E boxes, noncanonical E boxes, and exons in the human and mouse genomic sequences. Canonical E boxes in the mouse gene are indicated by black circles. The lead (+) or complement (−) sequences of noncanonical E boxes are also indicated. The regions that are amplified for the scanning ChIP assay are indicated by the lines above the human gene and labeled A to H. (B) The human GAPD gene was scanned by ChIP assay in the P493-6 cell system. P493-6 cells were either not treated (− Tet) or treated with tetracycline (+ Tet) for 72 h. ChIP was performed with anti-Myc or HGF, without antibody (no Ab), or mock control samples as indicated at the bottom of the graph.
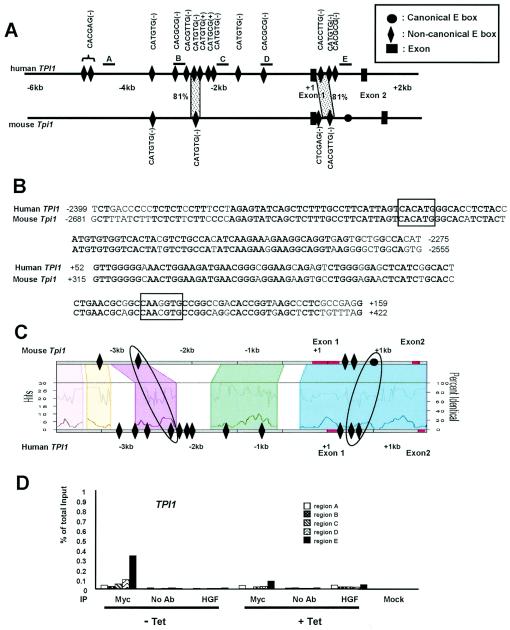
The human TPI1 gene was scanned by ChIP assay with five PCR primer pairs at approximately 1-kb intervals (Fig. 8A). Myc bound region E, which lies in intron 1 (approximately 800 bp downstream of the transcriptional start site) (Fig. 8D). We found 14 noncanonical E boxes in human TPI1 (11 in the promoter region and 3 in intron 1) and analyzed them by phylogenetic footprinting (Fig. 8A). Through dot plot analysis, we detected two highly conserved promoter and intron 1 regions. The conserved intron 1 region, however, does not contain perfectly matched noncanonical E box (5′-CAAGGTG-3′ in human TPI1). Both conserved regions extend over 100 bp with more than 80% sequence identity (Fig. 8A and B). However, in the Trafac analysis, an intronic canonical E box in the mouse Tpi1 gene is conserved and corresponds to two human noncanonical E boxes (Fig. 8C). It is intriguing that this conserved intronic region, but not the upstream region, was bound by Myc in the scanning ChIP assay (Fig. 8D, region E). Our data provide evidence that Myc could associate with noncanonical E boxes or unknown new binding sites in these genes. These studies indicate that GAPD and TPI1 are direct targets of Myc with binding sites that are largely unpredicted by phylogenetic footprinting.
FIG. 8.
Phylogenetic footprinting analysis and scanning ChIP assay of TPI1. (A) Noncanonical E boxes and exons are indicated in the human and mouse genomic sequences. The lead (+) or complement (−) sequences of noncanonical E boxes are also indicated. One canonical E box in mouse intron 1 is indicated as a black circle. The dotted areas represent the conservation of the noncanonical E box and its extended flanking region with more than 65% sequence identity for longer than 30 bp. The percentage of sequence identity is also shown. The regions that are amplified for the scanning ChIP assay are indicated as the lines above the human gene and labeled A to E. (B) Sequence alignments of the conserved E box and its extended flanking regions are shown. Noncanonical E boxes are boxed in each sequence alignment. Conserved nucleotides are shown in bold type. (C) Trafac analysis (regulogram) of human and mouse genomic sequences. (D) Scanning ChIP assay of human TPI1 in P493-6 cell system. P493-6 cells were either not treated (− Tet) or treated with tetracycline (+ Tet) for 72 h. ChIP was performed with anti-Myc or HGF, without antibody (no Ab), or mock control samples as indicated at the bottom of the graph.
Induction of glycolytic gene expression by Myc correlates with predicted canonical Myc binding sites.
To determine whether in vivo Myc binding correlates with the expression of glycolytic genes, we subjected tetracycline-treated and untreated P493-6 cells to quantitative real-time RT-PCR (for all 14 glycolytic genes) and Northern blot analysis (for HK2, GAPD, and PFKM [data not shown]). The changes in expression of 14 human glycolytic genes are listed in Table 1. The mRNA expression levels of all genes that showed strong Myc binding (ENO1, GAPD, HK2, LDHA, PFKM, and TPI1) were significantly elevated by Myc. Note that ENO1, HK2, and LDHA are among the highest Myc binding targets that were predicted by phylogenetic footprinting. However, the induction of gene expression is less well correlated with Myc binding in GAPD and TPI1, in which Myc binds noncanonical E-box regions. ALDOA, GPI, PFKM, PGK1, and PKM2 were induced by Myc, but we could not detect E-box conservation or Myc binding, suggesting that these genes are regulated by Myc indirectly or that Myc may bind ALDOA, GPI, PFKM, PGK1, and PKM2 in regions that we did not examine.
DISCUSSION
Our present studies using Myc and glycolytic genes as a model system provide not only a critical analysis of phylogenetic footprinting but also a comprehensive evaluation of Myc and the glycolytic target gene network. Our findings indicate that most glycolytic genes are directly regulated by Myc, supporting the notion that deregulated MYC can contribute to altered tumor glucose metabolism under normal oxygen levels.
Phylogenetic footprinting analyses by both manual alignment and Trafac analysis predicted evolutionarily conserved, canonical Myc binding sites in ENO1, HK2, and LDHA, which were all confirmed by ChIP assay results. We observed that in vivo, Myc binding correlated with the induction of glycolytic target gene expression by Myc. These genes belong to what we previously termed class I Myc target genes, which contain conserved canonical E boxes (18). Since promoter-reporter assays and EMSA do not reflect in situ Myc binding, we had not included these assays in the present study. Nevertheless, we previously demonstrated for LDHA that Myc binds the highly conserved LDHA E boxes in EMSA as well as in promoter-reporter assays and that mutation of the E boxes rendered the promoter unresponsive to Myc (37). Given the limitations of EMSA (18) and considering the fact that even artificial promoters bearing E boxes are responsive to Myc in transient-transfection assays (20), we have focused on ChIP as a measure of in vivo Myc binding.
All other glycolytic genes studied do not have evolutionarily conserved canonical E boxes, although most of the glycolytic genes we studied were up-regulated in the presence of high levels of Myc. Because human GAPD and TPI1 genes are induced by MYC but do not contain predicted Myc binding sites or canonical E boxes, we sought to determine whether Myc binds these genes by scanning ChIP assays. The ChIP assay identified strong Myc binding to the regulatory regions of GAPD and TPI1, providing evidence that GAPD and TPI1 are direct Myc targets. However, the correlation between Myc binding and phylogenetic conservation in GAPD and TPI1 is less clear. TPI1, which is induced by Myc, has a conserved noncanonical E box (5′-CACATG-3′) in the upstream region. However, Myc bound best to the intron 1 region that contains two noncanonical E boxes (5′-CACATG-3′ and 5′-CGCGTG-3′) that correspond to a mouse canonical E box. These intron 1 regions were aligned by Trafac analysis, but not by dot plotting. Instead, dot plotting identified different areas of noncanonical E-box conservation in the intron 1 regions (Fig. 8A and C). GAPD, which is also induced by Myc, has no conserved canonical or noncanonical E boxes. With GAPD, Myc bound best to the human promoter region F that is near two nonconserved, noncanonical E boxes (5′-CGCGTG-3′ and 5′-CGCGAG-3′). Hence, both GAPD and TPI1 belong to what we previously termed class II direct Myc target genes, whose Myc binding sites either drifted during evolution or are newly acquired (18).
The human PFKM gene contains a canonical E box in the promoter region, which is bound well by Myc, although this canonical E-box region is not conserved in the rat Pfkm gene. Most of the corresponding mouse sequence is currently unavailable. Myc slightly bound the human PGK1 region with one upstream and two intron 1 canonical E boxes. Myc bound the human GPI promoter region with one canonical E box. It is less clear whether Myc bound the human PKM2 region near an intron 1 canonical E box, when a control region several kilobases upstream is considered (Fig. 5B). No Myc binding was detected in the canonical E-box regions of human BPGM, PKLR, and PGAM2 genes. We were unable to identify suitable primers for quantitative PCR in the ALDOA and HK1 loci. In aggregate, these observations indicate that Myc binds weakly to glycolytic genes that do not contain conserved Myc canonical binding sites. Because we examined Myc binding only in canonical E-box regions for the remaining genes (Fig. 2A), we cannot exclude the possibility that Myc may bind noncanonical E boxes or unknown novel binding sites.
Since tetracycline treatment suppresses only ectopic Myc expression in P493-6 cells and does not affect endogenous Myc expression, this study provides a unique opportunity to examine endogenous Myc and ectopic Myc binding to the same genes. Only class I (ENO1, HK2, and LDHA) and class II (GAPD and TPI1) genes demonstrate binding by endogenous Myc (Fig. 5A, 7B, and 8D). Intriguingly, while both HK2 and LDHA displayed a corresponding enhanced binding of ectopic Myc to the same regions, ectopic Myc binding to ENO1 is higher in promoter region B than in the intronic region C that is bound well by endogenous Myc (Fig. 5A and 6). Through studying the human A1N4 breast epithelial cells, in which endogenous Myc is induced by the growth factor EGF, we also observed that intronic region C of ENO1 is better bound by endogenous Myc than the promoter region B. High basal Myc binding to ENO1 region C was observed compared to either LDHA or HK2. With GAPD, there was significant binding of ectopic Myc to region D, which was not bound by endogenous Myc, although this level of binding might be at the threshold of detection. With PFKM, PGK1, GPI, and PKM2, moderate to diminished binding was detected only with ectopic Myc. With endogenous Myc, phylogenetic footprinting was highly predictive of Myc binding to the conserved regions bearing canonical E boxes.
So why are these specific regions conserved in ENO1, HK2, and LDHA? As less than 20% of nonexonic sequence is conserved in the human and mouse genomes, significantly high level of conservation, which is more than 65% sequence identity at and around Myc binding regions extending 30 to more than 100 bp, shown in ENO1, HK2, LDHA, and TPI1 genes may have an impact on Myc-mediated transcriptional regulation. One possibility is that these extended conserved sequences may contain binding sites for other transcription factors or chromatin remodeling proteins, which could affect the transcriptional activity of Myc (12). Our preliminary Trafac analysis suggests that conserved binding sites for the transcription factors ETS, AP4, Sp1, CREB, and HIF-1 are found overrepresented near conserved Myc E boxes in glycolytic genes (A. G. Jegga, unpublished data). In particular, among the 10 glycolytic genes with more than three other conserved transcription factor binding sites near a conserved E box, only ENO1, HK2, and LDHA lack clusters containing AP4 sites with an E box (Jegga, unpublished). It will be intriguing to determine whether the lack of AP4 sites in ENO1, HK2, and LDHA contributes to their robust response to Myc. At this time, we are evaluating these candidate transcription factors, but the findings are beyond the scope of this report.
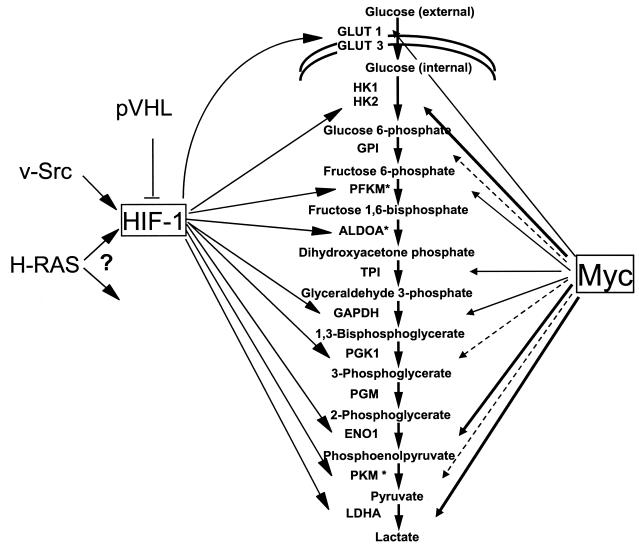
While previous studies reveal that hypoxia induces glycolysis through the HIF-1 transcription factor, our studies demonstrate that in normoxia, Myc can induce most glycolytic genes including ALDOA, ENO1, GAPD, GPI, LDHA, HK2, PFKM, PGK1, PKM, and TPI1 (Fig. 9). This supports the hypothesis that the propensity for some tumors to undergo aerobic glycolysis (Warburg effect), in which glucose is converted to lactate despite the availability of oxygen, could be the result of Myc activation of glycolytic gene expression independent of hypoxia. Of the Myc-induced genes, HK2, PFKM, and PKM were previously considered to be important rate-limiting regulatory points in glycolysis. However, according to metabolic control analysis, there are no so-called rate-limiting enzymes in a pathway. Rather, the control of metabolic flux through a pathway is shared between all enzymes in different proportions such that coordinate regulation of a number of enzymes in a specific pathway is sufficient to affect overall metabolic flux. Hence, the induction of genes encoding key enzymes by Myc appears sufficient for enhanced glycolytic flux.
FIG. 9.
Regulation of the glycolytic gene network by HIF-1 and Myc. Arrows emanating from HIF-1 indicate regulation of specific glycolytic genes by HIF-1 in response to hypoxia. Arrows emanating from Myc are shown with different thicknesses. The thickest arrows represent strong binding by Myc, whereas dashed arrows represent diminished binding. HIF-1 is shown downstream of oncogenic Ras and Src as well as being negatively regulated by von Hippel-Lindau protein (pVHL).
In addition to bearing evolutionarily conserved regulatory Myc binding sites, these glycolytic genes may share some other functional features that are important for mammalian cell growth and development. Recent studies suggest that glycolytic enzymes may not only be involved in glucose metabolism but they may also influence other biological processes. Several isoforms of hexokinase play a key role in mitochondrion-mediated apoptosis by modulating proapoptotic molecules including Bax and Bad, suggesting that the glycolytic pathway and apoptosis are integrated (9, 15, 33). The roles of LDHA and GAPD proteins in glycolysis are well-established, yet the nuclear localization of these proteins suggest additional biological functions (45). Recently, GAPD and LDH were both found in a transcriptional coactivator complex that assists the Oct-1 transcription factor in regulating histone H2B expression (44). GAPD, in particular, alters the coactivator activity as a function of NAD+/NADH ratio. Several studies suggest that ENO1 and MYC promoter-binding protein 1 (MBP-1) are encoded by the same gene (13, 38). MBP-1, which results from an internal translational initiation site of the ENO1 mRNA, is a negative transcriptional regulator of MYC transcription. These findings imply that a negative-feedback loop exists between MYC and ENO1. It is notable that while there is overlap between glycolytic genes that are responsive to hypoxia via the HIF-1 transcription factor and those that are regulated by Myc, a hierarchy of glycolytic genes preferentially regulated by HIF-1 is not apparent, since HIF-1 appears to uniformly affect genes encoding many enzymes of the entire pathway (13, 38). In aggregate, these observations suggest that glycolytic genes, which serve other biological functions, may have preferentially conserved regulatory sequences that are responsive to particular transcription factors or stimuli. In particular, we demonstrate here that Myc serves as a regulator of many key glycolytic genes, providing additional insight into the complexity of transcriptional control of glycolytic genes in normoxic conditions (Fig. 9).
Supplementary Material
Acknowledgments
This work was supported in part by NIH grants CA51497 (C.V.D.), CA57341 (C.V.D.), LM07515 (C.V.D.), T32HL07525 (K.I.Z.), and T32GM07819 (K.A.O.). J. Kim is a Howard Hughes Medical Institute Predoctoral Fellow.
We thank L. Lee, A. Chakravarti, and J. Yustein for helpful comments, D. Eick for P493-6 cells, and P. Farnham for ChIP protocols.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman, B. P., Y. Nibu, B. D. Pfeiffer, P. Tomancak, S. E. Celniker, M. Levine, G. M. Rubin, and M. B. Eisen. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. USA 99:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell, T. K., J. Huang, A. Ma, L. Kretzner, F. W. Alt, R. N. Eisenman, and H. Weintraub. 1993. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 13:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette, M., and M. Tompa. 2003. FootPrinter: a program designed for phylogenetic footprinting. Nucleic Acids Res. 31:3840-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, K. E., J. Wells, J. Gutman, S. M. Bartley, and P. J. Farnham. 1998. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc. Natl. Acad. Sci. USA 95:13887-13892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, M. D., and S. B. McMahon. 1999. The Myc oncoprotein: a critical evaluation of transactivation and target gene regulation. Oncogene 18:2916-2924. [DOI] [PubMed] [Google Scholar]
- 7.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang, C. V., and G. L. Semenza. 1999. Oncogenic alterations of metabolism. Trends Biochem. Sci. 24:68-72. [DOI] [PubMed] [Google Scholar]
- 9.Danial, N. N., C. F. Gramm, L. Scorrano, C. Y. Zhang, S. Krauss, A. M. Ranger, S. R. Datta, M. E. Greenberg, L. J. Licklider, B. B. Lowell, S. P. Gygi, and S. J. Korsmeyer. 2003. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature 424:952-956. [DOI] [PubMed] [Google Scholar]
- 10.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. [DOI] [PubMed] [Google Scholar]
- 11.Eilers, M., D. Picard, K. R. Yamamoto, and J. M. Bishop. 1989. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature 340:66-68. [DOI] [PubMed] [Google Scholar]
- 12.Elkon, R., C. Linhart, R. Sharan, R. Shamir, and Y. Shiloh. 2003. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 13:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feo, S., D. Arcuri, E. Piddini, R. Passantino, and A. Giallongo. 2000. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 473:47-52. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez, P. C., S. R. Frank, L. Wang, M. Schroeder, S. Liu, J. Greene, A. Cocito, and B. Amati. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlob, K., N. Majewski, S. Kennedy, E. Kandel, R. B. Robey, and N. Hay. 2001. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 15:1406-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 17.Grandori, C., and R. N. Eisenman. 1997. Myc target genes. Trends Biochem. Sci. 22:177-181. [DOI] [PubMed] [Google Scholar]
- 18.Haggerty, T. J., K. I. Zeller, R. C. Osthus, D. R. Wonsey, and C. V. Dang. 2003. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc. Natl. Acad. Sci. USA 100:5313-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jegga, A. G., S. P. Sherwood, J. W. Carman, A. T. Pinski, J. L. Phillips, J. P. Pestian, and B. J. Aronow. 2002. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 12:1408-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzner, L., E. M. Blackwood, and R. N. Eisenman. 1992. Myc and Max proteins possess distinct transcriptional activities. Nature 359:426-429. [DOI] [PubMed] [Google Scholar]
- 21.Lenhard, B., A. Sandelin, L. Mendoza, P. Engstrom, N. Jareborg, and W. W. Wasserman. 22 May 2003, posting date. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2:13. [Online.] http://jbiol.com/content/2/2/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Q., and C. V. Dang. 1999. c-Myc overexpression uncouples DNA replication from mitosis. Mol. Cell. Biol. 19:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Z., S. Van Calcar, C. Qu, W. K. Cavenee, M. Q. Zhang, and B. Ren. 2003. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc. Natl. Acad. Sci. USA 100:8164-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loots, G. G., I. Ovcharenko, L. Pachter, I. Dubchak, and E. M. Rubin. 2002. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 12:832-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao, D. Y., J. D. Watson, P. S. Yan, D. Barsyte-Lovejoy, F. Khosravi, W. W. Wong, P. J. Farnham, T. H. Huang, and L. Z. Penn. 2003. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr. Biol. 13:882-886. [DOI] [PubMed] [Google Scholar]
- 26.Markstein, M., P. Markstein, V. Markstein, and M. S. Levine. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 99:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99:6274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neiman, P. E., A. Ruddell, C. Jasoni, G. Loring, S. J. Thomas, K. A. Brandvold, R. Lee, J. Burnside, and J. Delrow. 2001. Analysis of gene expression during myc oncogene-induced lymphomagenesis in the bursa of Fabricius. Proc. Natl. Acad. Sci. USA 98:6378-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connell, B. C., A. F. Cheung, C. P. Simkevich, W. Tam, X. Ren, M. K. Mateyak, and J. M. Sedivy. 2003. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 278:12563-12573. [DOI] [PubMed] [Google Scholar]
- 30.Orian, A., B. van Steensel, J. Delrow, H. J. Bussemaker, L. Li, T. Sawado, E. Williams, L. W. Loo, S. M. Cowley, C. Yost, S. Pierce, B. A. Edgar, S. M. Parkhurst, and R. N. Eisenman. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oster, S. K., C. S. Ho, E. L. Soucie, and L. Z. Penn. 2002. The myc oncogene: marvelously complex. Adv. Cancer Res. 84:81-154. [DOI] [PubMed] [Google Scholar]
- 32.Osthus, R. C., H. Shim, S. Kim, Q. Li, R. Reddy, M. Mukherjee, Y. Xu, D. Wonsey, L. A. Lee, and C. V. Dang. 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 275:21797-21800. [DOI] [PubMed] [Google Scholar]
- 33.Pastorino, J. G., N. Shulga, and J. B. Hoek. 2002. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 277:7610-7618. [DOI] [PubMed] [Google Scholar]
- 34.Pennacchio, L. A., and E. M. Rubin. 2001. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2:100-109. [DOI] [PubMed] [Google Scholar]
- 35.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuhmacher, M., M. S. Staege, A. Pajic, A. Polack, U. H. Weidle, G. W. Bornkamm, D. Eick, and F. Kohlhuber. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9:1255-1258. [DOI] [PubMed] [Google Scholar]
- 37.Shim, H., C. Dolde, B. C. Lewis, C. S. Wu, G. Dang, R. A. Jungmann, R. Dalla-Favera, and C. V. Dang. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 94:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian, A., and D. M. Miller. 2000. Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc proto-oncogene. J. Biol. Chem. 275:5958-5965. [DOI] [PubMed] [Google Scholar]
- 39.Warburg, O. 1956. On the origin of cancer cells. Science 123:309-314. [DOI] [PubMed] [Google Scholar]
- 40.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 41.Watson, J. D., S. K. Oster, M. Shago, F. Khosravi, and L. Z. Penn. 2002. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 277:36921-36930. [DOI] [PubMed] [Google Scholar]
- 42.Zeller, K. I., T. J. Haggerty, J. F. Barrett, Q. Guo, D. R. Wonsey, and C. V. Dang. 2001. Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J. Biol. Chem. 276:48285-48291. [DOI] [PubMed] [Google Scholar]
- 43.Zeller, K. I., A. G. Jegga, B. J. Aronow, K. A. O'Donnell, and C. V. Dang. 11 September 2003, posting date. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4:R69. [Online.] http://genomebiology.com/2003/4/10/R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, L., R. G. Roeder, and Y. Luo. 2003. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114:255-266. [DOI] [PubMed] [Google Scholar]
- 45.Zhong, X. H., and B. D. Howard. 1990. Phosphotyrosine-containing lactate dehydrogenase is restricted to the nuclei of PC12 pheochromocytoma cells. Mol. Cell. Biol. 10:770-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.