Abstract
Pseudouridine, the most abundant modified nucleoside in RNA, is synthesized by posttranscriptional isomerization of uridines. In eukaryotic RNAs, site-specific synthesis of pseudouridines is directed primarily by box H/ACA guide RNAs. In this study, we have identified 61 novel putative pseudouridylation guide RNAs by construction and characterization of a cDNA library of human box H/ACA RNAs. The majority of the new box H/ACA RNAs are predicted to direct pseudouridine synthesis in rRNAs and spliceosomal small nuclear RNAs. We can attribute RNA-directed modification to 79 of the 97 pseudouridylation sites present in the human 18S, 5.8S, and 28S rRNAs and to 11 of the 21 pseudouridines reported for the U1, U2, U4, U5, and U6 spliceosomal RNAs. We have also identified 12 novel box H/ACA RNAs which lack apparent target pseudouridines in rRNAs and small nuclear RNAs. These putative guide RNAs likely function in the pseudouridylation of some other types of cellular RNAs, suggesting that RNA-guided pseudouridylation is more general than assumed before. The genomic organization of the new box H/ACA RNA genes indicates that in human cells, all box H/ACA pseudouridylation guide RNAs are processed from introns of pre-mRNA transcripts which either encode a protein product or lack protein-coding capacity.
Posttranscriptional covalent modification of ribonucleotides is an important step in the biosynthesis of stable cellular RNAs, including tRNAs, rRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) (40). Biochemical, biophysical, and genetic studies have shown that modified nucleotides are important for the appropriate function of mature RNAs; they facilitate correct RNA folding and contribute to the formation of appropriate RNA-RNA and RNA-protein interactions (reviewed in references 1, 7, 12, 15, and 44).
While in tRNAs, most modified nucleotides are synthesized by protein enzymes, in eukaryotic rRNAs and snRNAs, site-specific synthesis of the most prevalent modified ribonucleotides, the 2′-O-ribose-methylated nucleotides and the pseudouridines, is achieved by two distinct families of ribonucleoproteins (RNPs) (reviewed in references 14, 18, 27, 28, and 52). The modification guide RNPs consist of a sequence-specific guide RNA and a set of common proteins. Each 2′-O-ribose methylation guide RNA carries the conserved C, C′ (consensus, RUGAUGA), D, and D′ (CUGA) box motifs and possesses one or two 10- to 21-nucleotide-long antisense elements that are responsible for selection of the correct substrate ribonucleotides through the formation of double helices with the target RNAs (11, 32). The selected ribonucleotides are 2′-O-ribose methylated by the Nop1p/fibrillarin methyltransferase enzyme that, in addition to the Snu13 (15.5-kDa), Nop56p, and Nop58p RNP proteins, is associated with all box C/D RNAs (14, 18, 27, 52, 55).
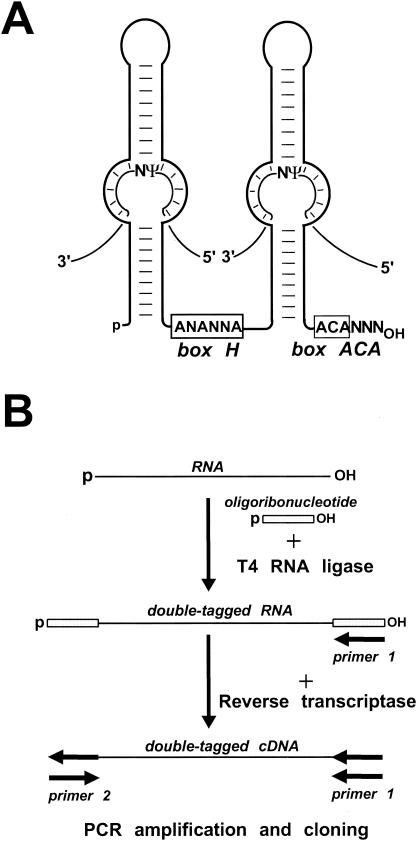
The pseudouridylation guide RNAs are composed of two major hairpin elements that are connected by a hinge and followed by a short tail region (Fig. 1A). The single-stranded hinge and tail region carry the conserved H (consensus, ANANNA) and ACA box motifs that are located at the bases of the 5′ and 3′ hairpins, respectively (6, 20). Two short antisense elements located in an internal loop of the 5′ and/or 3′ hairpins provide the sequence specificity for the pseudouridylation guide RNP by base pairing to the sequences that precede and follow the target uridine. This interaction creates the “pseudouridylation pocket,” in which the unpaired substrate uridine selected for pseudouridylation is located 14 or 15 bp upstream of the H or ACA motif of the guide RNA (19, 43). The dyskerin/Cbf5p pseudouridine synthase, together with the Nhp2, Nop10, and Gar1 RNP proteins, is an integral component of box H/ACA pseudouridylation guide RNPs (33, 56).
FIG. 1.
Schematic structure of box H/ACA RNAs and cDNA construction. (A) Selection of pseudouridylation sites by box H/ACA guide RNAs. For details, see the text. (B) Construction of a cDNA library of human box H/ACA RNAs. HeLa cell RNAs immunoprecipitated by an anti-GAR1 antibody were incubated with a phosphorylated oligoribonucleotide in the presence of T4 RNA ligase. RNA sequences tagged at both termini were converted into double-stranded DNA by a reverse transcription-PCR amplification approach. The amplified DNA was cloned into a plasmid vector, and individual clones were characterized by sequence analysis.
Vertebrate box H/ACA pseudouridylation guide RNAs are processed from removed and debranched pre-mRNA introns by exonucleolytic activities (18). The conserved H and ACA boxes together with the basal helices of the 5′ and 3′ hairpins provide the signals for correct RNA processing (6, 8, 20). The mature H/ACA RNAs accumulate either in the nucleolus (snoRNAs) or in nucleoplasmic Cajal bodies (small Cajal body-specific RNAs [scaRNAs]). The nucleolar accumulation of box H/ACA snoRNAs is supported by the conserved H and ACA boxes and the basal helix of the 3′ hairpin (34, 42). The box H/ACA scaRNAs carry a common Cajal body-specific localization signal, the CAB box (consensus, UGAG), that is found in the terminal loops of the 5′ and 3′ hairpins (47). In the nucleolus, most box H/ACA snoRNAs direct pseudouridylation of the 18S, 5.8S, and 28S rRNAs (19, 43), while the box H/ACA scaRNAs function in pseudouridylation of the RNA polymerase II (pol II)-transcribed spliceosomal snRNAs (13, 25, 26).
In the yeast Saccharomyces cerevisiae, most, if not all, ribosomal pseudouridines are synthesized by box H/ACA snoRNPs (49). Since only a limited number of box H/ACA RNAs have been identified in humans, it remains unknown to what extent guide RNAs participate in the pseudouridylation of human cellular RNAs (13, 20, 25, 26, 29, 31, 48, 54). A recent identification of partial sequences of several putative box H/ACA snoRNAs in mice suggested that mammalian cells express a large number of box H/ACA RNAs (23). In this study, the identification of 61 novel human box H/ACA RNAs provided us with new insights into the function and organization of the molecular machinery directing the pseudouridylation of human rRNAs, snRNAs, and probably other cellular RNAs.
MATERIALS AND METHODS
Construction and characterization of a cDNA library of human box H/ACA RNAs.
Preparation of HeLa cell extracts and immunoprecipitation by a GAR1 antibody of box H/ACA RNPs were performed essentially as described previously (20), except that HeLa cells were sonicated in 40 mM Tris-HCl (pH 7.5) buffer containing 200 mM NaCl and 0.05% Nonidet P-40. The anti-human GAR1 (hGAR1) antipeptide antibody was kindly provided by W. Filipowicz (Friedrich Miescher Institut, Basel, Switzerland). About 0.3 μg of RNA recovered by immunoprecipitation with the anti-hGAR1 antibody was mixed with 40 pmol of 5′-end-phosphorylated oligoribonucleotide (pAAUAAAGCGGCCGCGGAUCCAA) and incubated with 15 U of T4 RNA ligase (Promega) and 10 U of RNase inhibitor (Promega) as described previously (17). After phenol-chloroform extraction, the ligation products were recovered by ethanol precipitation, annealed with 40 pmol of oligodeoxynucleotide P1 (TTGGATCCGCGGCCGCTTTAT), and used as a template for cDNA synthesis with avian myeloblastosis virus reverse transcriptase (Promega). The resulting first-strand cDNA was used as a template for PCR amplification by Vent polymerase (Promega) with oligodeoxynucleotides P1 and P2 (AATAAAGCGGCCGCGGATCCAAA) as primers. The amplified DNA was digested with BamHI, inserted into the BamHI site of pBluescribe (Stratagene), and transformed into Escherichia coli DH5α cells. Plasmid purification and sequence analysis were performed according to standard laboratory protocols (50).
Mapping of pseudouridines.
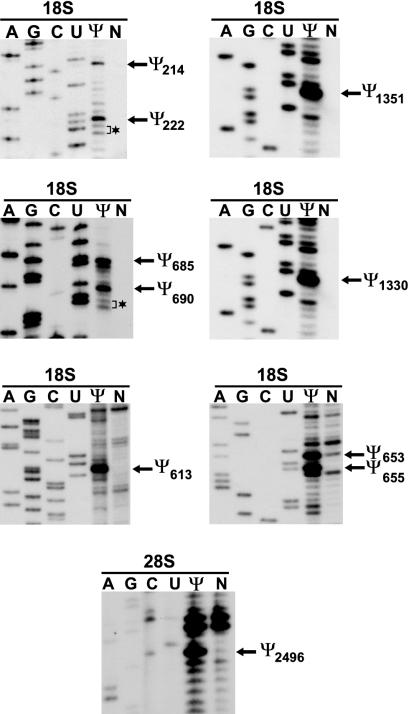
Isolation of RNA from human HeLa cells was performed by the guanidine thiocyanate-phenol-chloroform extraction procedure (21). Detection of pseudouridines in the 18S and 28S rRNAs was performed by primer extension analysis of carboxymethyl cellulose (CMC)-alkali-treated HeLa cell RNAs (5). 32P-labeled oligonucleotides complementary to the human 18S rRNA from positions C238 to U256 (Ψ222), U685 to G700 (Ψ613 and Ψ655), C762 to U784 (Ψ690), and A1374 to C1393 (Ψ1330 and Ψ1351) were used as primers. Mapping of Ψ2496 in the 28S rRNA was performed with a primer complementary to the 28S rRNA from positions A2531 to C2548. For numbering of human 18S and 28S rRNAs, see GenBank accession number U13369. The primer extension products were fractionated on 6% sequencing gels.
Expression constructs.
The ACA26, ACA35, and ACA57 scaRNAs were overexpressed in human HeLa cells. To this end, the coding regions of ACA26 (oligonucleotides ACTAATCGATTACATTTTGAAGTTAGTGG and TCTAACGCGTTTGAAATAAGTCAATAAG), ACA35 (oligonucleotides ACTAATCGATTAGACCTGAGATGTGCTTA and TCTAACGCGTACAGTCACTAAAGCCGTA), and ACA57 (oligonucleotides ACTAATCGATGTAAGTCTGCCTGTCCTAT and TCTAACGCGTCTTAGGACGGCCCTCCTA) were PCR amplified with HeLa cell genomic DNA as a template. The amplified fragments were digested with restriction endonucleases ClaI and XhoI and inserted into the same sites of the pCMV-globin expression construct (13). Transfection of HeLa cells was performed with Fugene 6 (Roche) transfection reagent according to the manufacturer's instructions.
Fluorescence in situ hybridization.
Synthesis and chemical conjugation of amino-modified oligodeoxynucleotides with FluoroLink Cy3 monofunctional dye (Amersham), fluorescence hybridization of transfected HeLa cells, and image acquisition and processing were performed as described elsewhere (http://singerlab.aecom.yu.edu) (13). The following oligonucleotide probes were used to detect transiently expressed human scaRNAs (asterisks indicate amino-allyl-modified T residues that are sites of attachment for the fluorescent label): ACA26, AT*CAGCAAAGTCTTACTT*CATCAGACTCAGCCT*T; ACA35, TT*CTTAAACCCAGCTAT*CACAACACATCACAAGCCTT*T; and ACA57, GT*GTGTCCTGCCAGACT*ACCCTGTTAGAACT*G. A polyclonal rabbit anti-p80-coilin antibody was kindly provided by A. Lamond. Nuclear DNA was stained with 0.1 μg of 4′,6′-diamidino-2-phenylindole/ml.
RESULTS AND DISCUSSION
Identification of novel human box H/ACA RNAs.
From a human HeLa cell extract, box H/ACA RNAs were isolated by immunoprecipitation with an antibody directed against the GAR1 box H/ACA RNP protein (16). Since vertebrate box H/ACA pseudouridylation guide RNAs are processed from pre-mRNA introns (18, 27), the mature RNAs carry a 5′-terminal monophosphosphate and a 3′-terminal hydroxyl group (31). To facilitate the synthesis of full-length cDNAs, the 5′ and 3′ termini of the immunoselected box H/ACA RNAs were extended by the addition of a phosphorylated oligoribonucleotide with the help of T4 RNA ligase (Fig. 1B). Synthesis of cDNA was performed with avian myeloblastosis virus reverse transcriptase and an oligodeoxynucleotide primer complementary to the oligoribonucleotide tag of the template RNA. The resulting doubly-tagged cDNA was PCR amplified, inserted into a plasmid vector, and transformed into E. coli. About 1,500 individual clones were analyzed by manual or automated plasmid sequencing.
We identified a total of 1,120 RNA sequences that defined 17 previously identified and 61 novel box H/ACA RNAs. As demonstrated by computer folding (57), the new RNAs displayed all the characteristic hallmarks of box H/ACA RNAs; they folded into a “hairpin-hinge-hairpin-tail” structure and carried H and ACA box motifs (data not shown). Since among the defining structural features of H/ACA RNAs, the presence of an ACA motif or, in a few instances, an AUA motif located 3 nucleotides from the RNA 3′ end was noted, the newly identified RNAs were designated ACA RNAs and numbered from 1 to 61. The expression and size of each RNA were confirmed by Northern blot analysis. The majority of recombinant plasmids (62%) carried cDNAs of full-length box H/ACA RNAs. In a few instances, 5′- and/or 3′-extended RNAs that apparently represented processing intermediates of mature box H/ACA RNAs were also identified. About one-fourth of the recombinant plasmids carried cDNAs corresponding to various fragments of the 18S and 28S rRNAs (15%) or representing full-length or partial sequences of the 5S and 5.8S rRNAs (8%). Less frequently (<2%), we also obtained plasmids carrying spliceosomal snRNA, tRNA, and mRNA sequences. Although about 20 box H/ACA RNAs were highly overrepresented in our cDNA library, we identified the sequences of 18 box H/ACA RNAs only once, suggesting that our survey was not saturated.
The computer-predicted two-dimensional structures of the new box H/ACA RNAs were scrutinized to find putative antisense target recognition elements and eventually to identify potential substrate RNAs. Based on their predicted functions, the new box H/ACA RNAs were divided into three groups. As expected, the majority of these RNAs (43 species) have been implicated in guiding the pseudouridylation of the 18S or 28S rRNAs. Another group of RNAs (6 species) have been assigned to the direction of pseudouridine synthesis for the major spliceosomal snRNAs. Finally, 12 box H/ACA RNAs classified in the third group lacked significant complementarities to any known stable cellular RNAs.
Guide RNAs directing the pseudouridylation of 18S rRNA.
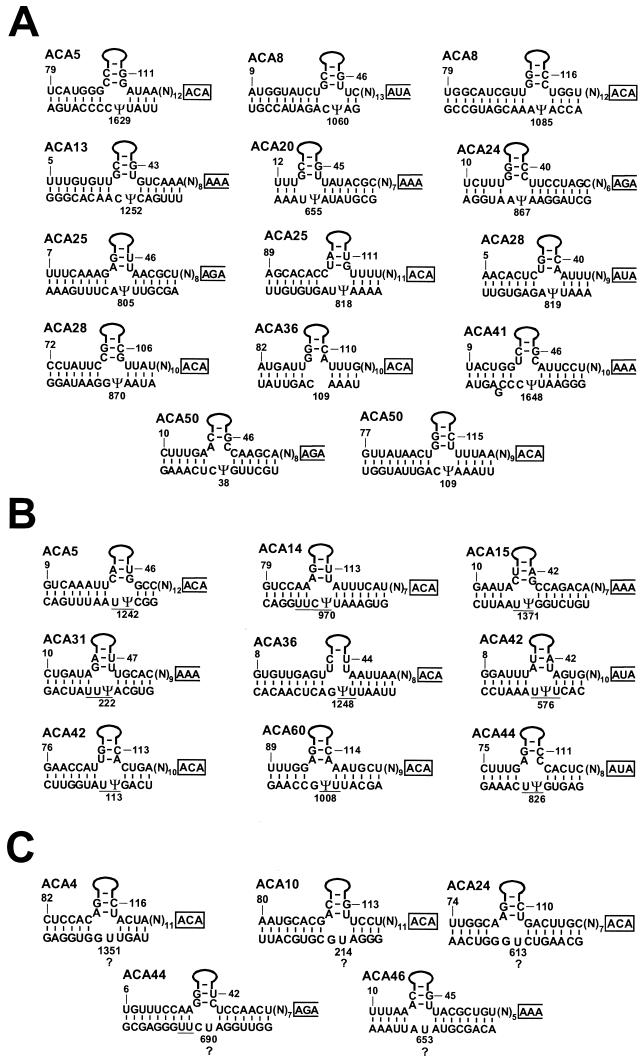
The human 18S rRNA is estimated to contain about 38 pseudouridine residues, 30 of which have been located either exactly or to within 2 or 3 nucleotides (35, 36). Of the newly identified box H/ACA RNAs, 19 have been predicted to function in 18S rRNA pseudouridylation (Fig. 2). The potential base-pairing interactions formed between these guide RNAs and 18S rRNA sequences perfectly conformed to the structural requirements defined for efficient RNA-guided pseudouridylation reactions (8, 19, 43). In the pseudouridylation pocket, the target uridines occupied an invariant position located 14 or 15 nucleotides upstream of the H or ACA box of the guide RNA.
FIG. 2.
Potential base-pairing interactions between box H/ACA RNAs and human 18S rRNA. (A) Selection of known pseudouridylation sites. The upper strands represent box H/ACA RNA sequences in a 5′-to-3′ orientation. Solid lines represent the upper parts of the 5′ or 3′ hairpins of guide RNAs. The ACA motifs are in closed boxes. The first three nucleotides of the putative H motifs are in open-ended boxes. The lower strands represent 18S rRNA sequences in a 3′-to-5′ orientation. The positions of pseudouridine residues were reported previously (36). The pseudouridine residues defined by interactions with guide RNAs are indicated (Ψ). The sequence of human 18S rRNA is from GenBank accession number U13369. (B) H/ACA RNAs distinguishing between two or three potential pseudouridylation sites. A bar below rRNA sequences indicates that one of the underlined uridines is pseudouridine. (C) Prediction of new pseudouridylation sites. Question marks indicate novel pseudouridylation sites revealed by identification of the corresponding guide RNAs.
Based on their predicted target sites, the newly discovered 18S rRNA-specific pseudouridylation guide RNAs could be divided into three groups. For ACA5 (Ψ1629), ACA8 (Ψ1060 and Ψ1085), ACA13 (Ψ1252), ACA20 (Ψ655), ACA24 (Ψ867), ACA25 (Ψ805 and Ψ818), ACA28 (Ψ819 and Ψ870), ACA36 (Ψ109), ACA41 (Ψ1648), and ACA50 (Ψ38 and Ψ109), the selected uridines already had been demonstrated to be pseudouridylated (35, 36) (Fig. 2A). The base-pairing capacity of some other guide RNAs, such as ACA5 (Ψ1242), ACA14 (Ψ970), ACA15 (Ψ1371), ACA31 (Ψ222), ACA36 (Ψ1248), ACA42 (Ψ113 and Ψ576), ACA44 (Ψ826), and ACA60 (Ψ1008), could distinguish between two or three possible pseudouridylation sites that had not been located to nucleotide resolution (36) (Fig. 2B). Finally, the uridine residues selected by the ACA4 (U1351), ACA10 (U214), ACA24 (U613), ACA44 (U690), and ACA46 (U653) putative pseudouridylation guide RNAs had not been reported to be pseudouridylated (36) (Fig. 2C).
Since not all pseudouridines had been located on the human 18S rRNA, the state of pseudouridylation of these uridine residues was examined by the CMC treatment-primer extension procedure (5) (Fig. 3). CMC reacts with N3 of pseudouridine, and the modified CMC-pseudouridine arrests reverse transcriptase 1 nucleotide before the pseudouridylation site. When 32P-labeled sequence-specific oligonucleotides were annealed to CMC-modified 18S rRNA and extended by reverse transcriptase, stop signals were observed 1 nucleotide before the U1351, U214, U613, U690, and U653 residues, indicating that they are pseudouridylated. Primer extension mapping of Ψ690 also revealed that, in contrast to previous reports (35, 36), neither U692 nor U693 is pseudouridylated in HeLa cell 18S rRNA. Likewise, mapping of Ψ222 showed that the 222-UUU-224 region of the human 18S rRNA contains only one and not two pseudouridines, as proposed before (35, 36). Thus, the characterization of new box H/ACA guide RNAs revealed new pseudouridylation sites and defined the correct positions of several previously detected pseudouridines in the human 18S rRNA.
FIG. 3.
Verification of pseudouridine residues in human 18S and 28S rRNAs predicted by guide RNA-rRNA interactions. CMC-alkali-modified (Ψ) or control (N) HeLa cell RNAs were analyzed by primer extension with 32P-labeled oligonucleotide primers complementary to the appropriate regions of the human 18S and 28S rRNAs. Lanes A, G, C, and U show dideoxy sequencing reactions performed on recombinant plasmids carrying the human 18S or 28S rRNA genes. Brackets and asterisks indicate uridines that were reported to be pseudouridylated.
Along with the previously characterized U23, U66, U67, U69, U70, and U71 snoRNAs (19), thus far 25 human box H/ACA guide snoRNAs have been implicated in 18S pseudouridylation (see Table S1 in the supplemental material). These guide RNAs together can define the correct positions of 33 pseudouridines. Provided that the human 18S rRNA contains a total of 38 pseudouridine residues (39), at this time only five pseudouridylation sites lack a potential guide snoRNA. In principle, it is possible that some of the pseudouridines missing a potential guide RNA, namely, Ψ684, Ψ922, Ψ1178 (36), Ψ1330 (Fig. 3), and one not yet placed, are synthesized by protein enzymes. However, it is also possible that pseudouridylation of the human 18S rRNA is achieved entirely by box H/ACA snoRNPs.
Pseudouridylation guide RNAs directing the modification of 28S rRNA.
Based on the estimated ratio of its pseudouridine content to its uridine content, the human 28S rRNA was predicted to carry about 57 pseudouridines (22, 35). Later, primer extension mapping located 54 pseudouridines to nucleotide resolution on the human 28S rRNA (45). At the outset of this study, six human box H/ACA guide snoRNAs (U19, U64, U65 U68, E2, and E3) had been implicated in the synthesis of nine pseudouridines in 28S rRNA (19) (see Table S2 in the supplemental material). Our survey identified 26 additional box H/ACA snoRNAs which could be linked to 35 reported pseudouridylation sites in the 28S rRNA (Fig. 4). Another snoRNA, ACA61, was predicted to position the U2496 residue for pseudouridylation. Indeed, primer extension mapping confirmed the presence of a novel pseudouridine at U2496 (Fig. 3), indicating that ACA61 is a genuine pseudouridylation guide RNA. Therefore, in total, 32 human box H/ACA snoRNAs have been implicated in 28S rRNA pseudouridylation. These guide RNAs can select 44 of the 57 pseudouridylation sites present in the human 28S rRNA (see Table S2 in the supplemental material).
FIG. 4.
Potential base-pairing interactions between box H/ACA RNAs and human 28S rRNA. The sequence of human 28S rRNA is from GenBank accession number U13369. The positions of pseudouridine residues were reported previously (45). See the legend to Fig. 2 for other details.
In summary, after identification of 56 putative human rRNA pseudouridylation guide RNAs (19; this study), we can attribute box H/ACA RNA-directed modification to 79 of the estimated 97 pseudouridylation sites present in the human 18S, 5.8S, and 28S rRNAs (see Tables S1 and S2 in the supplemental material). Even allowing the possibility that a few ribosomal pseudouridines are still unknown, we can conclude that the great majority of pseudouridines carried by the human rRNAs are synthesized by box H/ACA RNPs.
Of the 56 guide RNAs implicated in rRNA pseudouridylation, 22 are capable of directing two independent modification reactions. Usually, the two target sites of the “double pseudouridylation guides” are found on the same rRNA and, most frequently, are located close to each other in the primary rRNA sequence. The target pseudouridines selected by the 5′ hairpins of double guides can be located either upstream or downstream of the pseudouridylation sites determined by their 3′ hairpins. Less frequently, the 5′ and 3′ hairpins of a few double guides can function in the pseudouridylation of two rRNA species. The U69 snoRNA can direct the pseudouridylation of the 18S and 5.8S rRNAs, while the ACA10 and ACA31 snoRNAs are predicted to function in the modification of the 18S and 28S rRNAs.
Together, the two short helixes formed by the H/ACA guide RNA and rRNA sequences preceding and following the target uridine comprise a minimum of 9 bp or a maximum of 16 bp (Fig. 2 and 4). Most frequently (in 31% of the total instances), the rRNA-H/ACA RNA interaction involves 11 bp. In a few instances, mismatched (ACA24, ACA25, ACA15, and ACA58) or bulging (ACA41) nucleotides are also involved in the predicted rRNA-H/ACA RNA interaction. It is also noteworthy that G-U base pairs frequently occur in the predicted rRNA-H/ACA RNA helices, especially those composed of more than 10 bp.
Identification of the ACA2 and ACA34 snoRNAs provided us with new insights into the molecular mechanism of the evolution of modification guide RNAs. The ACA34 snoRNA and two sequence variants of the ACA2 snoRNA (ACA2a and ACA2b) are encoded within three different introns of a hypothetical protein gene, FLJ20436 (see Table S2 in the supplemental material). Interestingly, the ACA34 snoRNA shows a strong sequence similarity to both isoforms of ACA2 (66% identity). Therefore, it could be considered a third sequence variant of ACA2. Consistent with this notion, the 3′ hairpin of ACA34, similar to those of ACA2a and ACA2b, can position the U4283 residue in the 28S rRNA for pseudouridylation (Fig. 4). However, while the 5′ hairpins of ACA2a and ACA2b can direct the synthesis of Ψ4264, the 5′ hairpin of ACA34 selects Ψ4270 in the 28S rRNA. Apparently, the ACA2a, ACA2b, and ACA34 snoRNA genes have been generated by subsequent gene duplications during evolution. After the first duplication event, random point mutations in the target recognition motifs of the ACA34 gene or the parental gene of the contemporary ACA2a and ACA2b genes resulted in a novel snoRNA gene with new sequence specificity. The second gene duplication event generated the current ACA2a and ACA2b genes. Of course, random mutations in the ACA2a or ACA2b genes may produce another, functionally distinct pseudouridylation guide RNA gene during future evolution.
Guide RNAs implicated in the pseudouridylation of spliceosomal snRNAs.
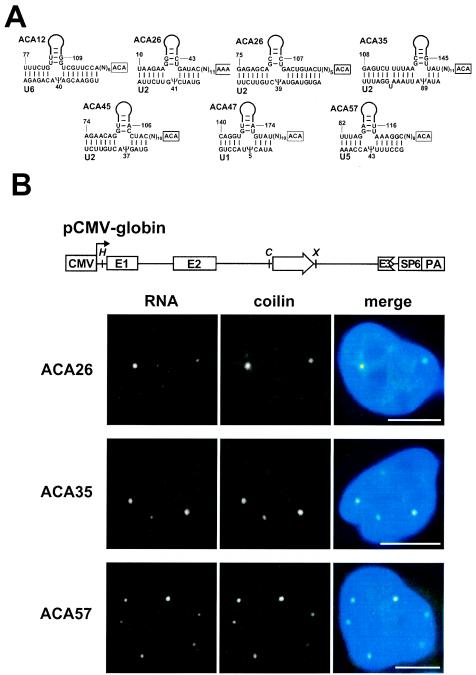
The human U1, U2, U4, U5, and U6 spliceosomal snRNAs together carry 21 pseudouridines (38). Earlier, we characterized four guide RNAs (U85, U89, U92, and U93) which were predicted to direct the synthesis of four pseudouridines in the U5 (Ψ85 and Ψ89) and U2 (Ψ92 and Ψ93) snRNAs (see Table S3A in the supplemental material). The current survey identified six additional box H/ACA RNAs implicated in the pseudouridylation of the U6 (ACA12), U2 (ACA26, ACA35, and ACA45), U1 (ACA47), and U5 (ACA57) snRNAs (Fig. 5A). Together, the above-described H/ACA guide RNAs can direct the synthesis of 11 pseudouridine residues in the U1, U2, U5, and U6 spliceosomal snRNAs. This finding indicates that the involvement of box H/ACA guide RNAs in snRNA pseudouridylation is more general than demonstrated before. However, at this time, we cannot exclude the possibility that protein enzymes also contribute to the pseudouridylation of spliceosomal snRNAs (25, 37).
FIG. 5.
Putative pseudouridylation guide RNAs directing the modification of spliceosomal snRNAs. (A) Proposed base-pairing interactions between box H/ACA guide RNAs and human spliceosomal snRNAs. (B) Fluorescence in situ localization of guide RNAs transiently overexpressed in HeLa cells. The schematic structure of the pCMV-globin expression construct is shown. The promoter region of cytomegalovirus (CMV), the exons in the human β-globin gene (E1 to E3), the polyadenylation region in the bovine growth hormone gene (PA), and the SP6 promoter are shown. The relevant restriction sites are indicated (H, HindIII; C, ClaI; X, XhoI). Fluorescence in situ hybridization with oligonucleotide probes specific for the ACA26, ACA35, and ACA57 scaRNAs was combined with indirect immunofluorescence with an antibody against the Cajal body marker protein, p80-coilin. The nuclear DNA was stained with 4′,6′-diamidino-2-phenylindole (blue). Bar, 10 μm.
Most of the previously identified modification guide RNAs implicated in pseudouridylation (U85 and U89) or 2′-O-ribose methylation (U87 and U88) of spliceosomal snRNAs are composed of a box H/ACA domain and a box C/D domain (13, 25). The U93 pseudouridylation guide RNA contains two tandemly arranged box H/ACA domains (26). In contrast, the new spliceosomal pseudoridylation guide RNAs, ACA12, ACA26, ACA35, ACA45, ACA47, and ACA57, possess the consensus hairpin-hinge-hairpin-tail structure of box H/ACA snoRNAs, indicating that canonical box H/ACA guide RNAs participate in the pseudouridylation of spliceosomal snRNAs more often than earlier observations suggested (13).
In human HeLa cells, the formerly characterized pseudouridylation and 2′-O-ribose methylation guide RNAs involved in modification of the RNA pol II-transcribed U1, U2, and U5 spliceosomal snRNAs were found to specifically accumulate in nucleoplasmic Cajal bodies (13, 24, 26). To further explore the subnuclear organization of the modification machinery of pol II-specific snRNAs, we investigated the localization of the newly identified ACA26, ACA35, and ACA57 RNAs that were implicated in the pseudouridylation of the U2 snRNA. Due to the detection limit of fluorescence in situ hydridization, the endogenous ACA26, ACA35, and ACA57 RNAs were not visible in HeLa cells (data not shown). Therefore, to facilitate detection, the ACA26, ACA35, and ACA57 RNAs were transiently overexpressed in HeLa cells by using the pCMV-globin expression construct, which had been developed to study the expression of intronic snoRNAs (13, 30) (Fig. 5B). Upon hybridization with fluorescent oligonucleotides specific for the ACA26, ACA35, and ACA57 RNAs, a few bright foci were observed in the nuclei of transfected HeLa cells. Staining of the same cells with an antibody against the Cajal body marker protein, p80-coilin (3), demonstrated that the foci accumulating the transiently expressed box H/ACA RNAs were Cajal bodies. These results further support the notion that the guide RNA machinery directing the modification of pol II-transcribed spliceosomal snRNAs is sequestered in Cajal bodies and that Cajal bodies are the nuclear locale for RNA-guided modification of pol II-specific spliceosomal snRNAs (13, 24, 26).
Putative pseudouridylation guide RNAs lacking substrate RNAs.
Our screen also identified 12 putative pseudouridylation guide RNAs which lacked the potential for guiding pseudouridine synthesis in human rRNAs, snRNAs, snoRNAs, scaRNAs, and YRNAs as well as in the U7, 7SK, MRP, RNase P, telomerase, and 7SL RNAs (see Table S3B in the supplemental material). The function of these so-called “orphan” guide RNAs remains unknown. According to the most obvious scenario, they may direct pseudouridine formation in some not-yet-identified RNAs. Thus, our data indicate that H/ACA RNA-directed RNA pseudouridylation is not restricted to rRNAs and snRNAs and is a more general phenomenon than assumed before. Alternatively, some of the orphan box H/ACA RNAs may also function in other aspects of RNA biogenesis. For example, the human U17 box H/ACA snoRNA and its yeast orthologue, snR30, play an essential role in the nucleolytic processing of 18S rRNA (4, 41). Likewise, a few box C/D snoRNAs play a crucial role in the nucleolytic processing of 18S (U3, U14, and U22) as well as 5.8S and 28S (U8) rRNAs (52). Therefore, it is possible that at least some of the newly identified orphan box H/ACA RNAs function in the nucleolytic processing of rRNAs or other cellular RNAs.
Northern blot analysis revealed that, in general, orphan H/ACA RNAs accumulate in HeLa cells at much lower levels than do snoRNAs involved in rRNA pseudouridylation (data not shown). In line with this observation, the majority of the new orphan RNAs appeared only once in our screen. This finding strongly suggests that the low-abundance orphan H/ACA RNAs, in contrast to the abundant rRNA modification guide RNAs, are highly underrepresented in our cDNA library. On the other hand, box H/ACA RNAs may also possess a tissue-specific expression pattern. The mouse and human HBI-36 snoRNAs that are encoded within introns of the brain-specific serotonin receptor gene accumulate only in brain tissues (10). Therefore, it is possible that many additional low-abundance and/or tissue-specific box H/ACA RNAs remain unidentified in human cells.
Genomic organization of human box H/ACA RNA genes.
Database searches revealed that the human genome carries one perfect and sometimes a few additional imperfect copies of the newly identified box H/ACA RNAs (see Tables S1, S2, and S3 in the supplemental material). The genomic loci that matched perfectly our cDNA sequences were considered bona fide H/ACA RNA genes. With no exception, all H/ACA RNA genes were found within introns of active transcription units known to produce spliced mRNAs. Moreover, all bona fide H/ACA RNA genes showed a parallel orientation with their host genes, indicating that the H/ACA RNAs and their host pre-mRNAs are synthesized cotranscriptionally. The majority of the H/ACA host genes are protein-coding genes, although in many instances, the functions of their predicted protein products remain unknown. Frequently, two or even more H/ACA RNAs are encoded within different introns of the same host gene. For example, the MGC5306 gene, which encodes a hypothetical protein, hosts at least six box H/ACA RNAs (ACA1, ACA8, ACA18, ACA25, ACA32, and ACA40) as well as two box C/D snoRNAs (mgh28S-2410 and mgh28S-2412) that are predicted to direct 2′-O-ribose methylation of the human 28S rRNA at positions C2410 and G2412 (unpublished results).
The rRNA pseudouridylation guide RNA genes are frequently located in ribosomal protein genes or in other genes connected to ribosome biogenesis (nucleolin) or protein synthesis (translation initiation and elongation factors and cytoplamic protein chaperones). Interestingly, the ACA36 and ACA56 genes lie within introns of the dyskerin gene, which encodes the common pseudouridine synthase of box H/ACA RNPs. Another snoRNA, ACA23, is hosted by the importin 7 gene, which encodes an import receptor for ribosomal proteins. Collectively, these observations further corroborate the notion that cotranscription is an important way of coordinating the regulation of factors required for ribosome synthesis and function. Therefore, it is possible that at least some of the snoRNA host genes that lack a function will be shown to participate in some aspects of protein synthesis. On the other hand, there are also a few host genes, for example, the cytochrome P450 oxidoreductase (ACA14a and ACA14b), methyl-CpG binding domain protein 2 (a DNA methyltransferase) (ACA37), and SRCAP (a transcriptional activator) (ACA30) genes, that cannot be directly linked to ribosome biogenesis or function.
Usually, the host genes of H/ACA RNAs directing the pseudouridylation of spliceosomal snRNAs (see Table S3A in the supplemental material) or lacking potential substrate RNAs (see Table S3B in the supplemental material) cannot be directly connected to ribosome biogenesis or translation. The ACA33 and ACA51 RNAs represent the only exceptions to this rule, since they are encoded within the S12 ribosomal protein and NOP56 box C/D snoRNP protein genes, respectively. The nonribosomal guide RNA genes frequently appear within genes encoding proteins involved in the functional maintenance of the human genome, such as the catalytic subunit of DNA polymerase alpha (ACA12), condensin subunit 1 (U85), nucleosome assembly protein 1-like protein (ACA54), and chromodomain helicase DNA binding protein 4 (ACA57). The functional significance of this observation is still unclear.
Several box H/ACA RNAs are encoded within genes that have little capacity for protein coding. The spliced and polyadenylated mRNA-like products of these genes contain no long open reading frames. Therefore, it appears that the only function of these genes is to express their intronic H/ACA RNAs (53). Of the 75 known human box H/ACA RNA genes, 15 seem to be located within introns of non-protein-coding genes, indicating that cotranscription within non-protein-coding pre-mRNAs is a rather common way to express pseudouridylation guide RNAs. Previously, four non-protein-coding snoRNA host genes were identified and shown to belong to the family of 5′-terminal oligopyrimidine (5′TOP) genes (9, 46, 51, 53). The sequence of 5′TOP mRNAs commences with a 5′-terminal C residue and is followed by a short pyrimidine tract that plays an important role in the upregulation of transcription and translation of 5′TOP mRNAs (2). The family of 5′TOP genes also includes ribosomal protein and translation elongation factor genes. The expression of rRNA modification guide snoRNAs within non-protein-coding 5′TOP pre-mRNAs therefore may provide a regulatory mechanism to coordinate the accumulation of snoRNAs and ribosomal proteins. Inspection of the 5′-terminal sequences of the expressed sequence tags of the newly identified non-protein-coding H/ACA snoRNA host genes revealed that at least two of them, named TOP1 (encoding ACA16, ACA44, and ACA61) and TOP2 (encoding ACA17 and ACA43), belong to the family of 5′TOP genes. Unfortunately, the correct 5′ ends of other non-protein-coding H/ACA host genes could not be inferred from their partial expressed sequence tags deposited in databases.
Besides the bona fide genes, most box H/ACA RNAs possess one or more imperfect genomic copies. These defective copies often represent 5′- or 3′-truncated versions of the authentic H/ACA RNA gene, indicating that they are apparently pseudogenes. Consistent with this conclusion, the sequences of the truncated H/ACA genes cannot be folded into the characteristic secondary structures of box H/ACA RNAs. Other genomic copies represent full-length H/ACA RNAs, but they contain numerous point mutations, short internal deletions, and/or insertions. These genomic sequences frequently fail to fold into a perfect hairpin-hinge-hairpin-tail structure or lack functional H and/or ACA motifs, suggesting that they do not code for functional RNAs. Supporting this notion, the default H/ACA sequences are frequently located in transcriptionally silent genomic loci or, alternatively, lie within known transcription units but in an opposite orientation. Both truncated and full-length pseudogene sequences are often followed by 8- to 15-nucleotide-long oligo(A) tracts and sometimes are flanked by short perfect repeats, indicating that they have been generated by retrotransposition (54). Finally, full-length H/ACA RNA sequences carrying a few point mutations are also found within introns of known genes in a parallel orientation. Since the sequences of these genes fold into the hairpin-hinge-hairpin-tail structure, they likely represent functional H/ACA RNA genes expressing sequence variants of the characterized RNAs.
Conclusions.
In this study, we identified 61 human box H/ACA snoRNAs and scaRNAs. The majority of these RNAs (49 species) are predicted to function as guide RNAs in the synthesis of 71 pseudouridine residues in the human 18S and 28S rRNAs and the U1, U2, U5, and U6 spliceosomal snRNAs. Some of the new H/ACA RNAs (12 species) lack potential target sites in human rRNAs, snRNAs, and snoRNAs. These orphan H/ACA RNAs either direct the pseudouridylation of some not-yet-identified RNAs or function in other aspects of cellular RNA biogenesis. As predicted by their genomic organization, all human box H/ACA RNAs are processed from pre-mRNA introns. The host genes of human H/ACA RNAs can be divided into three major groups. Most of them encode well-characterized proteins that frequently function in ribosome biogenesis or protein synthesis. Another group of H/ACA RNA host genes are predicted to encode proteins with unknown functions. Finally, the host genes of many H/ACA RNAs lack apparent protein-coding capacity and frequently belong to the family of 5′TOP genes.
Supplementary Material
Acknowledgments
We are grateful to Y. de Préval for the synthesis of oligodeoxynucleotides. We thank A. I. Lamond and W. Filipowicz for providing us with antibodies against p80-coilin and hGAR1. We thank M. Weber for critical reading of the manuscript and also for constructing a GenBank custom track of human H/ACA RNAs.
A.M.K. was funded by a short-term EMBO fellowship, a Hungarian State Eötvös fellowship, and l'Association pour la Recherche contre le Cancer. This work was supported by grants from CNRS, la Ligue Nationale contre le Cancer, and the Hungarian Research Foundation (OTKA, T31738).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agris, P. F. 1996. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acids Res. Mol. Biol. 53:79-129. [DOI] [PubMed] [Google Scholar]
- 2.Amaldi, F., and P. Pierandrei-Amaldi. 1997. TOP genes: a translationally controlled class of genes including those coding for ribosomal proteins. Prog. Mol. Subcell. Biol. 18:1-17. [DOI] [PubMed] [Google Scholar]
- 3.Andrade, L. E., E. K. Chan, I. Raska, C. L. Peebles, G. Roos, and E. M. Tan. 1991. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 173:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atzorn, V., P. Fragapane, and T. Kiss. 2003. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 24:1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakin, A., and J. Ofengand. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32:9754-9762. [DOI] [PubMed] [Google Scholar]
- 6.Balakin, A. G., L. Smith, and M. J. Fournier. 1996. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell 86:823-834. [DOI] [PubMed] [Google Scholar]
- 7.Björk, G. R., J. U. Ericson, C. E. D. Gustafsson, T. G. Hagervall, Y. H. Jönsson, and P. M. Wikstörm. 1987. Transfer RNA modification. Annu. Rev. Biochem. 56:263-287. [DOI] [PubMed] [Google Scholar]
- 8.Bortolin, M. L., P. Ganot, and T. Kiss. 1999. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 18:457-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bortolin, M. L., and T. Kiss. 1998. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4:445-454. [PMC free article] [PubMed] [Google Scholar]
- 10.Cavaillé, J., K. Buiting, M. Kiefmann, M. Lalande, C. I. Brannan, B. Horsthemke, J. P. Bachellerie, J. Brosius, and A. Hüttenhofer. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97:14311-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavaillé, J., M. Nicoloso, and J. P. Bachellerie. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383:732-735. [DOI] [PubMed] [Google Scholar]
- 12.Charette, M., and M. W. Gray. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49:341-351. [DOI] [PubMed] [Google Scholar]
- 13.Darzacq, X., B. E. Jády, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21:2746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decatur, W. A., and M. J. Fournier. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278:695-698. [DOI] [PubMed] [Google Scholar]
- 15.Decatur, W. A., and M. J. Fournier. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27:344-351. [DOI] [PubMed] [Google Scholar]
- 16.Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.England, T., A. Bruce, and O. Uhlenbeck. 1980. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 65:65-74. [DOI] [PubMed] [Google Scholar]
- 18.Filipowicz, W., and V. Pogacic. 2002. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 14:319-327. [DOI] [PubMed] [Google Scholar]
- 19.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 20.Ganot, P., M. Caizergues-Ferrer, and T. Kiss. 1997. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11:941-956. [DOI] [PubMed] [Google Scholar]
- 21.Goodall, G. J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148-161. [DOI] [PubMed] [Google Scholar]
- 22.Hughes, D. G., and B. E. Maden. 1978. The pseudouridine contents of the ribosomal ribonucleic acids of three vertebrate species. Numerical correspondence between pseudouridine residues and 2′-O-methyl groups is not always conserved. Biochem. J. 171:781-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hüttenhofer, A., M. Kiefmann, S. Meier-Ewert, J. O'Brien, H. Lehrach, J. P. Bachellerie, and J. Brosius. 2001. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 20:2943-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jády, B. E., X. Darzacq, K. E. Tucker, A. G. Matera, E. Bertrand, and T. Kiss. 2003. Modification of small nuclear RNAs occurs in the nucleoplasmic Cajal bodies following import from the cytoplasm. EMBO J. 22:1878-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jády, B. E., and T. Kiss. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiss, A. M., B. E. Jády, X. Darzacq, C. Verheggen, E. Bertrand, and T. Kiss. 2002. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 30:4643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20:3617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 29.Kiss, T., M. L. Bortolin, and W. Filipowicz. 1996. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol. Cell. Biol. 16:1391-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiss, T., and W. Filipowicz. 1995. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev. 9:1411-1424. [DOI] [PubMed] [Google Scholar]
- 31.Kiss, T., and W. Filipowicz. 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J. 12:2913-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss-László, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 33.Lafontaine, D. L., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange, T. S., M. Ezrokhi, F. Amaldi, and S. A. Gerbi. 1999. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell 10:3877-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maden, B. E. 1990. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acids Res. Mol. Biol. 39:241-303. [DOI] [PubMed] [Google Scholar]
- 36.Maden, E. H., and J. A. Wakeman. 1988. Pseudouridine distribution in mammalian 18 S ribosomal RNA. A major cluster in the central region of the molecule. Biochem. J. 249:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massenet, S., Y. Motorin, D. L. Lafontaine, E. C. Hurt, H. Grosjean, and C. Branlant. 1999. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 19:2142-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massenet, S., A. Mougin, and C. Branlant. 1998. Posttrancriptional modification in the U small nuclear RNAs, p. 201-227. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 39.McCallum, F. S., and B. E. Maden. 1985. Human 18 S ribosomal RNA sequence inferred from DNA sequence. Variations in 18 S sequences and secondary modification patterns between vertebrates. Biochem. J. 232:725-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCloskey, J. A., and P. F. Crain. 1998. The RNA modification database—1998. Nucleic Acids Res. 26:196-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrissey, J. P., and D. Tollervey. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell. Biol. 13:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narayanan, A., A. Lukowiak, B. E. Jády, F. Dragon, T. Kiss, R. M. Terns, and M. P. Terns. 1999. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 18:5120-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 44.Ofengand, J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514:17-25. [DOI] [PubMed] [Google Scholar]
- 45.Ofengand, J., and A. Bakin. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266:246-268. [DOI] [PubMed] [Google Scholar]
- 46.Pelczar, P., and W. Filipowicz. 1998. The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol. 18:4509-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richard, P., X. Darzacq, E. Bertrand, B. E. Jády, C. Verheggen, and T. Kiss. 2003. A common sequence motif determines the Cajal body-specific localisation of box H/ACA scaRNAs. EMBO J. 22:4283-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruff, E. A., O. J. Rimoldi, B. Raghu, and G. L. Eliceiri. 1993. Three small nucleolar RNAs of unique nucleotide sequences. Proc. Natl. Acad. Sci. USA 90:635-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samarsky, D. A., and M. J. Fournier. 1999. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 27:161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Smith, C. M., and J. A. Steitz. 1998. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol. 18:6897-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10:17-39. [PMC free article] [PubMed] [Google Scholar]
- 53.Tycowski, K. T., M. D. Shu, and J. A. Steitz. 1996. A mammalian gene with introns instead of exons generating stable RNA products. Nature 379:464-466. [DOI] [PubMed] [Google Scholar]
- 54.Vitali, P., H. Royo, H. Seitz, J. P. Bachellerie, A. Hüttenhofer, and J. Cavaillé. 2003. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 31:6543-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, H., D. Boisvert, K. K. Kim, R. Kim, and S. H. Kim. 2000. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 19:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zebarjadian, Y., T. King, M. J. Fournier, L. Clarke, and J. Carbon. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 19:7461-7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.