Abstract
NO regulates a variety of physiological processes, including cell proliferation, differentiation, and inflammation. S-nitrosylation, a NO-mediated reversible protein modification, leads to changes in the activity and function of proteins. In particular, the role of S-nitrosylation during adipogenesis is largely unknown. We hypothesized that the normal physiological levels of NO, but not the excess levels generated under severe conditions, such as inflammation, may be critically involved in the proper regulation of adipogenesis. We found that endogenous S-nitrosylation of proteins was required for adipocyte differentiation. By performing a biotin-switch assay, we identified FAS, a key lipogenic enzyme in adipocytes, as a target of S-nitrosylation during adipogenesis. Interestingly, we also observed that the dimerization of FAS increased in parallel with the amount of S-nitrosylated FAS during adipogenesis. In addition, we found that exogenous NO enhanced the dimerization and the enzymatic activity of FAS. Moreover, site-directed mutagenesis of three predicted S-nitrosylation sites indicated that S-nitrosylation of FAS at Cys1471 and Cys2091, but not at Cys1127, increased its enzymatic activity. Taken together, these results suggest that the S-nitrosylation of FAS at normal physiological levels of NO increases its activity through dimerization and may contribute to the proper regulation of adipogenesis.
Keywords: adipocyte, adipogenesis, nitric oxide
FAS, which is highly expressed in adipose tissue, liver, and lactating mammary glands, catalyzes the synthesis of palmitate from acetyl-CoA and malonyl-CoA in the presence of NADPH (1–3). FAS is active as a homodimer, and each monomer has seven separate functional domains, including malonyl/acetyltransferase, β-ketoacyl synthase, β-ketoacyl reductase, dehydrase, enoyl reductase, thioesterase, and acyl carrier protein (4). Although the monomer contains all the activities required for palmitate synthesis, dimer formation is essential for FAS function (5–9). Several studies have reported that FAS is involved in the regulation of adipose tissue mass (1, 10–18), a key enzyme regulating energy metabolism, and a metabolic oncogene (19, 20). Therefore, FAS has been considered as a potential target for the development of anti-obesity and anti-cancer drugs (21–25). Expression of FAS is transcriptionally regulated by the sterol regulatory element-binding protein-1c (SREBP-1c) and by upstream stimulatory factors 1 and 2 (USF1 and USF2) in response to feeding or to insulin (26, 27). So far, however, nothing is known about the posttranslational modification of FAS except that it is ubiquitinated (28, 29).
NO is a critical signaling molecule that is involved in a number of physiological and pathological processes. When overproduced, NO is known to have harmful effects on cell function and survival (30–32). In contrast, within the physiological concentration range, NO plays a critical role as a regulator of cellular signaling pathways (33–35). A major mechanism mediating the biological function of NO is protein S-nitrosylation, a posttranslational modification of proteins involving the addition of an NO+ to a cysteine thiol group of the proteins. Therefore, under physiological conditions, S-nitrosylation can affect a number of cellular signaling pathways by inducing conformational changes of the protein and affecting protein-protein interactions and protein functions (36–39). NO is reported to improve the β-oxidation of fatty acids through reversible protein S-nitrosylation (40). NO is also known to impair the anti-lipolytic action of insulin in obesity through S-nitrosylation (41) and to enhance adipogenesis in primary human preadipocytes (42). However, little is known about the target proteins of S-nitrosylation during adipogenesis.
In the present study, we showed that FAS is S-nitrosylated during adipogenesis and that the S-nitrosylation of FAS increases its activity by enhancing dimerization, indicating that S-nitrosylation of FAS contributes to adipogenesis.
MATERIALS AND METHODS
Reagents and antibodies
The NO scavengers, 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO) (P5084) and 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) (C221), were obtained from Sigma-Aldrich (St. Louis, MO). The NO donor, diethylamine diazeniumdiolate diethylammonium salt (DEA-NONOate) (D5431), and a NO synthase (NOS) inhibitor, Nω-nitro-L-arginine (N5501), were also obtained from Sigma-Aldrich. S-nitrosocysteine (SNOC) was prepared as described previously (43). Lipofectamine 2000 (11668019) and Lipofectamine LTX with Plus reagent (15338030) were obtained from Life Technologies (Carlsbad, CA). Anti-nitrosocysteine (ab94930) and anti-FAS (ab22759) antibodies were purchased from Abcam (Cambridge, UK), mouse monoclonal anti-FLAG (F3166) antibody from Sigma-Aldrich, and anti-GAPDH (SC-25778) from Santa Cruz Biotechnology (Dallas, TX).
Cell culture and adipose tissue
HEK293 cells were grown in DMEM (Life Technologies) containing 10% FBS at 37°C under 5% CO2/95% air. Human adipose-derived stem cells (ADSCs) were purchased from Lonza (Walkersville, MD). Confluent ADSCs were incubated in ADSC differentiation medium containing 1 μg/ml insulin, 1 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1 μM troglitazone (referred to as IDXT), and 10% FBS. The culture medium was changed every 2 days. Insulin (I5500), IBMX (I5879), dexamethasone (D8893), and troglitazone (T2573) were purchased from Sigma-Aldrich. Flash-frozen human subcutaneous adipose tissue (T-SQFX-FF) was purchased from Zen-Bio (Research Triangle Park, NC).
NO assay
For quantitative determination of the NO level, a NO assay kit (Colorimetric, ab65328) from Abcam was used. Total nitrate and nitrite levels were measured in a two-step process. The first step converted nitrate to nitrite with nitrate reductase. The second step used Griess reagents to convert nitrite to a deep purple azo compound. The amount of azochromophore reflected the amount of nitric oxide in the samples. The absorbance at 550 nm was immediately recorded and compared with the absorbance of a freshly prepared standard curve of sodium nitrite.
Immunocytochemistry
Adipocytes were fixed for 1 h in 10% formalin in PBS followed by permeabilization in 0.1% Triton X-100 for 10 min. Blocking was performed in PBS containing 10% normal goat serum for 1 h. The fixed cells were incubated using anti-FAS or anti-nitrosocysteine overnight in the blocking solution. The cells were washed and incubated for 1 h with either Alexa-Fluor-488-conjugated goat anti-mouse antibody (A10680) or Alexa-Fluor-594-conjugated goat anti-rabbit antibody (A11012) (Molecular Probes, Carlsbad, CA). After washing, the cells were mounted in a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI). Cells were observed using an EVOS FL cell imaging system (Life Technologies). Formalin solution (HT501128), goat serum (G9023), and Fluoroshield mounting medium with DAPI (F6057) were purchased from Sigma-Aldrich.
FAS constructs
A plasmid encoding human FAS (SC127829, NM_004104) was obtained from Origene (Rockville, MD) and subcloned into the pcDNA3.1 vector (Life Technologies). Point mutations were introduced to change each cysteine in the FAS gene (Cys161, Cys1127, Cys1471, or Cys2091) to an alanine. The primer sequences for PCR-directed mutagenesis of FAS Cys161 to Ala were: forward, 5′-CGCACTGGACACAGCCGCCTCCTCCAGCCTGATGGC-3′ reverse, 5′-GCCATCAGGCTGGAGGAGGCGGCTGTGTCCAGTGCG-3′. For mutagenesis of FAS Cys1127 to Ala, the primer sequences were: forward, 5′-CCACACGGAGGAGGGGGCCCTGTCTGAGCGCGCTG-3′ reverse, 5′-CAGCGCGCTCAGACAGGGCCCCCTCCTCCGTGTGG-3′. For mutagenesis of FAS Cys1471 to Ala, the primer sequences were: forward, 5′-CGGGAACCGCCTCCGGGCTGTGCTGCTCTCCAACC-3′ reverse, 5′-GGTTGGAGAGCAGCACAGCCCGGAGGCGGTTCCCG-3′. For mutagenesis of FAS Cys2091 to Ala, the primer sequences were: forward, 5′-CCAGCGCATGGCGTCCGCCCTGGAGGTGCTGGACC-3′ reverse, 5′-GGTCCAGCACCTCCAGGGCGGACGCCATGCGCTGG-3′.
Oil Red O staining
Adipocyte differentiation was assessed using an Oil Red O stain (O0625, Sigma-Aldrich) as an indicator of intracellular lipid accumulation. After ADSC differentiation to adipocytes, cells were washed twice with PBS, fixed with 10% formalin in PBS for 1 h, and then washed with 60% isopropanol, before being allowed to dry completely. Adipocytes were stained with 0.2% Oil Red O reagent for 10 min at room temperature and washed with water four times. Each sample was eluted with 100% isopropanol for 10 min and absorbance was measured at 500 nm using a spectrophotometer. To visualize the nucleus, adipocytes were counterstained with a hematoxylin reagent (H3136, Sigma-Aldrich) for 2 min and washed twice with water. The level of adipocyte differentiation was observed using an inverted phase microscope.
Biotin-switch assay
Cell lysates or adipose tissue lysates were prepared in HENTS buffer [100 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine, 1% Triton X-100, and 0.1% SDS (pH 7.4)]. The biotin-switch assay was carried out as described previously (44) with a slight modification. Briefly, free thiols in the sample were blocked by incubation with 10 mM methyl methanethiosulfonate (MMTS) at 50°C for 15 min. The MMTS was then removed by acetone precipitation and the pellet was resuspended in HENS buffer [100 mM HEPES, 1 mM EDTA, 0.1 mM neocuproine, and 0.1% SDS (pH 7.4)]. S-nitrosothiols were selectively reduced with 20 mM ascorbate and the reformed free thiols were labeled with 1 mM N-[6-(biotinamido)hexyl]3′-(2′-pyridyldithio)-propionamide (HPDP-biotin) for 1 h at room temperature. The biotinylated proteins were then collected on avidin agarose beads, which were then washed three times with neutralization buffer [20 mM HEPES-NaOH (pH 7.4), 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100]. Proteins were eluted from the beads by SDS-PAGE loading buffer and subjected to immunoblot analysis. Neocuproine (N1501), MMTS (64306), and ascorbate (A7506) were purchased from Sigma-Aldrich. EZ-Link HPDP-biotin and NeutrAvidin agarose beads were from Thermo Scientific (Waltham, MA).
FAS activity assay
The FAS activity assay was performed as previously described (45) with minor modifications. In brief, cell lysates were mixed with acetyl-CoA and NADPH in 0.2 M potassium phosphate buffer and 0.4 mM EDTA (pH 7.0), and incubated at 30°C for 10 min. The enzymatic reaction was initiated by adding 20 μl of malonyl-CoA solution (0.2 mM), and the decrease in optical density (OD) was measured every 1 min for 30 min via kinetic measurements obtained on a microplate reader set at 340 nm. Based on the results, the overall FAS enzyme activity was estimated by calculating the NADPH oxidation, using ε = 6,220 M−1cm−1. Acetyl-CoA (A2056), NADPH (N1630), and malonyl-CoA (M4263) were purchased from Sigma-Aldrich.
Statistical analysis
The statistical analyses of the data were analyzed with the Student’s t-test or by one-way ANOVA. Data are expressed as mean ± SD of at least three independent experiments. P < 0.05 was considered statistically significant.
RESULTS
Protein S-nitrosylation is required for adipocyte differentiation
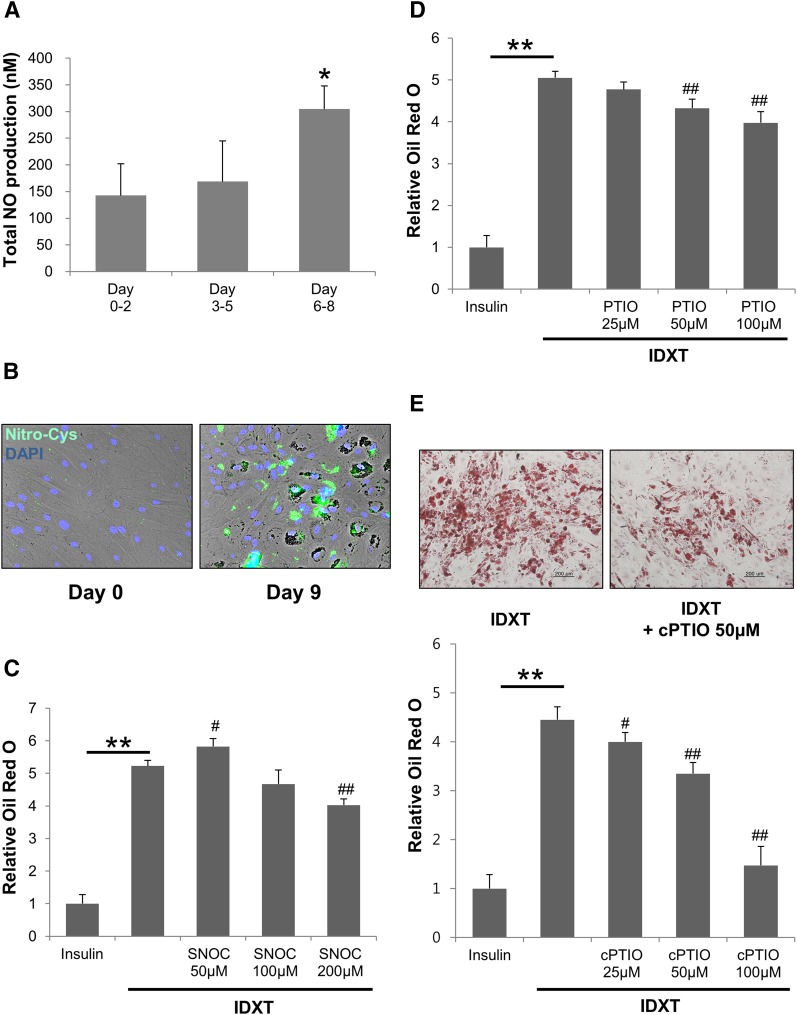
To investigate whether NO is involved in the process of adipocyte differentiation, we first evaluated NO production during adipocyte differentiation (Fig. 1A). The production of NO during adipocyte differentiation was estimated by measuring the total concentration of its oxidation metabolites, nitrite and nitrate, using a NO assay kit. Compared with the early differentiation period of adipocytes (days 0–2), there was a significant increase in total NO level in the supernatants of cells taken at the later differentiation period (days 6–8) (Fig. 1A). We next investigated whether protein S-nitrosylation, a major posttranslational modification by NO, might have a unique role in adipogenesis. Protein S-nitrosylation can occur even at low levels of NO and can affect the activity and stability of specific proteins (36–39). Immunofluorescence assay using an antibody against nitroso-cysteine showed that protein S-nitrosylation was higher in differentiated adipocytes (day 9) than in undifferentiated preadipocytes (Fig. 1B). SNOC, an NO donor, demonstrated a biphasic effect: it increased adipogenesis at a low concentration (50 μM) and decreased adipogenesis at a high concentration (200 μM) (Fig. 1C). cPTIO and PTIO, well-known NO scavengers, inhibited adipogenesis in a concentration-dependent manner (Fig. 1D, E). The better NO scavenger, cPTIO showed slightly more efficient inhibition of adipogenesis than PTIO at the same concentrations (Fig. 1D, E). To test the effect of NOS inhibitor on adipogenesis, we performed the same experiments with Nω-nitro-L-arginine (NNA), a broad NOS inhibitor. We found that there was no effect of NNA on adipogenesis (supplementary Fig. 1A). In agreement with a previous report (46), we could not see the expression of inducible NOS (iNOS) and neuronal NOS (nNOS) during adipogenesis, whereas the endothelial NOS (eNOS) protein band was detectable only at the later differentiation stage (day 14) (supplementary Fig. 1B).
Fig. 1.
Adipocyte differentiation requires protein S-nitrosylation. A: Total NO produced by differentiated adipocytes at each period (0–2 days, 3–5 days, or 6–8 days) was measured using an NO assay kit. *P < 0.05 compared with 0–2 days by t-test. C–E: ADSCs were grown in the IDXT condition and treated with the indicated concentration of SNOC (C), PTIO (D), or cPTIO (E). Lipid droplets in adipocytes were stained with Oil Red O dye and quantified. Data were normalized by setting the insulin control as 1 (mean ± SD). **P < 0.01 by t-test. #P < 0.05, ##P < 0.01 compared with IDXT by t-test. Data are representative of three individual experiments. B: On day 9 following IDXT incubation, immunofluorescence staining was performed on the control and the 9 day differentiated adipocyte samples using an anti-nitrosocysteine antibody.
Increased S-nitrosylation and dimerization of FAS in differentiated adipocytes
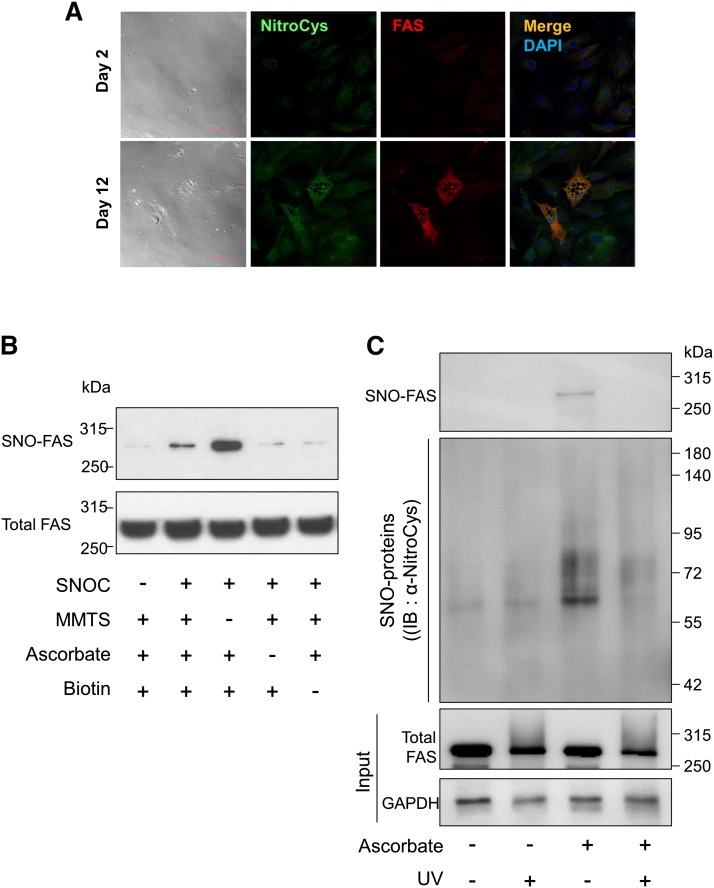
Following a previous report on fatty acid metabolism and S-nitrosylation (40), we analyzed the levels of FAS, one of the essential proteins for lipid droplet formation during adipogenesis, in differentiated adipocytes. We confirmed that the levels of nitrosocysteine and FAS in more differentiated adipocytes (day 12) were higher than those in less differentiated adipocytes (day 2) (Fig. 2A). The yellow color in the merged image of nitrosocysteine (green) and FAS (red) images implied that FAS was likely to be S-nitrosylated (SNO-FAS) (Fig. 2A). We also performed immunostaining for nitrosocysteine with negative controls to reaffirm antibody and detection specificity (supplementary Fig. 2). To confirm that the biotin-switch assay allowed detection of SNO-FAS, the lysate of undifferentiated adipocytes was subjected to the assay after incubation in SNOC with or without the assay components, including MMTS, ascorbate, and HPDP-biotin (Fig. 2B). In addition, we tested whether FAS in adipose tissue homogenates could be S-nitrosylated. We eliminated SNOC by treatment with UV-light and used this sample for negative control. We found that FAS was nitrosylated in human adipose tissue and the SNO-FAS in lysates was eliminated by UV-light (Fig. 2C).
Fig. 2.
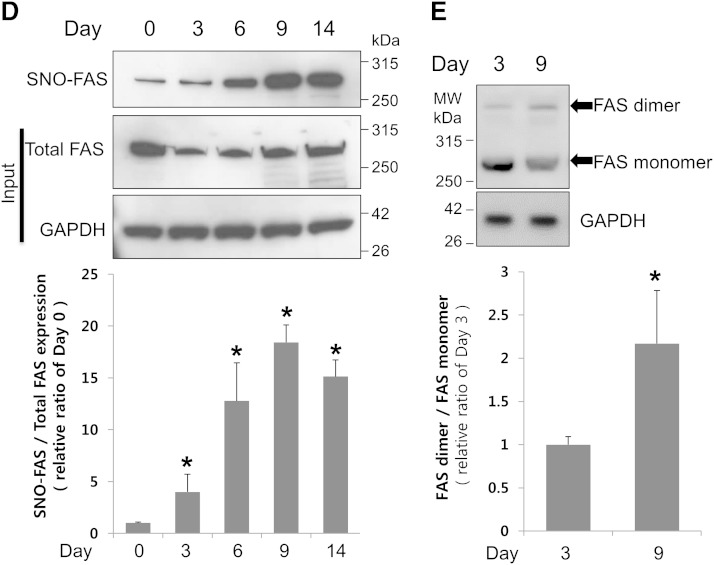
Increased S-nitrosylation and dimerization of FAS in differentiated adipocytes. A: Immunofluorescence to detect expression of FAS (red) and S-nitrosylated proteins (green) in differentiated adipocytes (on days 2 and 12). Merged images (yellow) demonstrate the overlap between the localization of FAS and S-nitrosylated proteins. DAPI staining (blue) was used to identify cell nuclei. Scale bar = 100 μm. B: To confirm the S-nitrosylation of FAS and that the biotin-switch assay detects SNO-FAS properly, the lysate of undifferentiated ADSCs was subjected to a biotin-switch assay with or without the components of the assay, as described in Materials and Methods, and immunoblotting (IB) following incubation with the NO donor SNOC or old SNOC. C: The lysates of human adipose tissue (200 mg) were irradiated with UV-light or incubated in the dark for 10 min. Then, the lysates were subjected to a biotin-switch assay with or without ascorbate. UV-irradiated sample was used as a negative control by eliminating SNOC. Data are representative of three individual experiments. D: The lysates of cells at different stages of adipocyte differentiation (days 0–14) were subjected to a biotin-switch assay followed by immunoblotting for SNO-FAS (top), total FAS (middle), and GAPDH (bottom), and the ratio of SNO-FAS/total FAS expression was quantified. Data were normalized by setting day 0 as 1 (mean ± SD). *P < 0.05 compared with day 0 by t-test. E: Immunoblotting of cell lysates carried out under nondenaturing conditions shows the dimerization of FAS during adipogenesis (days 3 and 9). The ratio of FAS dimer/FAS monomer was quantified. Data were normalized by setting day 3 as 1 (mean ± SD). *P < 0.05 compared with day 3 by t-test. Data are representative of three individual experiments.
To test whether SNOC-induced FAS nitrosylation is cPTIO sensitive, we incubated the cells with cPTIO before SNOC treatment and performed a biotin-switch assay. We found that SNOC-induced FAS nitrosylation was cPTIO sensitive (supplementary Fig. 3). It has been reported that DEA-NONOate, another NO donor, can nitrosylate proteins (47). Thus we tested the effect of DEA-NONOate on FAS nitrosylation. Similar to SNOC, DEA-NONOate nitrosylated FAS (supplementary Fig. 3). We then compared the relative amount of SNO-FAS at different stages of adipocyte differentiation (days 0–14). Interestingly, we found that the SNO-FAS level was increased during adipocyte differentiation (Fig. 2D). We also found that FAS dimer formation was increased during the later stage of adipocyte differentiation (day 9) (Fig. 2E).
Dimerization and activation of FAS are mediated by NO
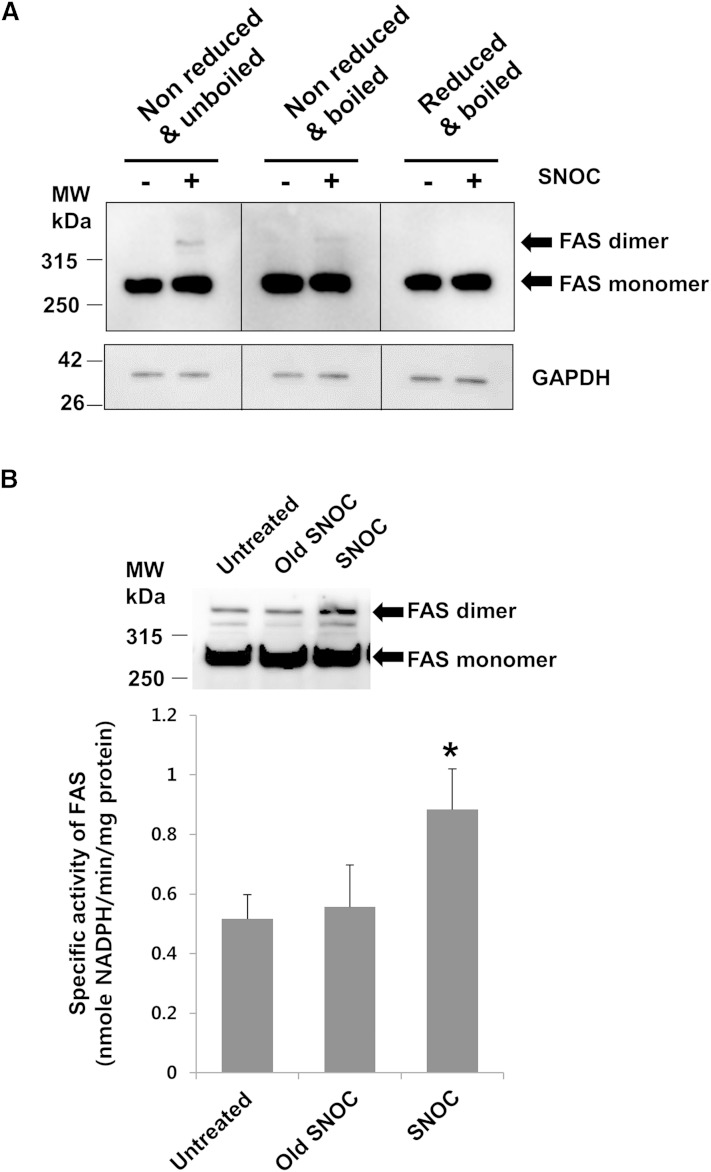
To identify whether FAS dimerization can be directly induced by NO, we examined the effect of exogenous NO on the dimerization level of FAS by nondenaturing PAGE and Western blot analysis. We found that SNOC significantly increased FAS dimerization (Fig. 3A). FAS dimer disappeared when cell lysates were boiled under reducing conditions, which suggested that FAS dimerization might be mediated through disulfide bonds. We then tested whether NO could influence FAS activity. Similar to the result with dimerization, the enzymatic activity of FAS was significantly higher in the lysate of ADSCs treated with fresh SNOC than in that treated with old SNOC, from which NO had dissipated (Fig. 3B).
Fig. 3.
Exogenous NO induces dimerization and activation of FAS. A: SNOC-induced FAS dimer formation in ADSCs was observed on a nondenaturing PAGE and by Western blot analysis. SNOC-induced dimer of FAS was well-detected when cell lysates were not boiled under nonreducing conditions (left) and still detected when cell lysates were boiled under nonreducing conditions (middle), but not detected when cell lysates were boiled under reducing conditions (right). B: The lysates of ADSCs were exposed to SNOC (25 μM) or old SNOC and subjected to nondenaturing PAGE (upper) or the FAS activity assay (lower). *P < 0.05 compared with old SNOC by t-test. Error bars indicate SD. Data are representative of three individual experiments.
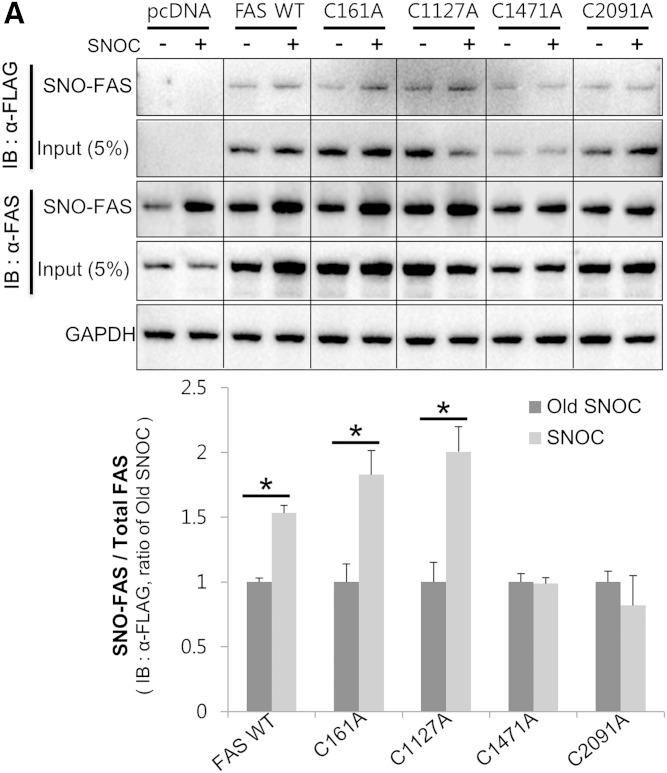
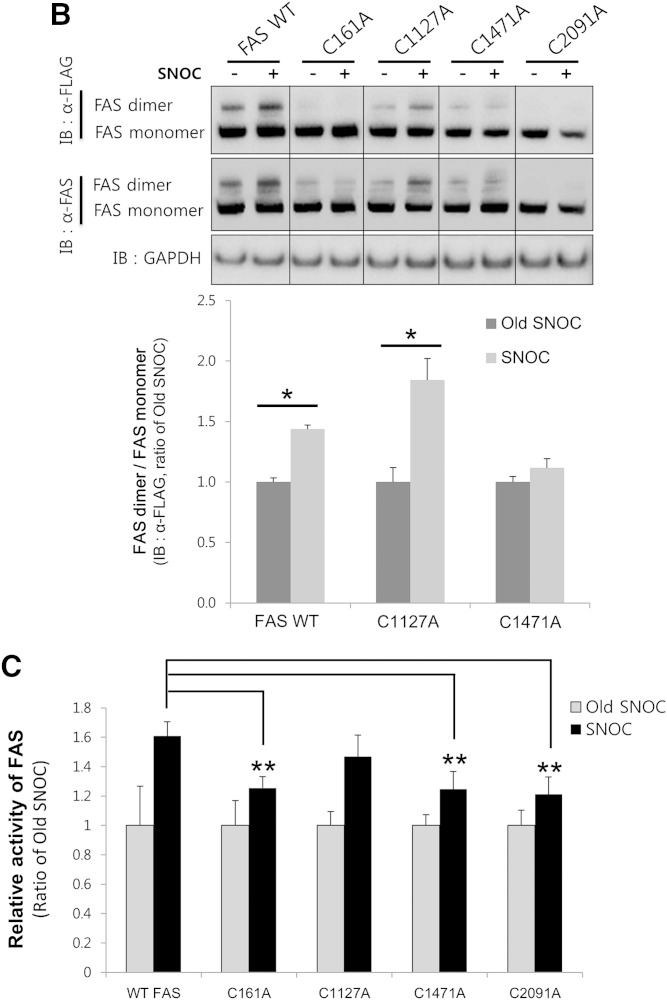
FAS is S-nitrosylated at Cys1471 and Cys2091
FAS contains several cysteines in its primary sequence. Recently, Cys1127, Cys1471, and Cys2091 have been predicted as candidate sites for FAS S-nitrosylation by a MS-based proteomics study (40). Thus, we examined whether these three cysteines might be involved in FAS S-nitrosylation. We generated mutants in which each of the cysteines was mutated to alanine (C1127A, C1471A, and C2091A). We also included Cys161, in the active site of FAS, as a negative control for FAS activity. HEK293 cells were transiently transfected with FLAG-tagged WT FAS (FAS-WT) or its cysteine to alanine mutants and were exposed to SNOC. Cell lysates were subsequently subjected to the biotin-switch assay. The treatment of cells with SNOC resulted in S-nitrosylation of FAS-WT and the C161A and C1127A mutant proteins. In contrast, neither the C1471A mutant nor the C2091A mutant was nitrosylated by SNOC (Fig. 4A). The dimerization of FAS-WT and the C1127A mutant was enhanced by SNOC, but the dimerization of C161A, C1471A, or C2091A did not increase (Fig. 4B), demonstrating that S-nitrosylation of FAS at Cys1471 or Cys2091 is important for FAS dimerization. Consistent with these results, the SNOC-induced increase in activity of the C161A, C1471A, or C2091A mutants was significantly lower than that of FAS-WT (Fig. 4C). In addition, we examined the effect of cysteine to alanine mutation itself upon the basal enzymatic activity, and there was no significant difference versus FAS-WT (supplementary Fig. 4).
Fig. 4.
FAS is S-nitrosylated on Cys1471 and Cys2091. A, B: HEK293 cells were transfected with FLAG-tagged FAS-WT or its cysteine mutants (C161A, C1127A, C1471A, and C2091A) and exposed to SNOC or old SNOC. The cell lysates were subsequently subjected to a biotin-switch assay (A) or to nondenaturing PAGE (B). After the blots were developed with a primary antibody against FLAG, they were stripped and reprobed with a primary antibody against FAS, and the ratio of SNO-FAS/total FAS expression (A) or FAS dimer/FAS monomer (B) was quantified. Data were normalized by setting old SNOC for each group as 1 (mean ± SD). *P < 0.05 by t-test. C: The lysates of cells transfected with FAS-WT and its cysteine mutants were exposed to SNOC (25 μM) or old SNOC and subjected to the FAS activity assay. Data were normalized by setting the old SNOC samples for each group as 1 (mean ± SD). **P < 0.01 compared with FAS-WT SNOC by t-test. Data are representative of three individual experiments. IB, immunoblot.
DISCUSSION
There are conflicting views regarding the role of NO in adipogenesis. Several articles have reported that NO suppresses adipocyte differentiation, but others have shown that NO can enhance adipogenesis (42, 46, 48–50). This discrepancy seems reasonable considering our results demonstrating that the production of NO was relatively low, even though it increased significantly during adipogenesis (Fig. 1A), and that SNOC has a biphasic effect on adipogenesis (Fig. 1C). Taken together, these data lead us to assume that a small amount of free NO is required for adipogenesis, whereas a larger amount of NO may suppress adipogenesis.
Protein S-nitrosylation is a major reversible posttranslational modification by NO that regulates protein function and stability (36–39, 51–56). Interestingly, it has been reported that protein S-nitrosylation is increased in adipose tissue in a model of obesity (41). To explore the biological role of S-nitrosylation in adipogenesis, we first focused on FAS, the key enzyme of fatty acid synthesis. We found that FAS was S-nitrosylated under physiological conditions and the level of SNO-FAS was gradually increased during adipogenesis (Fig. 2). FAS became more active following S-nitrosylation, indicating that S-nitrosylation plays an important role in adipogenesis. It has been reported that NO regulates mitochondrial fatty acid metabolism through protein S-nitrosylation regulation (40). Following a MS-based proteomics study, very long chain acyl-CoA dehydrogenase was proposed as a candidate S-nitrosylated protein involved in fatty acid metabolism. Similar to SNO-FAS, the activity of SNO-very long chain acyl-CoA dehydrogenase was found to be increased by S-nitrosylation, implying a stimulating role of S-nitrosylation for fatty acid oxidation. These studies suggest that S-nitrosylation might stimulate both adipogenesis and fatty acid oxidation. Therefore, the balance of the S-nitrosylation level might be a key signaling pathway regulating obesity.
Previous reports have shown that protein S-nitrosylation affects protein activity by enhancing protein dimerization of Drp1 (55) or interrupting protein dimerization of eNOS (57). FAS formed dimers following S-nitrosylation. So far, there is no cysteine residue which is known to be involved in FAS dimerization. Although we showed that the Cys1471 and Cys2091 residues of FAS are important for dimerization and activation by NO as targets for S-nitrosylation, neither cysteine residue is in the active site. However, interestingly, Cys1471 is in the center of the interdomain of FAS, which is known to be important for catalytically active dimer formation (58).
NO scavengers significantly inhibited adipogenesis. We suggest that the inhibition of SNO-FAS formation leading to FAS inactivation could be a possible signaling pathway by which NO scavengers inhibit adipogenesis. During adipocyte differentiation, the decrease in fatty acid synthesis is known to interrupt the cross-stimulation of C/EBPα and PPARγ, and leads to insufficient adipogenesis (17, 59, 60). We assume that these events may have occurred in our experimental system, at least in part, when exposed to NO scavengers. Based on the data showing no effect of NOS inhibitor on adipogenesis (supplementary Fig. 1A), we have a hypothesis that NO during adipogenesis in the present experiments could be coming from the intracellular “SNO pool,” such as S-nitrosoglutathione, SNOC, and other SNO-proteins, as suggested before (34). Though there are no previous reports about the higher thiol oxidation states of FAS, it is still possible that the further thiol oxidation of FAS can alter its activity in our experimental system. It is because reversible S-nitrosylation may facilitate further oxidation of the same cysteine thiol (61, 62). In addition to affecting adipogenesis, fatty acids produced by FAS are known to stimulate other signaling pathways, including autophagy and miRNA signaling (63, 64). FAS has also received much attention regarding the regulation of its activity during cancer development (19–23). As a key enzyme regulating energy metabolism and as a metabolic oncogene, FAS has been considered as a potential target for the development of anti-obesity and anti-cancer drugs. Taken together, our results suggest that the regulation of FAS activation by S-nitrosylation may contribute to the regulation of a variety of physiological or pathological events, including adipogenesis and cancer.
Supplementary Material
Footnotes
Abbreviations:
- ADSC
- adipose-derived stem cell
- cPTIO
- 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- DAPI
- 4′,6-diamidino-2-phenylindole
- DEA-NONOate
- diethylamine diazeniumdiolate diethylammonium salt
- HPDP-biotin
- N-[6-(biotinamido)hexyl]3′-(2′-pyridyldithio)-propionamide
- MMTS
- methyl methanethiosulfonate
- NOS
- NO synthase
- PTIO
- 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- SNOC
- S-nitrosocysteine
- SNO-FAS
- S-nitrosylated FAS
The authors state no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Loftus T. M., Jaworsky D. E., Frehywot G. L., Townsend C. A., Ronnett G. V., Lane M. D., and Kuhajda F. P.. 2000. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 288: 2379–2381. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z., Cha S. H., Chohnan S., and Lane M. D.. 2003. Hypothalamic malonyl-CoA as a mediator of feeding behavior. Proc. Natl. Acad. Sci. USA. 100: 12624–12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakil S. J. 1989. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 28: 4523–4530. [DOI] [PubMed] [Google Scholar]
- 4.Jayakumar A., Chirala S. S., and Wakil S. J.. 1997. Human fatty acid synthase: assembling recombinant halves of the fatty acid synthase subunit protein reconstitutes enzyme activity. Proc. Natl. Acad. Sci. USA. 94: 12326–12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakil S. J., Stoops J. K., and Joshi V. C.. 1983. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 52: 537–579. [DOI] [PubMed] [Google Scholar]
- 6.Stoops J. K., Ross P., Arslanian M. J., Aune K. C., Wakil S. J., and Oliver R. M.. 1979. Physicochemical studies of the rat liver and adipose fatty acid synthetases. J. Biol. Chem. 254: 7418–7426. [PubMed] [Google Scholar]
- 7.Singh N., Wakil S. J., and Stoops J. K.. 1984. On the question of half- or full-site reactivity of animal fatty acid synthetase. J. Biol. Chem. 259: 3605–3611. [PubMed] [Google Scholar]
- 8.Stoops J. K., Wakil S. J., Uberbacher E. C., and Bunick G. J.. 1987. Small-angle neutron-scattering and electron microscope studies of the chicken liver fatty acid synthase. J. Biol. Chem. 262: 10246–10251. [PubMed] [Google Scholar]
- 9.Stoops J. K., and Wakil S. J.. 1981. Animal fatty acid synthetase. A novel arrangement of the beta-ketoacyl synthetase sites comprising domains of the two subunits. J. Biol. Chem. 256: 5128–5133. [PubMed] [Google Scholar]
- 10.Makimura H., Mizuno T. M., Yang X. J., Silverstein J., Beasley J., and Mobbs C. V.. 2001. Cerulenin mimics effects of leptin on metabolic rate, food intake, and body weight independent of the melanocortin system, but unlike leptin, cerulenin fails to block neuroendocrine effects of fasting. Diabetes. 50: 733–739. [DOI] [PubMed] [Google Scholar]
- 11.Kumar M. V., Shimokawa T., Nagy T. R., and Lane M. D.. 2002. Differential effects of a centrally acting fatty acid synthase inhibitor in lean and obese mice. Proc. Natl. Acad. Sci. USA. 99: 1921–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mobbs C. V., and Makimura H.. 2002. Block the FAS, lose the fat. Nat. Med. 8: 335–336. [DOI] [PubMed] [Google Scholar]
- 13.Shimokawa T., Kumar M. V., and Lane M. D.. 2002. Effect of a fatty acid synthase inhibitor on food intake and expression of hypothalamic neuropeptides. Proc. Natl. Acad. Sci. USA. 99: 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacs P., Harper I., Hanson R. L., Infante A. M., Bogardus C., Tataranni P. A., and Baier L. J.. 2004. A novel missense substitution (Val1483Ile) in the fatty acid synthase gene (FAS) is associated with percentage of body fat and substrate oxidation rates in nondiabetic Pima Indians. Diabetes. 53: 1915–1919. [DOI] [PubMed] [Google Scholar]
- 15.Liu L. H., Wang X. K., Hu Y. D., Kang J. L., Wang L. L., and Li S.. 2004. Effects of a fatty acid synthase inhibitor on adipocyte differentiation of mouse 3T3-L1 cells. Acta Pharmacol. Sin. 25: 1052–1057. [PubMed] [Google Scholar]
- 16.Ronnett G. V., Kim E-K., Landree L. E., and Tu Y.. 2005. Fatty acid metabolism as a target for obesity treatment. Physiol. Behav. 85: 25–35. [DOI] [PubMed] [Google Scholar]
- 17.Schmid B., Rippmann J. F., Tadayyon M., and Hamilton B. S.. 2005. Inhibition of fatty acid synthase prevents preadipocyte differentiation. Biochem. Biophys. Res. Commun. 328: 1073–1082. [DOI] [PubMed] [Google Scholar]
- 18.Chakravarthy M. V., Zhu Y., Yin L., Coleman T., Pappan K. L., Marshall C. A., McDaniel M. L., and Semenkovich C. F.. 2009. Inactivation of hypothalamic FAS protects mice from diet-induced obesity and inflammation. J. Lipid Res. 50: 630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid A., Pizer E. S., Moga M., Milgraum L. Z., Zahurak M., Pasternack G. R., Kuhajda F. P., and Hamilton S. R.. 1997. Elevated expression of fatty acid synthase and fatty acid synthetic activity in colorectal neoplasia. Am. J. Pathol. 150: 201–208. [PMC free article] [PubMed] [Google Scholar]
- 20.Alò P. L., Visca P., Trombetta G., Mangoni A., Lenti L., Monaco S., Botti C., Serpieri D. E., and Di Tondo U.. 1999. Fatty acid synthase (FAS) predictive strength in poorly differentiated early breast carcinomas. Tumori. 85: 35–40. [DOI] [PubMed] [Google Scholar]
- 21.Sekiguchi M., Shiroko Y., Arai T., Kishino T., Sugawara I., Kusakabe T., Suzuki T., Yamashita T., Obara T., Ito K., et al. 2001. Biological characteristics and chemosensitivity profile of four human anaplastic thyroid carcinoma cell lines. Biomed. Pharmacother. 55: 466–474. [DOI] [PubMed] [Google Scholar]
- 22.Thupari J. N., Kim E-K., Moran T. H., Ronnett G. V., and Kuhajda F. P.. 2004. Chronic C75 treatment of diet-induced obese mice increases fat oxidation and reduces food intake to reduce adipose mass. Am. J. Physiol. Endocrinol. Metab. 287: E97–E104. [DOI] [PubMed] [Google Scholar]
- 23.Innocenzi D., Alò P. L., Balzani A., Sebastiani V., Silipo V., La Torre G., Ricciardi G., Bosman C., and Calvieri S.. 2003. Fatty acid synthase expression in melanoma. J. Cutan. Pathol. 30: 23–28. [DOI] [PubMed] [Google Scholar]
- 24.Gansler T. S., Hardman W., Hunt D. A., Schaffel S., and Hennigar R. A.. 1997. Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Hum. Pathol. 28: 686–692. [DOI] [PubMed] [Google Scholar]
- 25.Cha S. H., Hu Z., and Lane M. D.. 2004. Long-term effects of a fatty acid synthase inhibitor on obese mice: food intake, hypothalamic neuropeptides, and UCP3. Biochem. Biophys. Res. Commun. 317: 301–308. [DOI] [PubMed] [Google Scholar]
- 26.Paulauskis J. D., and Sul H. S.. 1989. Hormonal regulation of mouse fatty acid synthase gene transcription in liver. J. Biol. Chem. 264: 574–577. [PubMed] [Google Scholar]
- 27.Latasa M-J., Griffin M. J., Moon Y. S., Kang C., and Sul H. S.. 2003. Occupancy and function of the -150 sterol regulatory element and -65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol. Cell. Biol. 23: 5896–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Deng R., Zhu H. H., Zhang S. S., Zhu C., Montminy M., Davis R., and Feng G-S.. 2013. Modulation of fatty acid synthase degradation by concerted action of p38 MAP kinase, E3 ligase COP1, and SH2-tyrosine phosphatase Shp2. J. Biol. Chem. 288: 3823–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graner E., Tang D., Rossi S., Baron A., Migita T., Weinstein L. J., Lechpammer M., Huesken D., Zimmermann J., Signoretti S., et al. 2004. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 5: 253–261. [DOI] [PubMed] [Google Scholar]
- 30.Kilbourn R. G., and Griffith O. W.. 1992. Overproduction of nitric oxide in cytokine-mediated and septic shock. J. Natl. Cancer Inst. 84: 827–831. [DOI] [PubMed] [Google Scholar]
- 31.Dawson V. L. 1995. Nitric oxide: role in neurotoxicity. Clin. Exp. Pharmacol. Physiol. 22: 305–308. [DOI] [PubMed] [Google Scholar]
- 32.Stoclet J. C., Muller B., Andriantsitohaina R., and Kleschyov A.. 1998. Overproduction of nitric oxide in pathophysiology of blood vessels. Biochemistry (Mosc.). 63: 826–832. [PubMed] [Google Scholar]
- 33.Levonen A. L., Patel R. P., Brookes P., Go Y. M., Jo H., Parthasarathy S., Anderson P. G., and Darley-Usmar V. M.. 2001. Mechanisms of cell signaling by nitric oxide and peroxynitrite: from mitochondria to MAP kinases. Antioxid. Redox Signal. 3: 215–229. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T., and Lipton S. A.. 2013. Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxid. Redox Signal. 18: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei J., Vodovotz Y., Tzeng E., and Billiar T. R.. 2013. Nitric oxide, a protective molecule in the cardiovascular system. Nitric Oxide. 35: 175–185. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura T., Tu S., Akhtar M. W., Sunico C. R., Okamoto S-I., and Lipton S. A.. 2013. Aberrant protein s-nitrosylation in neurodegenerative diseases. Neuron. 78: 596–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura T., and Lipton S. A.. 2011. Redox modulation by S-nitrosylation contributes to protein misfolding, mitochondrial dynamics, and neuronal synaptic damage in neurodegenerative diseases. Cell Death Differ. 18: 1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan S. D., Dolatabadi N., Chan S. F., Zhang X., Akhtar M. W., Parker J., Soldner F., Sunico C. R., Nagar S., Talantova M., et al. 2013. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 155: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeon G. S., Nakamura T., Lee J-S., Choi W-J., Ahn S-W., Lee K-W., Sung J-J., and Lipton S. A.. 2014. Potential effect of S-nitrosylated protein disulfide isomerase on mutant SOD1 aggregation and neuronal cell death in amyotrophic lateral sclerosis. Mol. Neurobiol. 49: 796–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doulias P-T., Tenopoulou M., Greene J. L., Raju K., and Ischiropoulos H.. 2013. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci. Signal. 6: rs1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ovadia H., Haim Y., Nov O., Almog O., Kovsan J., Bashan N., Benhar M., and Rudich A.. 2011. Increased adipocyte S-nitrosylation targets anti-lipolytic action of insulin: relevance to adipose tissue dysfunction in obesity. J. Biol. Chem. 286: 30433–30443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hemmrich K., Gummersbach C., Paul N. E., Goy D., Suschek C. V., Kröncke K-D., and Pallua N.. 2010. Nitric oxide and downstream second messenger cGMP and cAMP enhance adipogenesis in primary human preadipocytes. Cytotherapy. 12: 547–553. [DOI] [PubMed] [Google Scholar]
- 43.Lei S. Z., Pan Z-H., Aggarwal S. K., Chen H-S. V., Hartman J., Sucher N. J., and Lipton S. A.. 1992. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 8: 1087–1099. [DOI] [PubMed] [Google Scholar]
- 44.Jaffrey S. R., and Snyder S. H.. 2001. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001: pl1. [DOI] [PubMed] [Google Scholar]
- 45.Kelley D. S., Nelson G. J., and Hunt J. E.. 1986. Effect of prior nutritional status on the activity of lipogenic enzymes in primary monolayer cultures of rat hepatocytes. Biochem. J. 235: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elizalde M., Rydén M., van Harmelen V., Eneroth P., Gyllenhammar H., Holm C., Ramel S., Olund A., Arner P., and Andersson K.. 2000. Expression of nitric oxide synthases in subcutaneous adipose tissue of nonobese and obese humans. J. Lipid Res. 41: 1244–1251. [PubMed] [Google Scholar]
- 47.Lohman A. W., Weaver J. L., Billaud M., Sandilos J. K., Griffiths R., Straub A. C., Penuela S., Leitinger N., Laird D. W., Bayliss D. A., et al. 2012. S-nitrosylation inhibits pannexin 1 channel function. J. Biol. Chem. 287: 39602–39612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nisoli E., Clementi E., Tonello C., Sciorati C., Briscini L., and Carruba M. O.. 1998. Effects of nitric oxide on proliferation and differentiation of rat brown adipocytes in primary cultures. Br. J. Pharmacol. 125: 888–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H., Aziz E., Shillabeer G., Wong A., Shanghavi D., Kermouni A., Abdel-Hafez M., and Lau D. C. W.. 2002. Nitric oxide promotes differentiation of rat white preadipocytes in culture. J. Lipid Res. 43: 2123–2129. [DOI] [PubMed] [Google Scholar]
- 50.Kawachi H., Moriya N. H., Korai T., Tanaka S., Watanabe M., Matsui T., Kawada T., and Yano H.. 2007. Nitric oxide suppresses preadipocyte differentiation in 3T3-L1 culture. Mol. Cell. Biochem. 300: 61–67. [DOI] [PubMed] [Google Scholar]
- 51.Stamler J. S., Lamas S., and Fang F. C.. 2001. Nitrosylation. The prototypic redox-based signaling mechanism. Cell. 106: 675–683. [DOI] [PubMed] [Google Scholar]
- 52.Hess D. T., Matsumoto A., Kim S-O., Marshall H. E., and Stamler J. S.. 2005. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6: 150–166. [DOI] [PubMed] [Google Scholar]
- 53.Benhar M., Forrester M. T., Hess D. T., and Stamler J. S.. 2008. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 320: 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsushita K., Morrell C. N., Cambien B., Yang S. X., Yamakuchi M., Bao C., Hara M. R., Quick R. A., Cao W., O’Rourke B., et al. 2003. Nitric oxide regulates exocytosis by S-nitrosylation of N-ethylmaleimide-sensitive factor. Cell. 115: 139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho D. H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., and Lipton S. A.. 2009. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 324: 102–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunico C. R., Nakamura T., Rockenstein E., Mante M., Adame A., Chan S. F., Newmeyer T. F., Masliah E., Nakanishi N., and Lipton S. A.. 2013. S-Nitrosylation of parkin as a novel regulator of p53-mediated neuronal cell death in sporadic Parkinson’s disease. Mol. Neurodegener. 8: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erwin P. A., Lin A. J., Golan D. E., and Michel T.. 2005. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J. Biol. Chem. 280: 19888–19894. [DOI] [PubMed] [Google Scholar]
- 58.Chirala S. S., Jayakumar A., Gu Z. W., and Wakil S. J.. 2001. Human fatty acid synthase: role of interdomain in the formation of catalytically active synthase dimer. Proc. Natl. Acad. Sci. USA. 98: 3104–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosen E. D., Sarraf P., Troy A. E., Bradwin G., Moore K., Milstone D. S., Spiegelman B. M., and Mortensen R. M.. 1999. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell. 4: 611–617. [DOI] [PubMed] [Google Scholar]
- 60.Wu Z., Rosen E. D., Brun R., Hauser S., Adelmant G., Troy A. E., McKeon C., Darlington G. J., and Spiegelman B. M.. 1999. Cross-regulation of C/EBPα and PPARγ controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol. Cell. 3: 151–158. [DOI] [PubMed] [Google Scholar]
- 61.Gu Z., Kaul M., Yan B., Kridel S. J., Cui J., Strongin A., Smith J. W., Liddington R. C., and Lipton S. A.. 2002. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 297: 1186–1190. [DOI] [PubMed] [Google Scholar]
- 62.Uehara T., Nakamura T., Yao D., Shi Z. Q., Gu Z., Ma Y., Masliah E., Nomura Y., and Lipton S. A.. 2006. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 441: 513–517. [DOI] [PubMed] [Google Scholar]
- 63.Komiya K., Uchida T., Ueno T., Koike M., Abe H., Hirose T., Kawamori R., Uchiyama Y., Kominami E., Fujitani Y., et al. 2010. Free fatty acids stimulate autophagy in pancreatic β-cells via JNK pathway. Biochem. Biophys. Res. Commun. 401: 561–567. [DOI] [PubMed] [Google Scholar]
- 64.Yang W. M., Jeong H-J., Park S. Y., and Lee W.. 2014. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 588: 3939–3946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.