Abstract
Human positive cofactor 4 (PC4) is a transcriptional coactivator with a highly conserved single-strand DNA (ssDNA) binding domain of unknown function. We identified PC4 as a suppressor of the oxidative mutator phenotype of the Escherichia coli fpg mutY mutant and demonstrate that this suppression requires its ssDNA binding activity. Saccharomyces cerevisiae mutants lacking their PC4 ortholog Sub1 are sensitive to hydrogen peroxide and exhibit spontaneous and peroxide-induced hypermutability. PC4 expression suppresses the peroxide sensitivity of the yeast sub1Δ mutant, suggesting that the human protein has a similar function. A role for yeast and human proteins in DNA repair is suggested by the demonstration that Sub1 acts in a peroxide resistance pathway involving Rad2 and by the physical interaction of PC4 with the human Rad2 homolog XPG. We show that XPG recruits PC4 to a bubble-containing DNA substrate with a resulting displacement of XPG and formation of a PC4-DNA complex. We discuss the possible requirement for PC4 in either global or transcription-coupled repair of oxidative DNA damage to mediate the release of XPG bound to its substrate.
Oxidative DNA damage and the mutations it causes have been implicated in a number of human diseases, including cancer and neurodegenerative diseases, and are a contributing factor to aging (6, 12, 35, 36). Thus, a thorough understanding of genes involved in the prevention and repair of oxidative DNA damage and its mutagenic consequences is important to our understanding of the mechanisms mitigating these diseases and exacerbating normal degenerative processes associated with aging.
Oxidative DNA damage results from the interaction of reactive oxygen species (ROS) with DNA. ROS are produced as by-products of normal aerobic metabolism and by exogenous factors, such as ionizing radiation and chemical oxidants (6, 12, 35, 36). The deleterious consequences of ROS to an organism's genetic material are held in check by proteins that prevent or repair oxidative DNA damage (7, 12, 13, 15, 21). Unrepaired oxidative lesions result in increased mutagenesis, lethality, and apoptosis (23, 27). A balance between DNA repair and damage prevention mechanisms and ROS production is required to maintain a low spontaneous mutation rate. Factors that increase ROS production, reduce ROS detoxification, or factors that affect repair of oxidative DNA lesions result in increased mutagenesis. This is best demonstrated in Escherichia coli; mutations that inactivate the fpg and mutY genes, whose products repair the predominant oxidative lesion 8-oxoguanine (8-oxoG) and its mispaired intermediate 8-oxoG:A, respectively, result in a mutator phenotype that specifically increases GC→ TA transversion mutagenesis (37).
In this study, we screened a human cDNA library and describe the isolation and characterization of the human transcription positive cofactor 4 (PC4) gene as a suppressor of oxidative mutagenesis in the E. coli fpg mutY strain. We demonstrate that PC4 and its Saccharomyces cerevisiae ortholog SUB1 are required for resistance to hydrogen peroxide and function to prevent spontaneous and induced oxidative mutagenesis. The oxidation resistance function of PC4 requires its single-strand DNA (ssDNA) binding activity, which is not required for the transcription coactivator function of PC4 (56, 57). A function of PC4 in repair of oxidative DNA damage is suggested by its physical and biochemical interactions with the multifunctional human DNA repair protein XPG, the structure-specific endonuclease activity of which is essential for nucleotide excision repair (NER) but which also plays important nonenzymatic roles in base excision repair (BER) and transcription-coupled repair (TCR) of oxidative damage (52a).
MATERIALS AND METHODS
Bacterial strains.
The lacZ mutant E. coli strain cc104 and its isogenic derivatives MV4705 (cc104 Δfpg::Tn10), MV4707 (cc104 ΔmutY::Catr), and MV4709 (cc104 Δfpg::Tn10 ΔmutY::Catr) were constructed by P1 transduction selecting for the appropriate antibiotic resistance marker.
Bacterial mutagenesis assays.
The papillation assay medium (39) contains 0.2% d-glucose, 1× A salts, 1 mM MgSO4, 5 μg of thiamine hydrochloride/ml, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml, 0.5 mg of phenyl-β-d-galactopyranoside (P-Gal)/ml, 50 μg of carbenicillin/ml, and 2% agar. For pBAD24 induction, 0.2% l-arabinose (20) was added and the glucose concentration was reduced to 0.05%. Papillation was scored 5 to 6 days after plating. Spontaneous mutation frequencies (revertants/total cells) were quantitated by plating dilutions of the overnight cultures of individual transformants on lactose and glucose minimal media and incubating 3 days at 37°C. LacZ revertants were detected on lactose plates, total colonies on glucose plates.
Construction of Fpg, hOGG1, and hMYH expression vectors.
To overexpress the E. coli Fpg protein, a 1-kbp EcoRI/HindIII fragment from V243 (39) was subcloned into pTrc 99A (Pharmacia, Inc.) (pTrc-Fpg). To construct pTrc-hOGG1, a 5.7-kbp XbaI/HindIII fragment from pET8c-OGG1-1a (42) (Y. Nakabeppu, Kyushu University, Fukuoka, Japan) was subcloned into pTrc 99A. To construct pTrc-hMYH, a 4.5-kbp StuI/BamHI fragment from pT7blue-hMYHα3-2 (43) (Y. Nakabeppu, Kyushu University) was subcloned into SmaI/BamHI-linearized pTrc 99A.
Construction of wild-type and mutant PC4 expression vectors.
Plasmids carrying the full-length coding sequences of the wild type (PC4-wt) or the ssDNA binding-defective mutants (PC4-W89A and PC4-β2β3) (57) were obtained from M. Meisterernst (Munich, Germany). The full-length PC4 wild-type and mutant protein coding sequences were isolated as NdeI and EcoRI fragments and subcloned into the corresponding sites in pET-28b(+) to attach the six-histidine tag to their amino termini. The insert containing NcoI and SalI fragments from the pET-28(+) derivatives were subcloned into the corresponding sites of pBAD24 for araBAD promoter-dependent expression. Expression of the six-histidine-tagged PC4 proteins was verified by Western blotting using the anti-Penta His antibody (QIAGEN).
Construction of yeast strains and expression vectors.
The S. cerevisiae wild-type strain FY833 (MATa his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52) and the sub1Δ mutant strain YMH476 (FY833 sub1Δ::hisG) (59) were obtained from M. Hampsey (Rutgers University). Additional sub1Δ mutants were constructed by PCR-mediated one-step gene replacement methods (8). Cells were cultured either in YPD (1% yeast extract, 2% peptone, 2% dextrose) or, for plasmid-bearing strains, in synthetic complete medium lacking leucine or uracil (SC-leu or SC-ura). To express PC4 in yeast strains, the coding sequence for amino acids (aa) 40 to 127 was amplified by PCR using the PC4 cDNA clone initially isolated from the genetic screen as the template with primers PC4-N (ACGCGTCGACATGCAAAAGACAGGTGAGACTTCGAGAGCTCTG) and PC4-C (CCGCTCGAGTCATCTTACAAATTCCTCTGC). The ATG initiation codon shown in boldface along with the italicized sequences for SalI and XhoI restriction sites were added for protein expression and cloning purposes. The SalI- and XhoI-treated PCR product was inserted into pMV611, which carries the LEU2 gene from pRS315 (49) and the GPD promoter from p426-GPD (41). To express the Sub1 protein, the SUB1 open reading frame was amplified from the FY833 genomic DNA by PCR using primers X001-SUB1f (GCTCTAGATGTCATATTACAACAGGTATAGG) and E292-SUB1r (GCGAATTCTTATTCTTCTTCACTTATGTCG). The italicized sequences for XbaI and EcoRI restriction sites were introduced to the ends of the PCR product for cloning into p416-GPD, which carries the URA3 gene for selection (41). MVY219 (rad2Δ::TRP1) and MVY221 (sub1Δ rad2Δ::TRP1) RAD2 deletion strains were constructed by transforming FY833 (wt) and YMH476 (sub1Δ) with SalI-digested pWS521 (W. Siede, University of North Texas Health Science Center).
H2O2 sensitivity and induced mutagenesis.
Yeast strains were grown to mid-log phase at 30°C, washed, and resuspended in phosphate-buffered saline. Cells were then treated with H2O2 by shaking at 30°C for 1 h as indicated. Cells were harvested by centrifugation, washed, resuspended in sterile deionized H2O, diluted, and plated on YPD for survival analysis of non-plasmid-bearing strains or SC-leu or SC-ura for plasmid-bearing strains. Survival experiments were repeated from 3 to 10 times, and representative data indicating the reproducible differences between strains are shown. To measure the can1r mutagenesis, cells were plated on synthetic medium lacking arginine but containing 60 μg of canavanine/ml and incubated at 30°C for 3 to 4 days for survival and 4 to 5 days for mutagenesis.
UV and MMS sensitivity tests.
Yeast strains were grown to mid-log phase at 30°C, washed with water, suspended in phosphate-buffered saline, incubated with the indicated concentrations of methyl methanesulfonate (MMS; Sigma Chemical) for 1 h at 30°C, diluted, and plated on YPD plates. Surviving colonies were counted after 3 to 4 days of incubation at 30°C. For UV dose response tests, approximately 1,000 log-phase cells were placed in each spot on YPD plates. Spots were irradiated with UV (λ = 254 nm) in 30-J/m2 increments using a Stratalinker UV Crosslinker (Stratagene). Plates were then incubated in the dark at 30°C for 3 to 4 days.
Expression and purification of PC4 and XPG.
Full-length wild-type PC4 was expressed in the E. coli BL21(DE3) strain and purified according to the methods of Ge et al. (18). The cDNA for human XPG (a generous gift from Stuart Clarkson) was inserted into a pFASTBAC vector, expressed in High5 insect cells, and purified to 95% homogeneity essentially as described previously (16). For use only in the far-Western assays, a heart muscle kinase (HMK) recognition sequence tag, RRASV, was added at the C terminus.
Protein-protein interaction assays.
For far-Western analysis, human PC4 protein (1.5 μg), human Nth1 protein (2.1 μg), and E. coli EndoIII (3 μg) were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (precast 4 to 20% gel; Bio-Rad), transferred to a polyvinylidene difluoride membrane, and stained with Ponceau S to visualize the proteins on the membrane. The membrane was then blocked with a blocking buffer (25 mM HEPES-KOH [pH 7.7], 25 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 1% nonfat milk, 0.1% NP-40) for 2 h at 4°C and probed overnight at 4°C on a rocker with blocking buffer containing 150 mM KCl and 32P-labeled XPG with a HMK tag. The membrane was washed with washing buffer (25 mM HEPES-KOH [pH 7.7], 25 mM NaCl, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 1% nonfat milk, 0.5% NP-40), and interactions were visualized by both phosphorimager and autoradiography. For slot blot analysis, 1 μg each of PC4, hNth1, and EndoIII was applied to a nitrocellulose membrane, and the membrane was processed as described above.
Electrophoretic mobility shift assay.
The sequences of the oligonucleotides used to form the bubble DNA substrate for the electrophoretic mobility shift assay were as follows (with the central unpaired region of the substrates in bold): 10T-strand, 5′-GGGCAGACAACGTGGCGCTGTTTTTTTTTTGTGTCCTAGCACAGCGTATG-3′; and 10C-strand, 5′-CATACGCTGTGCTAGGACACCCCCCCCCCCCAGCGCCACGTTGTCTGCCC-3′.
The 10T-strand was 5′-end labeled with T4 polynucleotide kinase and [γ-32P]ATP and annealed with the complementary 10C-strand by heating for 3 min at 90°C and cooling to room temperature. The resulting DNA bubble substrate was gel purified. Fifty femtomoles of 32P-labeled bubble DNA was incubated at room temperature with XPG, PC4, or both for 20 min in a 20-μl reaction mixture containing 10 mM HEPES-KOH (pH 7.5), 110 mM KCl, 1 mM EDTA, 1 mM DTT, 4% glycerol, and 0.2 μg of poly(dI · C-dI · C). After the incubation, the samples were loaded on a 4.5% nondenaturing polyacrylamide gel (19:1, acrylamide to bisacrylamide). Electrophoresis was performed under refrigeration at 150 V for 2 h in 0.5× Tris-borate-EDTA buffers. The gels were analyzed by phosphorimager and ImageQuant software (Molecular Dynamics).
RESULTS
Suppression of the E. coli oxidative mutator phenotype.
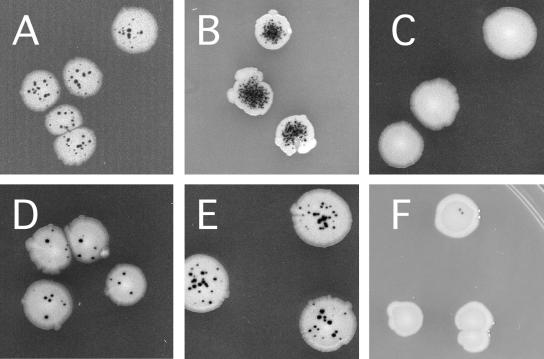
The fpg mutY strain of E. coli was used to screen for human cDNAs that express proteins that either prevent spontaneous DNA oxidation or repair the DNA lesions produced. This E. coli strain is a spontaneous mutator because Fpg repairs 8-oxoG, the predominant oxidative DNA lesion, and MutY removes A mispaired with 8-oxoG, a high-frequency mispairing event during replication of template 8-oxoG lesions resulting in GC→TA transversion mutations (37, 38, 40). Production of ROS by normal metabolic processes leads to accumulation of 8-oxoG lesions in DNA and results in elevated spontaneous mutagenesis (37). The GC→ TA transversions are detected using the cc104 allele of lacZ, which reverts only by GC→ TA transversion (14). On indicator medium, E. coli cells carrying lacZ(cc104) produce distinctive white LacZ− colonies containing dark blue LacZ+ revertant microcolonies, or papillae (Fig. 1) (37). Figure 1 compares the mutator phenotype of the fpg mutY double mutant strain (Fig. 1B) with that of wild-type E. coli (Fig. 1A) and shows the suppression of mutagenesis resulting from the expression of E. coli and human DNA repair genes. Complete suppression of the fpg mutY strain's mutator phenotype is seen upon high-level expression of the bacterial fpg gene (Fig. 1C), reducing the frequency of transversions to levels below that seen in the wild type (Fig. 1A). Since most of the transversion mutations seen in the wild-type strain can also be prevented by fpg overexpression (60), GC→TA transversions can be considered a signature of oxidative DNA damage. Suppression of the fpg mutY mutator phenotype is also seen upon expression of the human 8-oxoG glycosylase, OGG1 (Fig. 1D), or the human MutY ortholog, MYH (Fig. 1E) (1, 3, 43, 46, 48, 50).
FIG. 1.
Mutator phenotype of E. coli fpg mutY and its suppression by bacterial and human DNA repair genes. Upper panels show the phenotype of (A) wild-type E. coli cc104 carrying the GC→TA transversion-specific allele of lacZ and (B) its isogenic fpg mutY double mutant derivative. The antimutator activity resulting from (C) expression of bacterial fpg protein (E. coli fpg mutY/pfpg), (D) expression of the human 8-oxoG DNA glycosylase OGG1 (E. coli fpg mutY/phOGG1), (E) expression of the human MutY ortholog hMYH (E. coli fpg mutY/phMYH), and (F) expression of the truncated form of PC4 isolated in this study (E. coli fpg mutY/pSE380-PC4).
Isolation of the human PC4 gene.
The PC4 gene was isolated by transforming the E. coli fpg mutY strain with a human cDNA library constructed in a bacterial expression vector (45, 54) and then screening individual transformed colonies for suppression of the spontaneous mutator phenotype. PC4 is one of the strong antimutators identified (Fig. 1F). Expression of PC4 provides complete mutation suppression under these conditions, as confirmed by a quantitative mutagenesis assay (Table 1).
TABLE 1.
Mutation frequency of fpg mutY strains expressing bacterial and human DNA repair genes
| Strain | Genotype | Plasmid | Mutation frequency (mutants/108 cells)a |
|---|---|---|---|
| MV4724 | Wild type | pTrc99A vector | 3 |
| MV4755 | fpg mutY | pTrc99A vector | 2,750 |
| MV4763 | fpg mutY | pTrc99A-fpg | 0 |
| MV4761 | fpg mutY | pTrc99A-hOGG1 | 11 |
| MV4762 | fpg mutY | pTrc99A-hMYH | 58 |
| MV4722 | fpg mutY | pSE380b-PC4 | 0 |
Representative data are shown.
pSE380 is identical to pTrc99A except for the presence of additional cloning sites in the vector.
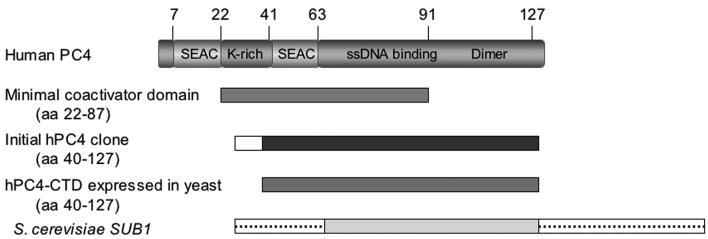
PC4 is a transcription cofactor mediating activator-dependent transcription by RNA polymerase II (19, 32) through interactions with sequence-specific activators and TFIIA of the basal transcription machinery (19, 32). PC4 encodes a polypeptide of 127 aa (Fig. 2). Functional deletion analyses revealed a bipartite structure of PC4 comprising an amino-terminal regulatory domain (aa 1 to 62) and a carboxyl-terminal, ssDNA binding and dimerization domain (CTD; aa 63 to 127) (10, 28, 56, 57). The X-ray crystal structure of the PC4-CTD shows that it forms a dimer with two ssDNA binding channels running in opposite directions (10). Previously, in vitro transcription studies showed that a peptide comprised of aa 22 to 87 of PC4 is necessary and sufficient for coactivation (28, 32, 56, 57) and that the lysine-rich motif between aa 22 and 41 is required for transcription activation (26, 28). It has also been demonstrated that inactivation of the ssDNA binding activity does not affect the ability of PC4 to function in transcription activation (57).
FIG. 2.
Structure of PC4 and its derivatives. The upper panel shows the domains of wild-type PC4. The protein region designated aa 22 to 87 is the minimal coactivator clone described by Kaiser et al. (28), and it is shown for comparison. The initial PC4 clone is the form of PC4 initially isolated in our screen. The white amino-terminal box indicates the in-frame vector sequence fused to the 40 to 127 aa region of PC4. The PC4-CTD expressed in yeast was constructed by adding an ATG codon 5′ to sequences encoding PC4 aa residues 40 through 127. The S. cerevisiae SUB1 gene is also shown, the boxes containing the dotted lines (not to scale) depict the heterologous 39-aa amino-terminal and 187-aa carboxyl-terminal domains of unknown function.
The PC4 cDNA clone isolated in this study is truncated at its 5′ end, lacking the codons for the first 39 amino acid residues but containing a short heterologous DNA sequence of unknown origin encoding MPSNSAPAHGTSS fused to glutamine 40 of PC4. The N-terminal truncation of PC4 removes the lysine-rich motif required for coactivation (28) but leaves intact the ssDNA binding and dimerization domains (10, 56). This suggests that the ssDNA binding and dimerization motifs alone are sufficient for PC4 to function as an oxidative antimutator protein in E. coli. To examine this further and to rule out effects of the heterologous upstream sequence, we obtained the cloned full-length wild-type PC4 gene, transferred it into the l-arabinose-inducible pBAD24 bacterial expression vector (20), and tested its ability to function as an antimutator. Figure 3A shows that wild-type PC4 is able to fully suppress the mutator phenotype of the E. coli fpg mutY strain, indicating that the full-length and truncated fusion forms of PC4 isolated in this study are functionally similar.
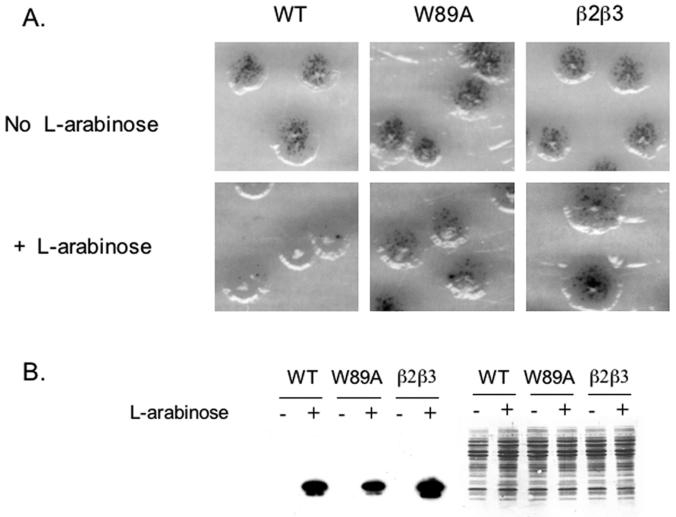
FIG. 3.
ssDNA binding activity of PC4 is required for mutation suppression in E. coli fpg mutY. Histidine-tagged forms of PC4 and its ssDNA binding-defective mutants W89A and β2β3 are expressed from the l-arabinose-inducible araBAD promoter present on the pBAD24 vector. (A) Upper panels show the mutator activity in the absence of l-arabinose, lower panels show the mutator activity of wild-type and mutant forms of PC4 after induction by l-arabinose. (B) Left section shows the Western blotting using antihistidine antibody to determine levels of wild-type (WT) and mutant protein expression; the right section shows the same gel stained with Coomassie brilliant blue.
ssDNA binding activity of PC4 is required for suppression of oxidative mutagenesis.
Since the PC4 clone we originally isolated contained only the ssDNA binding and dimerization domains of PC4, we tested if ssDNA binding activity is required for the antimutator activity in the fpg mutY strain of E. coli by comparing the β2β3 and W89A ssDNA binding-deficient mutants of PC4 constructed by Werten et al. (56) with the full-length, wild-type PC4. In the absence of l-arabinose, no protein expression was detectable by Western blotting (Fig. 3B, left panel) and no mutation suppression was detected (Fig. 3A, upper panels). In the presence of inducer, all forms of PC4 were expressed equally well (Fig. 3B). However, compared to wild-type PC4, the ssDNA binding-deficient mutants were severely impaired in their ability to suppress the oxidative mutator phenotype when expressed in the fpg mutY strain (Fig. 3A), thus demonstrating that the ssDNA binding activity is required for the antimutator function of PC4 in the bacterial assay and that the wild-type protein functions as well as the truncated form of PC4 in this assay.
Yeast sub1Δ mutants are hypersensitive to hydrogen peroxide.
To determine if PC4 functions to prevent oxidative mutagenesis in eukaryotes, we turned to yeast genetics. Sequence analysis reveals that PC4 orthologs exist in all sequenced eukaryotic genomes and that the most conserved region is the C-terminal ssDNA binding and dimerization domains. The S. cerevisiae PC4 ortholog, termed Sub1 (31) or Tsp1 (22), shows 48% identity and 58% similarity when compared with the C-terminal region (aa residues 63 to 127) of PC4 (2). Like its human ortholog, Sub1 is involved in various aspects of transcription but is not essential for viability (11, 22, 31, 59).
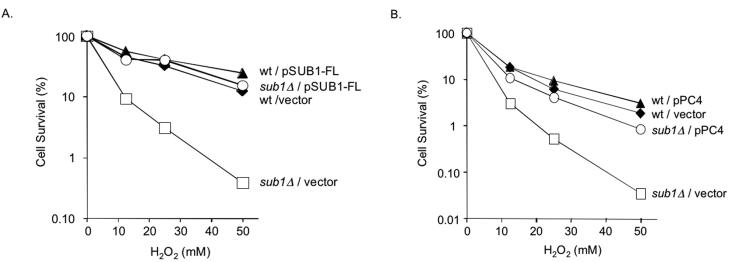
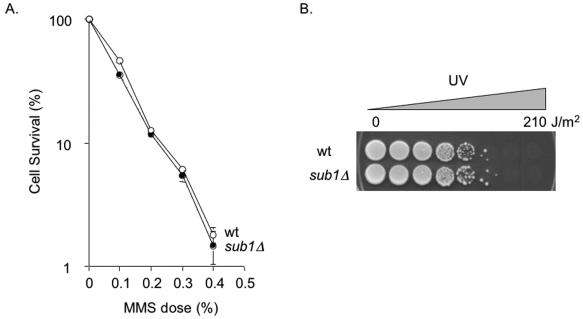
To determine if Sub1 plays a role in oxidation resistance, we tested yeast sub1Δ mutants for phenotypes associated with oxidative stress, DNA damage, or repair. Figure 4A shows that the yeast sub1Δ mutant (59) obtained from M. Hampsey (Rutgers University) is extremely hypersensitive to hydrogen peroxide compared to its wild-type parent. To confirm this, we disrupted the SUB1 gene in two other laboratory strains, W303-1B (lab strain) and RDKY3023 (R. Kolodner, University of California, San Diego), and found similar degrees of sensitization (data not shown). Reintroduction of the wild-type SUB1 gene on the p416-GPD yeast expression vector (41) fully restores peroxide resistance to the sub1Δ mutant, demonstrating that the observed peroxide sensitivity is solely due to the sub1Δ mutation (Fig. 4A). In contrast, the sub1Δ mutant strain does not cause increased sensitivity to methylation or UV treatments (Fig. 5), suggesting that Sub1 does not play a role in either NER or BER of alkylation damage but rather is specific for protection from oxidative DNA damage.
FIG. 4.
Peroxide sensitivity of the yeast sub1Δ mutant strain, its suppression by yeast SUB1, and truncated PC4 gene expression. (A) ⧫, S. cerevisiae wild-type (wt) yeast carrying the vector p416-GPD; □, S. cerevisiae sub1Δ mutant carrying the vector p416-GPD; ▴, wild type carrying the full-length SUB1 gene; ○, S. cerevisiae sub1Δ mutant carrying the full-length SUB1 gene expression plasmid. (B) ⧫, S. cerevisiae wild-type yeast carrying the vector pMV611; □, S. cerevisiae sub1Δ mutant carrying the vector pMV611; ▴, wild type carrying the truncated PC4 gene expression plasmid; ○, S. cerevisiae sub1Δ mutant carrying the truncated PC4 gene expression plasmid.
FIG. 5.
MMS and UV survival in the wild type (wt) and sub1Δ mutant of S. cerevisiae. (A) MMS survival of wild-type yeast (○) and the sub1Δ mutant (•). Data shown represent the averages of three experiments. Error bars indicate standard errors of the mean and are shown when they extend beyond the symbol. (B) UV spot test. Overnight cultures, diluted to inoculate each spot with approximately 1,000 cells of the wild type (upper row) and the sub1Δ mutant (lower row), were placed on YPD agar plates and exposed to increasing doses of UV.
Human PC4 can suppress the peroxide sensitivity of the yeast sub1Δ mutant.
To determine if the human PC4 gene can function to protect against oxidative DNA damage, we tested if it suppresses the peroxide sensitivity of the yeast sub1Δ mutant strain. We constructed a plasmid that expresses the truncated form of PC4 by adding an ATG codon to the 5′ end of PC4 beginning with the glutamine 40 codon. This construct corresponds to the PC4 coding sequence of the truncated form of PC4 originally isolated (Fig. 2). Expression of this clone in yeast results in a complete restoration of peroxide resistance (Fig. 4B), indicating that the truncated form of PC4 can fully substitute for the yeast SUB1 gene and strongly suggesting that the human PC4 gene, like its yeast counterpart, functions in oxidation resistance.
The yeast sub1Δ mutation increases spontaneous and induced mutagenesis.
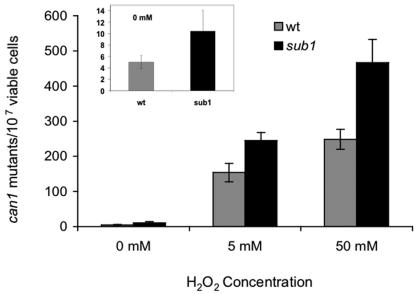
Since many mutations affecting peroxide sensitivity also affect spontaneous and peroxide-induced mutagenesis, we tested if the sub1Δ mutation has such effects by measuring the forward mutation frequency to canavanine resistance. Canavanine resistance can result from a wide variety of genetic changes including GC→TA transversions, and OGG1 mutants of yeast show a sevenfold increase in spontaneous canavanine resistance mutagenesis (52). Figure 6 shows that the sub1Δ mutant exhibits a twofold increase in both spontaneous and peroxide-induced mutagenesis, indicating that Sub1 protects against mutations arising from the low spontaneous production of endogenous ROS and exposure to high levels of exogenous hydrogen peroxide.
FIG. 6.
The yeast sub1Δ mutation results in elevated mutagenesis. Spontaneous and induced mutagenesis in the wild type (wt) and the sub1Δ mutant of S. cerevisiae. The insert expands the view of the mutagenesis in the absence of exogenous peroxide addition.
rad2Δ partially suppresses peroxide sensitivity of sub1Δ.
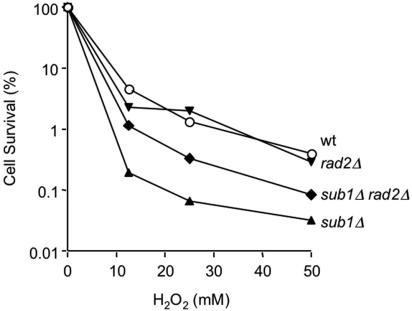
A nonenzymatic function of XPG is essential for TCR of oxidative damage (34) and has also been implicated in transcription per se (9, 33). XPG also stimulates initiation of BER by the NTH1 glycosylase that removes oxidized pyrimidines, again in a nonenzymatic capacity (4, 30, 34). Furthermore, XPG also interacts with and stimulates APE1, the major human AP endonuclease activity in BER, and appears to coordinate NTH1 and APE1 function in vitro (B. Haltiwanger and P. K. Cooper, unpublished results). Because PC4 and Sub1 also have roles in both transcription and resistance to oxidative damage, we wondered whether they might interact with XPG and its yeast homolog Rad2, respectively. We therefore constructed a sub1Δ rad2Δ double mutant strain in order to see if these mutations are epistatic or result in increased peroxide sensitivity. Interestingly, the rad2Δ mutation partially rescues the sub1Δ mutant strain, reducing its peroxide sensitivity (Fig. 7). This finding suggests an interplay between Rad2 and Sub1 in minimizing oxidative damage. In particular, it raises the possibility that in the absence of Sub1, an activity of Rad2 responding to oxidative damage is deleterious.
FIG. 7.
Partial suppression of sub1Δ hydrogen peroxide sensitivity by rad2Δ. ○, wild type (wt); ▴, sub1Δ mutant strain; ▾, rad2Δ mutant strain; ⧫, sub1Δ rad2Δ double mutant strain. Representative data are shown.
Direct interaction of PC4 with XPG.
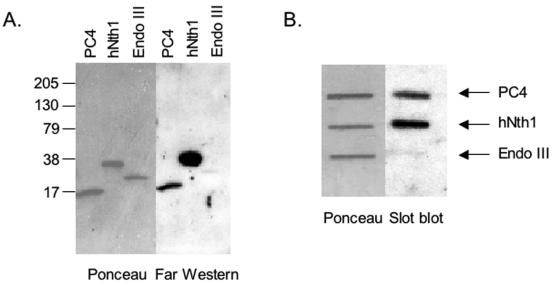
Since Sub1 evidently functions in a pathway involving Rad2, we tested the possibility of direct interaction between the human PC4 and XPG proteins by far-Western analysis. To test PC4-XPG interaction, 32P-labeled XPG was used as a probe for PC4 transferred onto a membrane from a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. A strong signal appears at the position corresponding to PC4, indicating that XPG and PC4 directly interact in the absence of DNA (Fig. 8A). Under these conditions, XPG bound the positive-control human NTH1 but did not bind its E. coli homolog EndoIII, the negative control. To eliminate the possibility that the interaction might be due to denaturation of PC4 during the electrophoresis step, we also tested native PC4 bound directly to a membrane in a slot blot assay (Fig. 8B). Notably, the interaction of PC4 and XPG is preserved, providing additional evidence for its specificity.
FIG. 8.
Interaction of PC4 and XPG. Slot blot and far-Western tests were performed as described in Materials and Methods. Full-length hNTH1 and E. coli EndoIII were used as the positive and negative controls, respectively. The left portions of the panels show Ponceau S-stained images of the membrane, and right portions show the results of incubation of membranes with 32P-labeled XPG-HMK. The position of the protein in the membrane is indicated.
PC4 binding to bubble substrates is enhanced by XPG.
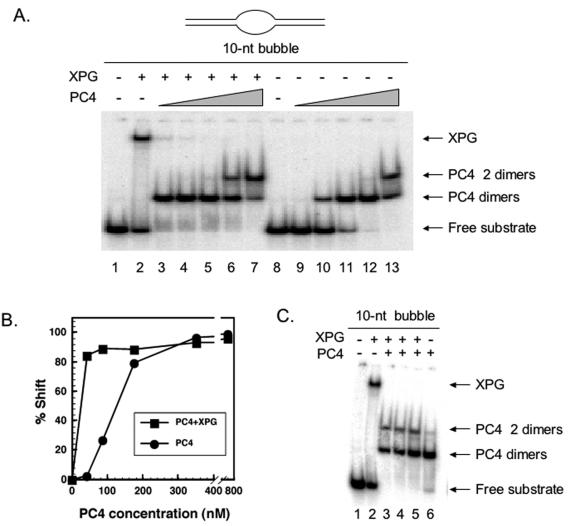
The preferred DNA substrates for binding by PC4 are DNA bubble structures (56). Perhaps not coincidentally, XPG also binds stably, specifically, and with high affinity to DNA bubbles resembling in size the open regions associated with transcription (A. H. Sarker, S. E. Tsutakawa, and P. K. Cooper, unpublished results). In order to determine if these two proteins compete for this substrate or interact synergistically, we tested if the presence of XPG affects binding by full-length PC4 protein to a DNA bubble substrate having a central 10-bp unpaired region. XPG alone bound to this substrate to form a slowly migrating complex (Fig. 9A, lane 2). In the absence of XPG, PC4 also bound specifically to the 10-bp DNA bubble (Fig. 9A, lanes 9 to 13), with complete binding at 352 nM (lane 12). We also observed an additional shifted band that appears only at the highest concentration assayed, 704 nM (Fig. 9A, lane 13). Since it is known that PC4 binds as a dimer (10, 56), we postulate that the slower-migrating shifted species represents a double complex of PC4 dimers bound to DNA, as has been previously observed by others (56, 57). To determine if XPG affects PC4 interaction with DNA bubbles, we preincubated the 10-bp bubble DNA with XPG and then added PC4 at various concentrations (Fig. 9A, lanes 3 to 7). XPG strongly stimulated PC4 binding, with complete binding achieved at the lowest PC4 protein concentration analyzed (44 nM, lane 3), compared to 352 nM in the absence of XPG. Quantitation from the phosphorimage in Fig. 9A of the amount of substrate shifted as a function of PC4 concentration reveals dramatically increased binding of PC4 in the presence of XPG (Fig. 9B). Correspondingly, the PC4 tetramer-bound complex appears at lower concentrations (352 nM, lane 6) in the presence of XPG. Interestingly, in no case did we observe a trimeric complex of XPG, PC4, and DNA. At approximately equimolar ratios of PC4 and XPG (44 nM, lane 3), there appears to be a complete replacement of XPG by PC4 in the protein-bound DNA complexes. This observation is significant, since XPG itself remains stably bound to bubble DNA substrate even in the presence of high concentrations of competitor or other DNA binding proteins with which XPG interacts (A. H. Sarker, S. E. Tsutakawa, and P. K. Cooper, unpublished data). To test if PC4-mediated displacement of XPG is dependent upon the order of addition, the proteins were added either in order or simultaneously. In all cases, only the complexes migrating to the positions detected when PC4 alone is added are seen (Fig. 9C, lanes 3 to 5). These results strongly suggest that XPG recruits PC4 to bubble-containing DNA substrates and that binding of PC4 displaces XPG from the bubble substrate.
FIG. 9.
(A) XPG protein enhances PC4 binding to DNA bubble substrates. Portions (2.5 nM) of the 10-nucleotide (nt) bubble DNA substrate were incubated with 41 nM purified XPG (lanes 2 to 7) or without XPG (lanes 9 to 13) as described in Materials and Methods. Reaction mixtures were supplemented with 44 (lanes 3 and 9), 88 (lanes 4 and 10), 176 (lanes 5 and 11), 352 (lanes 6 and 12), and 704 nM (lanes 7 and 13) human PC4 protein. Samples were loaded onto a 4.5% native gel, and electrophoresis was conducted at 150 V for 2 h at 4°C. The gel was dried and exposed on a phosphorimager screen. No protein was added in lanes 1 and 8. (B) Quantitative representation of the effect of XPG on PC4 binding. (C) Displacement of XPG by PC4 does not depend on the order of addition of the proteins. In all experiments, 44 nM XPG and 352 nM PC4 protein and the same DNA bubble substrates shown in Fig. 8 were used. Lanes 2 and 3, XPG was first incubated with the DNA bubble substrate (lane 2) and PC4 protein was then added (lane 3); lane 4, PC4 protein was first incubated with the DNA bubble substrate followed by XPG addition; lane 5, XPG and PC4 were mixed first and then added to the DNA bubble substrate; lane 6, PC4 alone was incubated with the substrate. Samples were run in a 4.5% native gel, and electrophoresis was conducted at 150 V for 2 h in the cold. The gel was dried and exposed on a phosphorimager screen.
DISCUSSION
In this study we identified PC4 as a human protein capable of suppressing the oxidative mutator phenotype of the fpg mutY strain of E. coli. We characterized the PC4 gene and its yeast ortholog, demonstrating that a yeast mutant devoid of its PC4 ortholog, SUB1, is sensitive to hydrogen peroxide, exhibits a spontaneous mutator phenotype, and is hypermutable upon treatment with hydrogen peroxide. These results indicate that Sub1 is a eukaryotic peroxide resistance protein. The result demonstrating that expression of the human gene can restore peroxide resistance to the sub1Δ mutant of yeast suggests a similar function for the human PC4 protein. These observations, combined with previous results (54), demonstrate the utility of the bacterial screen for the identification of unique human oxidation resistance proteins.
The previously identified transcriptional coactivator function of PC4 does not require the ssDNA binding activity (56, 57) contained in the most highly conserved region of this family of proteins. A novel function in DNA repair for the ssDNA binding activity of PC4 is indicated by the result that the truncated form of human PC4, lacking sequences required for transcription coactivation, can function to suppress oxidative mutagenesis in bacteria and can complement the peroxide sensitivity of a yeast sub1Δ mutant. This conclusion is further supported by the observation that the DNA binding-defective mutant forms of human PC4 are incapable of functioning as antimutators in the bacterial oxidative mutagenesis assays. Thus, we propose that PC4 functions both in transcription and in repair of oxidative DNA damage. These two functions are genetically separable; mutations in PC4's amino terminal domain primarily affect transcription coactivation, and mutations in its ssDNA binding domain primarily affect DNA repair.
Taken together, the observations that Sub1 functions in a repair pathway involving Rad2 and that PC4 directly and functionally interacts with the DNA repair protein XPG suggest a role for PC4 in the repair of oxidative damage. XPG functions in multiple DNA repair pathways in human cells. Both its enzymatic activity as a structure-specific endonuclease and a nonenzymatic function evidently involve interactions with other proteins required in NER (16, 55). In addition, a nonenzymatic function of XPG is required for TCR of oxidative DNA damage, evidently at an early step presumably involving recognition of RNA polymerase stalled at a lesion (34). XPG also both stimulates BER enzymes in vitro through direct interactions (4, 30; B. M. Haltiwanger and P. K. Cooper, unpublished data) and stimulates removal of oxidative lesions in the cell (34). A role for PC4 in NER is unlikely, because we have shown that deletion of its yeast homolog does not affect sensitivity to UV. Thus, the interaction of PC4 with XPG in repair of oxidative damage could conceivably affect either global BER or TCR. The stable, specific binding of XPG to double-strand DNA containing unpaired regions is functionally separate from its structure-specific endonuclease activity (25; Sarker, Tsutakawa, and Cooper, unpublished), and an attractive possibility is that its preferential binding to the transcription-sized bubble is relevant to the TCR function of XPG. In TCR, rapid preferential repair is initiated on transcribed strands after an RNA polymerase is stalled at a DNA lesion, and it is thought that the RNA polymerase must be removed or remodeled in order for the repair enzymes to gain access to the lesion (for a review, see reference 51). XPG is apparently required along with CSB and TFIIH for this early step in TCR (34; S. E. Tsutakawa and P. K. Cooper, unpublished). Our finding that XPG bound to a DNA bubble substrate recruits PC4 to the complex with a resulting displacement of XPG suggests the possibility that PC4 may be involved in TCR at a step immediately following XPG.
The observed reduction of peroxide sensitivity seen when the sub1Δ rad2Δ double mutant is compared with the sub1Δ single mutant strain is consistent with the idea that Rad2 produces a potentially lethal intermediate in repair of oxidative damage that requires Sub1 for efficient further processing. Inability to release Rad2 from the partially repaired lesion (or lesion plus stalled RNA polymerase) may block access to proteins required to complete subsequent steps of repair. According to this model, the accumulation of unrepaired or inefficiently repaired intermediates leads to increased lethality in the sub1Δ mutant. Blocking production of the intermediates by eliminating Rad2 improves survival because the initial damage is not as lethal as the DNA repair intermediate. It should be noted that the effect of the rad2Δ mutation is only partial, suggesting that Sub1 may either have additional functions in DNA repair or may perform similar functions for other DNA repair enzymes.
In the context of this model, our finding that PC4 displaces XPG that is stably bound to a DNA bubble structure suggests the possibility that PC4 is required in TCR for release of XPG to allow subsequent processing either of the lesion itself or of the stalled RNA polymerase. Significantly, a requirement for an XPG release factor in NER has recently been suggested by the results of Riedl et al., who found that XPG was not released from the DNA substrate after excision of the lesion in vitro without the addition of an unknown factor present in nuclear extracts (47). While this NER release factor is presumably not PC4, since deletion of SUB1 does not result in sensitivity to UV, it is possible that XPG similarly is not released on its own after its function in TCR but requires PC4 in order to turn over. The observation that PC4 stimulates XPG release is particularly intriguing in light of results demonstrating that PC4 can stimulate DNA synthesis via an interaction with replication protein A (44). A possible explanation combining all these results is that PC4 may function in TCR as an intermediary between RNA polymerase removal or remodeling by XPG together with other TCR proteins, possibly clearing the initial TCR machinery from the damaged region, and subsequent steps including recruitment of repair enzymes and synthesis of the repair patch. The observation that PC4 blocks RNA polymerase elongation in vitro and the ability of TFIIH to alleviate this block (17, 57) raise the possibility that PC4 may also have additional functions in TCR that affect the resumption of transcription. Clearly, the close association of PC4 with other transcriptional processes and components of the transcription machinery makes a possible role for PC4 in transcription-coupled DNA repair processes particularly interesting. However, it is presently unclear if PC4 functions in XPG-stimulated BER, XPG-dependent TCR of oxidative DNA damage, or both, and further experimentation will be required to elucidate its possible roles in these processes. In this connection, it should be noted that the lack of sensitivity of the sub1Δ mutant to UV does not rule out a requirement for PC4 in TC-NER (TCR of UV damage), since loss of TCR by deletion of RAD26 (the yeast homolog of CSB) does not render yeast sensitive to UV (53). Thus, the postulated role for PC4 in TCR as a release factor for XPG could conceivably apply to TCR of UV and oxidative lesions.
The function of PC4 in XPG-related DNA repair processes does not readily explain the ability of this protein to function in the E. coli oxidative antimutator assay. However, since the highly sensitive E. coli antimutator assay requires only a small number of repair events per cell per generation to reveal suppression of spontaneous mutagenesis, the repair-enhancing activity of PC4 need not be very efficient. The activity we observed could be due to a general effect of PC4 binding to damaged DNA or to unpaired DNA regions produced as intermediates of repair, creating a more accessible environment for additional E. coli DNA repair factors. It is unlikely that protein-protein interactions between human PC4 protein and bacterial DNA repair proteins would be functional, and moreover, E. coli does not encode any proteins homologous to PC4. While the interaction of PC4 with XPG implicates it in an XPG-dependent DNA repair pathway in human cells, it may also function in a more general fashion to stimulate, stabilize, or assist DNA repair by other factors via a direct interaction with DNA, as it evidently can do in E. coli. Expression of human PC4 in either the wild type or the fpg mutY double mutant strain of E. coli does not result in enhanced resistance to the lethal effects of hydrogen peroxide exposure (data not shown). However, since fpg and mutY mutations have little or no effect on peroxide lethality (unpublished observations), it is unclear if this observation indicates a specificity of PC4 for repair of nonlethal lesions, such as 8-oxoG, or that the DNA repair activity required for mutation suppression when PC4 is expressed in bacteria is weak.
Sub1 mutations in yeast result in increased spontaneous and induced oxidative mutagenesis and lethality. Since expression of the PC4 protein in yeast can restore the wild-type phenotype, it suggests that the human PC4 protein may also function to prevent mutations resulting from oxidative DNA damage in human cells. Mutations that cause increased spontaneous and damage-induced mutagenesis cause an increased risk of cancer (24). PC4 maps to chromosome 15p13, a region that frequently suffers from loss of heterozygosity in bladder and lung tumors (5, 58). This has been interpreted to suggest that the regulatory properties of PC4 may be important in tumor suppression, and support for this hypothesis has been presented previously (29). However, our findings suggest an alternative, or additional, mechanism for PC4 in tumor suppression. Loss of PC4 function may also increase the rate of mutagenesis resulting from spontaneous or induced oxidative DNA damage in humans. This can increase the level of mutations leading to transformation or secondary mutations within tumors leading to tumor promotion.
Acknowledgments
We thank Michael Hampsey for the yeast sub1Δ mutant and its corresponding wild type, Michael Meisterernst for the full-length mutant and wild-type human PC4 clones, Yusaka Nakabeppu for the human OGG1 and MYH clones, Patricia Foster for the cloned bacterial fpg gene used to construct the expression vector, Wolfram Siede for the vectors used to construct the rad2Δ mutants, and Rabindra Roy for human NTH1 protein. We thank Nathan Elliott and Susan Tsutakawa for critical comments and helpful suggestions on the manuscript.
This study was supported by a grant from the Worcester Foundation for Biomedical Research to M.R.V. and by NIH grants GM56420 to M.R.V. and CA63503 to P.K.C.
REFERENCES
- 1.Aburatani, H., Y. Hippo, T. Ishida, R. Takashima, C. Matsuba, T. Kodama, M. Takano, A. Yasui, K. Yamamoto, M. Asano, K. Fukasawa, T. Yoshinari, H. Inoue, E. Ohtsuka, and S. Nisimura. 1997. Cloning and characterization of mammalian 8-hydroyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 57:2151-2156. [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arai, K., K. Morishita, K. Shinmura, T. Kohno, S. R. Kim, T. Nohmi, M. Taniwaki, S. Ohwada, and J. Yokota. 1997. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene 14:2857-2861. [DOI] [PubMed] [Google Scholar]
- 4.Bessho, T. 1999. Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 27:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohm, M., H. Kirch, T. Otto, H. Rubben, and I. Wieland. 1997. Deletion analysis at the DEL-27, APC and MTS1 loci in bladder cancer: LOH at the DEL-27 locus on 5p13-12 is a prognostic marker of tumor progression. Int. J. Cancer 74:291-295. [DOI] [PubMed] [Google Scholar]
- 6.Bohr, V. A. 2002. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 32:804-812. [DOI] [PubMed] [Google Scholar]
- 7.Bohr, V. A., and G. L. Dianov. 1999. Oxidative DNA damage processing in nuclear and mitochondrial DNA. Biochimie 81:155-160. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Bradsher, J., J. Auriol, L. Proietti de Santis, S. Iben, J. L. Vonesch, I. Grummt, and J. M. Egly. 2002. CSB is a component of RNA pol I transcription. Mol. Cell 10:819-829. [DOI] [PubMed] [Google Scholar]
- 10.Brandsen, J., S. Werten, P. C. van der Vliet, M. Meisterernst, J. Kroon, and P. Gros. 1997. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat. Struct. Biol. 4:900-903. [DOI] [PubMed] [Google Scholar]
- 11.Calvo, O., and J. L. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 12.Croteau, D. L., and V. A. Bohr. 1997. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J. Biol. Chem. 272:25409-25412. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham, R. P. 1997. DNA glycosylases. Mutat. Res. 383:189-196. [DOI] [PubMed] [Google Scholar]
- 14.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demple, B., and L. Harrison. 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63:915-948. [DOI] [PubMed] [Google Scholar]
- 16.Evans, E., J. Fellows, A. Coffer, and R. D. Wood. 1997. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. EMBO J. 16:625-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukuda, A., S. Tokonabe, M. Hamada, M. Matsumoto, T. Tsukui, Y. Nogi, and K. Hisatake. 2003. Alleviation of PC4-mediated transcriptional repression by the ERCC3 helicase activity of general transcription factor TFIIH. J. Biol. Chem. 278:14827-14831. [DOI] [PubMed] [Google Scholar]
- 18.Ge, H., E. Martinez, C. M. Chiang, and R. G. Roeder. 1996. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 274:57-71. [DOI] [PubMed] [Google Scholar]
- 19.Ge, H., and R. G. Roeder. 1994. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78:513-523. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henle, E. S., and S. Linn. 1997. Formation, prevention and repair of DNA damage by iron/hydrogen peroxide. J. Biol. Chem. 272:19095-19098. [DOI] [PubMed] [Google Scholar]
- 22.Henry, N. L., D. A. Bushnell, and R. D. Kornberg. 1996. A yeast transcriptional stimulatory protein similar to human PC4. J. Biol. Chem. 271:21842-21847. [DOI] [PubMed] [Google Scholar]
- 23.Hockenbery, D. M., Z. N. Oltvai, X. M. Yin, C. L. Milliman, and S. J. Korsmeyer. 1993. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell 75:241-251. [DOI] [PubMed] [Google Scholar]
- 24.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 25.Hohl, M., F. Thorel, S. G. Clarkson, and O. D. Scharer. 2003. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J. Biol. Chem. 278:19500-19508. [DOI] [PubMed] [Google Scholar]
- 26.Holloway, A. F., F. Occhiodoro, G. Mittler, M. Meisterernst, and M. F. Shannon. 2000. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. J. Biol. Chem. 275:21668-21677. [DOI] [PubMed] [Google Scholar]
- 27.Jian, D., Z. Hatahet, J. O. Blaisdell, R. J. Melamede, and S. S. Wallace. 1997. Escherichia coli endonuclease VIII: cloning, sequencing and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol. 179:3773-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser, K., G. Stelzer, and M. Meisterernst. 1995. The coactivator p15 (PC4) initiates transcriptional activation during TFIIA-TFIID-promoter complex formation. EMBO J. 14:3520-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kannan, P., and M. A. Tainsky. 1999. Coactivator PC4 mediates AP-2 transcriptional activity and suppresses ras-induced transformation dependent on AP-2 transcriptional interference. Mol. Cell. Biol. 19:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klungland, A., I. Rosewell, S. Hollenbach, E. Larsen, G. Daly, B. Epe, E. Seeberg, T. Lindahl, and D. E. Barnes. 1999. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA 96:13300-13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knaus, R., R. Pollock, and L. Guarente. 1996. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 15:1933-1940. [PMC free article] [PubMed] [Google Scholar]
- 32.Kretzschmar, M., K. Kaiser, F. Lottspeich, and M. Meisterernst. 1994. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell 78:525-534. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. K., S. L. Yu, L. Prakash, and S. Prakash. 2002. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. Implications for Cockayne syndrome. Cell 109:823-834. [DOI] [PubMed] [Google Scholar]
- 34.Le Page, F., E. E. Kwoh, A. Avrutskaya, A. Gentil, S. A. Leadon, A. Sarasin, and P. K. Cooper. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159-171. [DOI] [PubMed] [Google Scholar]
- 35.Loft, S., and H. E. Poulsen. 1996. Cancer risk and oxidative damage in man. J. Mol. Med. 74:297-312. [DOI] [PubMed] [Google Scholar]
- 36.Marnett, L. J. 2000. Oxyradicals and DNA damage. Carcinogenesis 21:361-370. [DOI] [PubMed] [Google Scholar]
- 37.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaels, M. L., L. Pham, C. Cruz, and J. H. Miller. 1991. MutM, a protein that prevents GC→TA transversions, is formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 19:3629-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaels, M. L., J. Tchou, A. P. Grollman, and J. H. Miller. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964-10968. [DOI] [PubMed] [Google Scholar]
- 41.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 42.Nishioka, K., T. Ohtsubo, H. Oda, T. Fujiwara, D. Kang, K. Sugimachi, and Y. Nakabeppu. 1999. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohtsubo, T., K. Nishioka, Y. Imaiso, S. Iwai, H. Shimokawa, H. Oda, T. Fujiwara, and Y. Nakabeppu. 2000. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 28:1355-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan, Z. Q., H. Ge, A. A. Amin, and J. Hurwitz. 1996. Transcription-positive cofactor 4 forms complexes with HSSB (RPA) on single-stranded DNA and influences HSSB-dependent enzymatic synthesis of simian virus 40 DNA. J. Biol. Chem. 271:22111-22116. [DOI] [PubMed] [Google Scholar]
- 45.Perkins, E. L., J. F. Sterling, V. I. Hashem, and M. A. Resnick. 1999. Yeast and human genes that affect the Escherichia coli SOS response. Proc. Natl. Acad. Sci. USA 96:2204-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radicella, J. P., C. Dherin, C. Desmaze, M. S. Fox, and S. Boiteux. 1997. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:8010-8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riedl, T., F. Hanaoka, and J. M. Egly. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22:5293-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roldan-Arjona, T., Y. F. Wei, K. C. Carter, A. Klungland, C. Anselmino, R. P. Wang, M. Augustus, and T. Lindahl. 1997. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl. Acad. Sci. USA 94:8016-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slupska, M. M., C. Baikalov, W. M. Luther, J. H. Chiang, Y. F. Wei, and J. H. Miller. 1996. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J. Bacteriol. 178:3885-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svejstrup, J. Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3:21-29. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, D., A. D. Scot, R. Barbey, M. Padula, and S. Boiteux. 1997. Inactivation of OGG1 increases the incidence of G · C→T · A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 254:171-178. [DOI] [PubMed] [Google Scholar]
- 52a.Tsutakawa, S. K., and P. K. Cooper. 2000. Transcription-coupled repair of oxidative DNA damage in human cells: mechanisms and consequences. Cold Spring Harbor Symp. Quant. Biol. 65:201-215. [DOI] [PubMed] [Google Scholar]
- 53.van Gool, A. J., R. Verhage, S. M. Swagemakers, P. van de Putte, J. Brouwer, C. Troelstra, D. Bootsma, and J. H. Hoeijmakers. 1994. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 13:5361-5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volkert, M. R., N. A. Elliott, and D. E. Housman. 2000. Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc. Natl. Acad. Sci. USA 97:14530-14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wakasugi, M., J. T. Reardon, and A. Sancar. 1997. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J. Biol. Chem. 272:16030-16034. [DOI] [PubMed] [Google Scholar]
- 56.Werten, S., F. W. Langen, R. van Schaik, H. T. Timmers, M. Meisterernst, and P. C. van der Vliet. 1998. High-affinity DNA binding by the C-terminal domain of the transcriptional coactivator PC4 requires simultaneous interaction with two opposing unpaired strands and results in helix destabilization. J. Mol. Biol. 276:367-377. [DOI] [PubMed] [Google Scholar]
- 57.Werten, S., G. Stelzer, A. Goppelt, F. M. Langen, P. Gros, H. T. Timmers, P. C. Van der Vliet, and M. Meisterernst. 1998. Interaction of PC4 with melted DNA inhibits transcription. EMBO J. 17:5103-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wieland, I., M. Bohm, K. C. Arden, T. Ammermuller, S. Bogatz, C. S. Viars, and M. F. Rajewsky. 1996. Allelic deletion mapping on chromosome 5 in human carcinomas. Oncogene 12:97-102. [PubMed] [Google Scholar]
- 59.Wu, W. H., I. Pinto, B. S. Chen, and M. Hampsey. 1999. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153:643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyrzykowski, J., and M. R. Volkert. 2003. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J. Bacteriol. 185:1701-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]