Abstract
The high degree of size heterogeneity of apo(a), the distinct protein component of lipoprotein (a) [Lp(a)], renders the development and selection of specific antibodies directed to apo(a) more difficult and poses significant challenges to the development of immunoassays to measure its concentration in plasma or serum samples. Apo(a) is extremely variable in size not only between but also within individuals because of the presence of two different, genetically determined apo(a) isoform sizes. Therefore, the antigenic determinants per particle available to interact with the antibodies will vary in the samples and the calibrators, thus contributing to apo(a) size-dependent inaccuracy of different methods. The lack of rigorous validation of the immunoassays and common means of expressing Lp(a) concentrations hinder the harmonization of results obtained by different studies and contribute to the lack of common cut points for identification of individuals at risk for coronary artery disease or for interventions aimed at reducing Lp(a) levels. The aim of our review is to present and critically evaluate the issues surrounding the measurements of Lp(a), their impact on the clinical interpretation of the data, and the obstacles we need to overcome to achieve the standardization of Lp(a) measurements.
Keywords: apolipoprotein (a), cardiovascular disease, analytical methods, assay standardization
Lp(a) STRUCTURAL CHARACTERISTICS AND THEIR IMPACT ON Lp(a) MEASUREMENT
Lipoprotein (a) [Lp(a)], is the most complex and polymorphic of the lipoprotein particles. Despite more than 50 years of intense research that has elucidated many aspects of Lp(a)’s structure and biochemistry, its physiological and pathological roles are still poorly understood. Lp(a) is composed of a lipoprotein particle quite similar in protein and lipid composition to LDL, containing one molecule of apoB wrapped around a particle that has primarily a core of cholesteryl ester and triglyceride with phospholipids and unesterified cholesterol at its surface. The presence of a unique hydrophilic, highly glycosylated protein referred to as apo(a), covalently attached to apoB-100 by a single disulfide bridge, differentiates Lp(a) from LDL (1, 2). Apo(a) is part of the plasminogen gene superfamily, and its presence imparts distinctive synthetic and catabolic properties to Lp(a) along with a marked size heterogeneity (3).
Treatment of purified Lp(a) with a reducing agent dissociates apo(a) from the particle yielding a lipoprotein particle that is similar to LDL in physical and chemical properties. However, Lp(a) particles have been reported to associate noncovalently with triglyceride-rich lipoproteins in hypertriglyceridemic individuals or after a fatty meal (4). This association may result in overestimation of Lp(a) measured by ELISA methods based on the apo(a) capture/apoB detection approach.
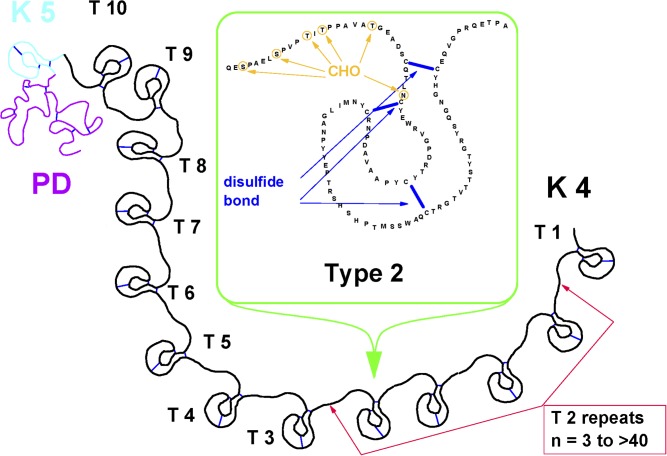
Apo(a), shares a high amino acid sequence homology to several regions of the serine protease zymogen plasminogen, including the protease domain, and the so-called kringle 4 (K4) and 5 domains, which are tri-loop polypeptides stabilized by three internal disulfide bridges. Apo(a) is thus formed by an inactive carboxy-terminal protease-like domain and by a kringle 5 domain, both of which exhibit ∼85% homology with plasminogen, and multiple copies of the plasminogen-like K4 domain (Fig. 1). Based on amino acid sequence differences, the K4 domain of apo(a) is divided into 10 similar but distinct K4 types (1 through 10), having 75% to 85% amino acid homology with the K4 of plasminogen (5, 6). Each of the K4 types, except K4 type 2, is present as a single copy, whereas the identical K4 type 2 repeats vary from a minimum of 3 to as many as 40 (3, 7). As a consequence, apo(a) has the unique characteristic of being highly polymorphic in size, and the variable numbers of the K4 type 2 domains are primarily responsible for the size heterogeneity of Lp(a). Apo(a) is also heterogeneous in its glycosylation, which occurs both within the core of K4 motifs and within the linker sequences that join individual kringles (8), thus additionally contributing to the size heterogeneity of Lp(a).
Fig. 1.
Schematic representation of apo(a). Based on amino acid sequence differences, the plasminogen-like K4 domain of apo(a) is formed by 10 distinct K4 types. K4 type 1 and types 3 to 10, are present as a single copy, whereas the K4 type 2 is present in a variable number of identical copies ranging from 3 to >40. In the figure insert is the complete amino acid sequence of K4 type 2.
A variety of immunochemical methods, such as ELISA, nephelometry, immunoturbidimetry, and dissociation-enhanced lanthanide fluorescent immunoassay, are used to measure Lp(a) in human plasma or sera. The peculiar structural characteristics of Lp(a), including the high degree of size heterogeneity, the covalent association of apo(a) with apoB in the Lp(a) macromolecular complex, and the high sequence homology between apo(a) and plasminogen, constitute a significant challenge to the development of suitable immunoassays for the accurate measurement of Lp(a). The first hurdle is to develop suitable antibodies that are specific for apo(a) in the Lp(a) molecular complex. One possible approach is to make antibodies against purified apo(a), obtained by dissociating it from Lp(a) with a reducing agent. However, antibodies against apo(a) prepared in this manner generally react poorly with apo(a) in Lp(a), presumably because the reducing agent not only cleaves the disulfide bond between apoB and apo(a), but also cleaves the three intrachain disulfide bonds in each of the kringle domains, contributing to altered apo(a) conformation and to changes in its immunoreactivity. An alternate approach is to raise antibodies against purified Lp(a). Polyclonal antibodies produced by this approach would require absorptive removal of antibodies reacting with apoB and plasminogen. Similarly, monoclonal antibodies (MAbs) made against Lp(a) need to be selected for antigenic determinants specific for apo(a) and therefore not present in plasminogen or apoB.
To correctly frame the issues related to the measurement of Lp(a), it is appropriate to provide some basic information on the use of immunoassays to measure the concentration of different proteins in human samples. Measurements by immunoassay are based on the antibody-antigen reaction whereby the measurement of a signal is generated by the formation of the antigen-antibody complex. A value is obtained by comparison of the signal in samples with that generated by a standard containing a known concentration of the analyte. For an assay to be accurate, 1) the antibody needs to be specific for the analyte being measured, 2) the analyte being measured in the sample should have the same structural characteristics as the analyte in the assay calibrator to achieve the same degree of immunoreactivity per particle, 3) an accuracy-based target value should be assigned to the assay calibrators using an appropriate reference material to guarantee consistency and comparability of results, and 4) common protocols should be available for transferring an accurate value from the reference material to the assay calibrators and to verify that accurate results are obtained on test samples.
Considering the intra- and interindividual high degree of size variation in apo(a) due to the variable number of K4 type 2 repeats, it is practically impossible to select assay calibrators with the same apo(a) size present in individual samples to be analyzed. Because a greater number of antibodies directed to K4 type 2 will react with the larger than the smaller Lp(a) particles, Lp(a) molecules in the samples larger than those in the calibrator will give a higher signal than that of the calibrator, resulting in overestimation of Lp(a) values. In contrast, in the samples with Lp(a) molecules smaller than those in the calibrator, Lp(a) values will be underestimated. Therefore, the mass of the measured particles will not reflect the number of Lp(a) particles. Furthermore, the degree of inaccuracy in the samples will vary depending on the choice of the apo(a) sizes in the assay calibrators, their lot-to-lot changes, and the different approaches taken to assign their target value. Considering that calibrators of turbidimetric and immunonephelometric assays are usually selected to have high Lp(a) levels, the isoforms present in the calibrators may be predominantly constituted by small apo(a) sizes with the consequence that the majority of samples will have apo(a) isoforms larger than those in the calibrator resulting in overestimation of Lp(a) values.
The different approaches taken to assign the target value to the assay calibrators and to express the Lp(a) values are other major factors contributing to Lp(a) method inaccuracy and to the lack of comparability of values obtained by commercially available methods. In the early 1970s, the first immunochemical methods developed to measure Lp(a) concentrations were based on antibodies generated using purified Lp(a), and the values assigned to the calibrators and consequently to the samples were expressed in milligrams per deciliter of the total lipoprotein mass obtained by summation of the major components of Lp(a) preparations purified from plasma (9, 10). The majority of the assays subsequently developed expressed Lp(a) values in milligrams per deciliter of the total lipoprotein particle even though no common approaches were followed by manufacturers or research laboratories to assign the target values to the assay calibrators. Even though using modern immunoassays, Lp(a) levels are measured using antibodies specific to apo(a), the distinct protein component of Lp(a), many commercial methods and research laboratories continue to use standards with values assigned in milligrams per deciliter or grams per liter of the total mass of Lp(a) using different and poorly defined “master calibrators.” However, it is not possible to accurately determine the total mass of the heterogeneous Lp(a) particle because it requires the quantification of all the independent Lp(a) constituents, not only the protein but also the multiple lipid and carbohydrate components. Even if all the Lp(a) components could be accurately measured and the summation value of the components transferred to assay calibrators, the value would not be accurate because most individuals express two forms of Lp(a) that differ in apo(a) size, Lp(a) mass, and composition. The major lipid components include triglycerides, phospholipids, and cholesteryl esters with each containing numerous fatty acid species that vary in molecular mass due to differences in chain length and the degree of saturation. Lp(a) also contains variable types and amounts of sphingolipids and other fat-soluble molecules. The carbohydrate component of Lp(a) is also quite variable. Thus, the Lp(a) mass cannot be accurately computed from its constituent components because of inaccuracies in the determination of the mass of the major components and the failure to measure all components.
Lp(a) mass can also be estimated by physical chemical methods such as sedimentation and flotation equilibrium (11). Unfortunately, estimation of purified Lp(a) mass by physical chemical methods requires a number of assumptions and approximations that contribute to inaccuracy in the molecular mass estimations of the Lp(a) macromolecular complex. Another approach that has been followed is to first determine the mass of the protein components of Lp(a), apo(a), and apoB and then assume that the nonprotein components of Lp(a) are similar to that of LDL. Although the apoB component of Lp(a) presumably contains carbohydrate similar to that of LDL, the apo(a) component contains a considerable amount of additional carbohydrate, and the amount varies with the size of apo(a). Furthermore, no rigorous studies have been performed to evaluate to what extent LDL and Lp(a) may also differ in the highly heterogeneous and variable lipid component. Computation of the mass of Lp(a) requires an accurate quantification of each of the different lipid, protein, and carbohydrate components of Lp(a). Highly specialized analytical approaches, each with its own margin of error, are required to quantify the different Lp(a) constituents. As a result, computation of the mass of Lp(a) is fraught with errors. Also, using conventional purification procedures, the amount of Lp(a) recovered from plasma is usually only ∼30% of the Lp(a) initially present. Therefore, the composition of the Lp(a) particles isolated by these procedures may not be representative of the total Lp(a) particles in plasma, and the relative proportion of the two Lp(a) species present in plasma of heterozygous individuals may not be the same as that in the purified Lp(a) preparation. Detailed physical/chemical analyses of Lp(a) mass purified by more advanced approaches such as immunoaffinity purification are not yet available. In any case, for analytical purposes, even if Lp(a) mass could be accurately determined in a primary standard, the Lp(a) mass and the relative concentration of each of the components would be different from that in the assay calibrators or in the test samples.
Additionally, the weight ratio of the protein mass of apo(a) to apoB in Lp(a) varies widely depending on the size of apo(a). For example, an Lp(a) containing 15 apo(a) K4 units has an apo(a)/apoB weight ratio of 0.44, whereas an Lp(a) containing 32 apo(a) K4 units has a weight ratio of 0.85. Thus, it is not possible for the primary standard, secondary reference material, assay calibrators, and human samples to have the same Lp(a) composition. Another major point to consider is that there are no other lipoproteins where the values are expressed in total mass, but instead the values are expressed either in milligrams per deciliter or in SI units on the basis of which lipoprotein component is directly measured, being it a specific protein or a specific lipid.
IMPACT OF apo(a) SIZE HETEROGENEITY ON THE ACCURACY OF Lp(a) MEASUREMENTS
To evaluate the contribution of the apo(a) size polymorphism on the inaccuracy of Lp(a) measurements, a variety of MAbs were generated in our laboratory, selected, and carefully characterized for their apo(a) domain specificity, high affinity, and immunochemical properties (12). Using an ELISA sandwich format, an MAb (MAb a-6) was selected to be coated on the ELISA plate wells. The epitope recognized by this antibody is located in apo(a) K4 type 2. As depicted in Fig. 1, the K4 type 2 is present in a variable number of identical repeats, and the selection of this MAb was made to achieve the capture of all Lp(a) particles present in the plasma samples. To mimic the immunochemical properties of polyclonal antibodies, MAb a-5, directed to an epitope present in both K4 type 1 and 2 (Fig. 1), was selected as the detecting antibody in one ELISA format. MAb a-40, specific for a unique apo(a) epitope located in K4 type 9 (Fig. 1), was selected as the detecting antibody in a second ELISA format (12).
We have demonstrated that Lp(a) contains 1 mol of apo(a) and 1 mol of apoB (13), and therefore, we determined by amino acid analysis the protein concentration of an Lp(a) preparation isolated from human plasma containing an apo(a) with 21 K4 motifs. This purified Lp(a) was then used as a primary standard. To circumvent the problems of the apo(a) size variability, the Lp(a) protein concentration was calculated and expressed in nanomoles per liter, thus reflecting the number of Lp(a) particles. The value obtained in the primary standard was then transferred to the assay calibrator. This value transfer is performed by using the primary standard to calibrate the assay and by performing multiple analyses of the calibrator over a period of several days. The mean of the values constitutes the assigned value of the assay calibrator. A final check of the accuracy of the value transfer is performed by calibrating the assay with the calibrator material and by analyzing multiple times the primary standard. The value transfer is considered accurate if the mean value obtained on the primary standard is within 2% of the expected value. Samples used in this study were selected from subjects who demonstrated a single apo(a) isoform by a high-resolution phenotype system (14). We have shown that the log of the number of apo(a) K4-encoding sequences in the apo(a) gene are linearly related to the relative mobility of the apo(a) isoforms on agarose gel, providing the basis for a standardized isoform nomenclature where each isoform is defined by its number of K4 domains (15).
The selected 723 samples were analyzed in parallel with the same ELISA conditions using the two different detecting MAbs (12). The average bias between the a-5 MAb and a-40 MAb formats was highly correlated with the number of apo(a) K4 domains in the sample. Thus, Lp(a) values measured using MAb a-5, which mimics the immunochemical reactivity of a polyclonal antibody, were higher than those using MAb a-40 in samples containing >21 K4 units and were lower in samples containing <21 K4 units (Fig. 2). For example, by using MAb a-5, Lp(a) values in samples containing 18 K4 were underestimated by 10%, whereas the values were overestimated by ∼20% in samples containing 25 K4 as compared with the values obtained by MAb a-40. The results of this experiment clearly show that the apo(a) size heterogeneity has a significant impact on the accuracy of the measurement of Lp(a).
Fig. 2.
Comparison of Lp(a) values obtained by ELISA using the same MAb (a-6) to capture Lp(a) in the samples and different detecting MAbs (a-5 and a-40) and a polyclonal antibody to B-100. Mean Lp(a) levels obtained by each detecting antibody were compared as a function of the predominantly expressed apo(a) isoform size.
The 723 samples were also analyzed by the same ELISA conditions, but using a polyclonal antibody against apoB as detecting antibody (12). Very similar Lp(a) values, regardless of apo(a) size, were obtained by using either MAb a-40 or the polyclonal antibody directed against apoB as detecting antibody (Fig. 2). These results clearly indicate that harmonization of Lp(a) values can be achieved using a common calibrator and antibodies unaffected by the size polymorphism of apo(a). More recently, Lp(a) values obtained by the MAb a-40 ELISA method on 80 samples spanning a large range of levels and apo(a) isoform size showed high correlation and excellent agreement of absolute values with an ultraperformance liquid chromatography/mass spectrometry method (16), again confirming that using calibrator values traceable to a common reference material, comparable Lp(a) values can be obtained by different methods not affected by apo(a) size variation.
EFFECT OF ASSAY INACCURACY ON THE INTERPRETATION OF CLINICAL DATA
To directly evaluate the impact that method inaccuracy may have on the classification of patients at high risk for CVD based on their Lp(a) levels, we used results of analyses performed on the Framingham offspring cohort during the fifth cycle (17). Lp(a) levels and apo(a) isoforms were determined in our laboratory on 2,940 samples. Lp(a) levels were also determined in another laboratory by a commercially available turbidimetric method on 2,556 samples. Individuals with Lp(a) values above the 75 percentile of the Framingham cohort were considered at an increased risk for CVD. The turbidimetric assay as compared with results obtained by the reference ELISA method misclassified 136 individuals as being at increased risk for CVD (false positive) and 23 individuals as being not at risk (false negative). All the false-positive results obtained by the turbidimetric assay were explained by overestimation of Lp(a) values based on the predominant apo(a) size in the sample (17). These findings are consistent with the fact that the size of apo(a) in the calibrator was quite small and most samples in this population had apo(a) sizes larger than the apo(a) in the calibrator. The few false-negative results by this assay were explained by the relatively few samples with apo(a) smaller than the apo(a) in the calibrator.
To further evaluate the impact of Lp(a) method differences on the interpretation of clinical data, Lp(a) levels were measured by the ELISA reference method and by a commercially available nephelometric method on samples from 195 participants in the Physician Health Study who subsequently developed angina and on 195 gender- and age-matched controls (18). Previously published results from the Physician Health Study found no association between Lp(a) levels, as measured by a nephelometric method, and risk of future myocardial infarction (MI), stroke, or peripheral vascular disease (19–21). The results obtained with the reference ELISA indicated that the median Lp(a) concentration was significantly higher in cases as compared with controls and baseline Lp(a) values were predictive of future angina. A 2-fold risk was found in study participants with Lp(a) concentrations in the 80th percentile and a 4-fold risk was found in those with Lp(a) above the 95th percentile. In contrast, when using the results obtained by the nephelometric method, median Lp(a) levels did not differ significantly between cases and controls, and Lp(a) levels were not significantly associated with the development of angina. Based on the results of this study (18), the authors in the discussion concluded that “it seems likely that the Lp(a) method affected by apo(a) size that was previously used in the Physician Health Study may have underestimated or even obscured the true relationship between Lp(a) concentration and CVD.”
DEVELOPMENT AND USE OF REFERENCE MATERIAL FOR STANDARDIZATION OF Lp(a) ASSAYS
In an attempt to standardize the measurement of Lp(a), the major aim of the International Federation of Clinical Chemistry (IFCC) working group on Lp(a), was to select and characterize a secondary reference material to be used by manufacturers of commercially available methods to assign an accuracy-based Lp(a) target value to their assay calibrators (22). Among the different preparations evaluated, a proposed reference material (PRM) was selected to be further characterized. To circumvent the strong limitations in the determination of the total mass of the highly heterogeneous Lp(a) previously discussed and taking into consideration the size polymorphism of apo(a), which is the Lp(a) constituent usually directly measured by the immunoassays, the members of the IFCC working group decided that the target value to PRM be assigned in nanomoles per liter of Lp(a) protein.
We have previously demonstrated that there is 1 mol of apo(a) and 1 mol of apoB in Lp(a) particles (13), and therefore, by determining the molar concentration of the two proteins, we can circumvent the variable molecular mass of apo(a) because the molar units indicate the number of Lp(a) particles independently of their relative mass. The first step in this process was the preparation of a primary reference material. Lp(a) was isolated from plasma of an individual with high Lp(a) levels and a single apo(a) formed by 19 K4 domains by using two different isolation procedures (23). The molar concentration of apo(a) and apoB in the two preparations was determined by amino acid analysis performed in duplicate for each preparation. The apoB amino acid composition was constant, but because the amino acid composition of apo(a) varied by size, a single apo(a) isoform of a known size was required to compute the amino acid composition of apo(a). The amino acid composition of Lp(a) derived from the amino acid analysis was compared with the combined expected amino acid composition derived from apo(a) and apoB sequence data obtained from the Protein Information Resource database. A very low average bias was observed between the expected molar percentage for each amino acid and that observed by amino acid analysis. We can reasonably state that an accurate absolute mass of the Lp(a) protein in molar units was obtained by this approach.
The Lp(a) primary preparation with an accuracy-based assigned value was used to calibrate the ELISA method, and PRM was analyzed in duplicate, multiple times per day on multiple plates over the period of a week yielding a total of 144 values. From these data, a target value of 107 nM was assigned to PRM (23). The transfer of target values in nanomoles per liter from PRM to assay calibrators is the only available approach to circumvent the problem associated with the intra- and interindividual size variability of apo(a). However, for different methods to provide comparable and accurate results on patient samples, methods that are not affected by the variation in apo(a) size are required.
The reference material underwent an extensive evaluation by the members of the IFCC working group demonstrating excellent stability and commutability properties. The complete documentation was submitted to the World Health Organization (WHO) Committee of Biological Standards, and PRM was accepted as the first WHO/IFCC International Reference Reagent for Lp(a) immunoassays (24).
After the selection of the reference material, experiments were conducted to evaluate to what extent its use would improve the comparability of Lp(a) values obtained by different commercially available assays (23). PRM was used to calibrate the 22 difference systems participating in the study and to assign a target value to the different assay calibrators. Uniformity of calibration was demonstrated among the 22 evaluated systems by the high concordance of values obtained on PRM with an among-method coefficient of variation (CV) of 2.8%. Lp(a) was then measured on 30 fresh-frozen samples covering a wide range of Lp(a) values and apo(a) sizes with values assigned by the reference ELISA method. The among-laboratory CVs for these samples were considerably higher than those obtained for PRM and ranged from 6% to 31%. A significant apo(a) size-dependent bias on the fresh-frozen samples was also observed, and the number of samples with Lp(a) values under- or overestimated varied among the different methods as a function of apo(a) size in the assay calibrators. Other factors, such as differences in antibody properties, assay precision and robustness, and sensitivity of the different analytical methods to sample handling and storage conditions, may have also contributed to the lack of comparability of the obtained Lp(a) values. In conclusion, despite the use of a common reference material, harmonization of results obtained by different methods was not achieved, indicating the serious impact that variation in apo(a) size has on the immunochemical measurement of Lp(a).
Among the evaluated systems, only one latex-enhanced turbidimetric method produced by Denka Seiken, Japan, demonstrated an excellent concordance of values with those obtained by the ELISA method with most of the inaccuracy being due to the overestimation of Lp(a) levels in samples with large apo(a) isoforms (23). This demonstrates that the use of the reference material can result in harmonized Lp(a) values if Lp(a) is measured by suitable methods.
AN APPROACH TO Lp(a) IMMUNOASSAY OPTIMIZATION
Because of the low bias between the Lp(a) values obtained by the Denka method and those obtained by ELISA, further evaluation of this kit was performed in our laboratory. The polyclonal antibodies demonstrated a lack of reactivity with apoB and plasminogen and a high immunoreactivity with the K4 type 2 domain of apo(a) (unpublished observation). Therefore, the antibodies used in this assay are sensitive to apo(a) size variability even though their binding to latex particles and the consequent formation of very large immunocomplexes may somewhat reduce the impact of apo(a) size variability. The distinctive feature of this kit is that instead of using serial dilutions of a single standard with a high level of Lp(a) as the assay calibrator, five independent standards with values ranging from low to high are used to calibrate the different instruments.
Considering the inverse relationship between apo(a) size and Lp(a) values, there is a high probability that the apo(a) isoforms in the five calibrators range from large to small thus minimizing the impact of apo(a) size on the accuracy of Lp(a) levels. To confirm this hypothesis, an experiment was conducted in our laboratory using a commercially available turbidimetric assay (25). Analyses were performed in parallel on a large number of samples by a Roche 917 chemistry analyzer using the assay calibrator diluted according to the manufacturer’s instructions or by calibrating the instrument using five samples selected to have different apo(a) sizes ranging from large to small and Lp(a) levels ranging from low to high. A target value in nanomoles per liter was assigned to the manufacturer-provided calibrator and to the five calibrators selected in our laboratory. A consistent size-dependent bias was observed in the samples analyzed with the original assay calibrator while the effect of apo(a) size variability was substantially minimized when the assay was calibrated by the five independent calibrators (25).
We can therefore conclude that the low level of impact of apo(a) size variation observed by the Denka method is primarily due to the use of five independent calibrators, each containing a suitable distribution of apo(a) isoforms and accurately assigned values. Technically, it is not entirely correct to state that the Denka assay is insensitive to apo(a) size because there is not a direct consistent inverse relationship between Lp(a) levels and apo(a) isoform size and high or low Lp(a) levels may be observed in samples with a relatively large range of apo(a) size (26), and therefore, the impact of apo(a) size may not be adequately minimized in all the samples. Additionally, great care needs to be used in the selection of the pool of samples that are used to form each calibrator, in the lot-to-lot variability and in the assignment of the target values. Considering that standard curves with different slopes can be obtained in different instruments, the assignment of target values to each of the five calibrators needs to be evaluated in each specific type of instrument to demonstrate that the Lp(a) assay’s sensitivity to apo(a) size variation has been adequately minimized.
For this purpose, a multistep protocol for the transfer of target values from the WHO/IFCC reference material to the assay calibrators was established and used by the participants in the standardization project (24). As the distributor of the WHO/IFCC reference material, we developed a multistep standardization protocol to be used by manufacturers and clinical or research laboratories for the evaluation of their kits. The first step is the transfer of target values from the WHO/IFCC reference material to the assay calibrator (24), followed by the verification of the accuracy of results on 80 samples from individual donors selected to have a large range of Lp(a) values and apo(a) isoforms. Specific criteria were established to evaluate the acceptability of the absolute bias between the observed and the target values and the contribution of the apo(a) isoform variability on the obtained results.
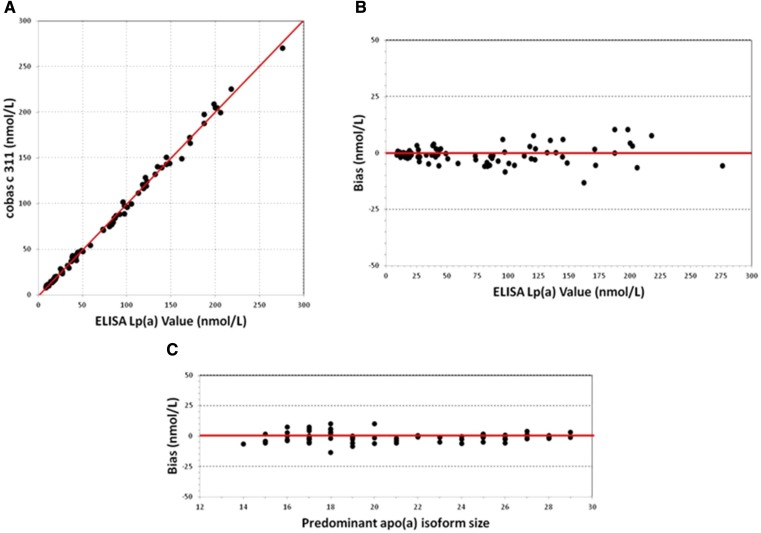
As an example, Fig. 3 illustrates the evaluation of the results obtained using the Denka kit on a Roche Cobas c311 instrument after transfer of the target value from the WHO/IFCC reference material to the five-point assay calibrator. The values obtained on the set of 80 samples were highly correlated with those assigned by the reference ELISA method (Fig. 3A), and a low bias was generally observed between the two methods as a function of Lp(a) assigned values (Fig. 3B). The evaluation of the bias as a function of the predominant apo(a) isoform in the sample did not show a significant impact of apo(a) size variability on the measured values (Fig. 3C).
Fig. 3.
Evaluation of Lp(a) values obtained by the use of Denka reagent on a Roche Cobas c311 instrument after transfer of the target value from the WHO/IFCC reference material to the five-point assay calibrator. A: Correlation between the Lp(a) values obtained by the Cobas c311 (y-axis) with Lp(a) value determined by ELISA (x-axis). B: Bias (nM) as a function of Lp(a) concentration. C: Bias (nM) as a function of apo(a) isoform size.
Since April 2012, when the set of 80 samples was prepared, 42 analytical systems were evaluated and certified for traceability of Lp(a) values to the WHO/IFCC reference material and for lack of significant apo(a) isoform-dependent bias. All the systems with the exception of one were based on the use of different lots of the Denka reagent and different lots of the five-point calibrators. In the same period, six manufacturers, one using a five-point calibrator set and the remaining using single-level calibrators, asked to participate in the method standardization protocol using in-house developed reagents. All six methods demonstrated a high apo(a) size-dependent bias and, therefore, did not meet the certification criteria.
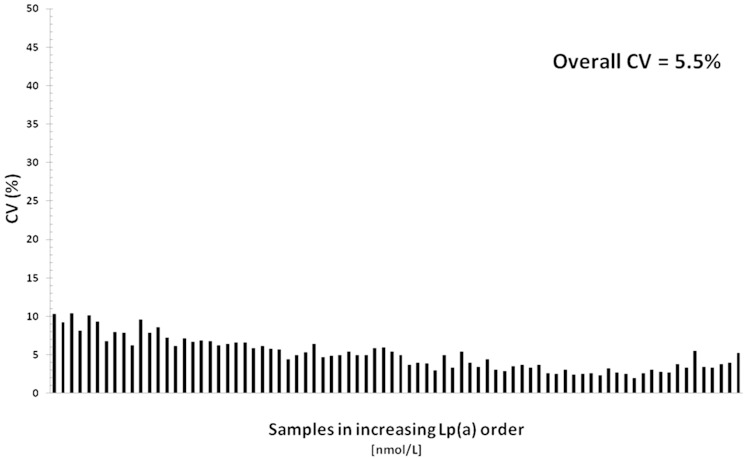
To evaluate the comparability of Lp(a) results obtained by the 42 systems after uniform calibration with the WHO/IFCC reference material, we evaluated the among-method CV on each of the 80 samples with Lp(a) levels ranging from 8.7 nM to 276 nM. The among-method CVs, sorted by increasing sample Lp(a) concentrations, are presented in Fig. 4. The overall CV was 5.5% and ranged from 10.5% to 2.1% demonstrating an excellent harmonization of results obtained by a variety of different instruments and different calibrator lots.
Fig. 4.
CV of Lp(a) values obtained by 42 validated systems on each of 80 samples sorted by increasing Lp(a) concentrations ranging from 8.7 nM to 276.0 nM.
Denka reagent kits are available through different distributors with calibrator values assigned in nanomoles per liter to be traceable to the WHO/IFCC reference material and verified for accuracy by our laboratory. However, Denka kits are also distributed by manufacturers with calibrator values traceable to different internal master calibrators and expressed in milligrams per deciliter of total Lp(a) mass. None of the kits with values assigned in milligrams per deciliter were evaluated by our laboratory. Therefore, we recently conducted an experiment with the aim to evaluate the comparability of Lp(a) values obtained by two different Denka kit distributors using different calibrator lots with values assigned in milligrams per deciliter.
Analyses were performed on 80 samples by a manufacturer in Europe using Denka reagent and five-point calibrator set with values assigned using the manufacturer’s internal master calibrator. The laboratory in the United States performed analyses using Denka reagent and calibrator obtained by a U.S. distributor. Values to the calibrator set were in milligrams per deciliter with no information provided on how the value assignment was performed by the assay distributor. For each sample, we calculated the ratio between the value obtained by each of the Denka kits calibrated in milligrams per deciliter and the target value assigned by ELISA in nanomoles per liter. As expected, the ratio between the milligrams per deciliter and the nanomoles per liter value was not constant in the samples for either kit but largely varied as a function of the predominant apo(a) size in the sample. For the data provided by the European manufacturer, the mean ratio was 2.12 for samples with apo(a) isoforms ≤21 K4 motifs and 1.72 for samples with isoforms >21 K4 motifs with an overall mean ratio of 2.01.
For the data provided by the U.S. clinical laboratory, the mean ratio was 1.88 for samples with isoforms ≤21 K4 and 1.53 for samples >21 K4, and the overall mean ratio was 1.64. Almost identical results were obtained by the use of the two Denka kits for samples with values <10 mg/dl, whereas the U.S. laboratory obtained results that were significantly and consistently higher for all the other samples. In both assays, the impact of apo(a) size variation was not adequately minimized. These findings clearly confirm that the Denka assay’s ability to minimize the impact of apo(a) size variability on the accuracy of Lp(a) values is strictly dependent not only on the use of the five-point calibrators, but also on a properly assigned target value to each of the calibrators. The lack of comparability of data obtained by the two kits also demonstrates the lack of a uniform approach to assign the target values to the calibrators of the two kits. In fact, even though the Lp(a) values obtained by the two methods were highly correlated (r = 0.997), the results obtained by the U.S. laboratory were overall 17% higher than those obtained in Europe.
Another important conclusion from this experiment is that the factor to convert Lp(a) values from nanomoles per liter of apo(a) to milligrams per deciliter of Lp(a) mass is highly imprecise, not only because it is dependent on the size of apo(a) in the samples, but also because it is greatly dependent on how the target values in milligrams per deciliter to the assay calibrators are assigned. In fact, using the same reagent, with one calibrator, the mean overall factor to convert the values from nanomoles per liter to milligrams per deciliter was 2.02, whereas it was 1.64 for the other calibrator. These different conversion factors for samples with identical isoform distribution clearly emphasize that Lp(a) values traceable to the WHO/IFCC reference material and expressed in nanomoles per liter cannot be converted to milligrams per deciliter when values are obtained by kits using calibrators with values not traceable to a common reference material and not verified to provide comparable results on patient samples.
Based on Lp(a) mass composition analyses performed in our laboratory, we found that by taking the average of the Lp(a) nonprotein component, the conversion factor for transforming Lp(a) values from nanomoles per liter to milligrams per deciliter varied from 2.85 for a small apo(a) size to 1.85 for a large one (27). To familiarize the physicians with the use of nanomoles per liter, we suggested the use of a mean conversion factor of 2.4 even though we cautioned the users about the limitation and the inaccuracy of such a factor. Based on different computations of the Lp(a) mass, several higher conversion factors were suggested (28, 29). Based on the data we obtained using the same method but different calibrators, we found that to harmonize the Lp(a) values in milligrams per deciliter to those obtained by the ELISA reference method in nanomoles per liter, the mean conversion factor was 2.02 for one assay and 1.67 for the other, substantially lower than the 2.4 factor we suggested. However, if we would apply the proposed higher conversion factors of 3.17 or 3.3 to the values obtained in milligrams per deciliter, the resulting values in nanomoles per liter would be almost double the values traceable to the WHO/IFCC reference material. In conclusion, we have demonstrated that the practice of using a conversion factor, irrespectively of how it is calculated, should be discontinued.
Lp(a) AND RISK OF CVD
A large number of retrospective case-control and prospective studies have evaluated the association of elevated Lp(a) levels with the risk of CVD, and this topic will be covered in depth in another thematic review in this series. Here, we will discuss this issue mainly from a methodological standpoint. The vast majority of studies found that elevated Lp(a) is an independent risk factor for CVD in both men and women (30–33). Meta-analysis of long-term prospective studies were performed to assess the relationship of Lp(a) concentration with risk of major vascular and nonvascular outcomes. In the evaluated studies, Lp(a) concentrations were measured using a variety of analytical methods. Results of the meta-analysis studies indicated a clear though modest association between Lp(a) and CVD (34–36) and a nearly continuous increase in the risk of CVD with increasing level of Lp(a) and with limited changes in risk at lower Lp(a) levels (31, 36). To evaluate the hypothesis that genetically elevated Lp(a) levels are not only associated with but are the cause of increased risk of MI, Kamstrup et al. (37) studied three large cohorts of white individuals of Danish descent. Using a “Mendelian randomization” approach, this elegant study has provided the first evidence for a casual role of Lp(a) in MI.
The lack of risk or a reduced risk for CVD of Lp(a) levels observed in some prospective studies may be related to inappropriate or nonstandardized methods (18) or inappropriate handling or storage of samples used to measure Lp(a) levels (38). In some studies, it was found that the CVD risk attributable to Lp(a) may also, in part, depend on the presence of other risk factors, such as elevated LDL cholesterol (LDL-C) in the individuals evaluated. For example, Lp(a) levels were no longer predictive of CVD risk in men whose LDL was substantially decreased by lipid-lowering therapy (39). Another study also suggested that LDL-C reduction with statin attenuates the CVD risk of Lp(a) (40). Furthermore, the level of CVD risk associated with elevated Lp(a) has been reported to be higher in individuals with high global CVD risk (41). However, a Danish study failed to find any evidence of interaction between Lp(a) levels and LDL-C levels on CVD risk (31).
Differences in population, study design, length of follow-up, statistical evaluation of the data, methods used to measure Lp(a) concentration, and the methods’ sensitivity to temperature and length of storage of samples may account for the differences in the observed findings. Our ELISA method has been extensively evaluated in terms of sample storage conditions. Practically identical results were obtained between fresh samples and samples frozen at −70°C. Upon thawing, Lp(a) values were stable for 3–4 days with a subsequent decrease up to 30%, particularly in samples with high Lp(a). No significant difference in Lp(a) levels was found in samples stored at 4°C for up to 6 days. The largest, sample-dependent Lp(a) decrease was found in samples stored at −20°C even when analyzed immediately after thawing or after multiple freezing-thawing cycles of samples stored at −70°C (unpublished observations). Despite the profound impact that variation in sample storage conditions may have on Lp(a) values obtained by different methods, very little information on this topic is provided in the literature reporting results from clinical trials.
To minimize the effect of methodological differences in the interpretation of the data in meta-analysis studies (35, 36), cases were compared directly with controls within each of the evaluated studies, thus avoiding the bias due to use of different analytical methods. However, this approach will not correct the under- or overestimation of values obtained by methods affected by apo(a) size variability, and this will impact the interpretation of the results if the apo(a) isoform distribution varies between cases and controls and may, in part, explain why the association of Lp(a) concentration with CVD risk was found to be only modest, albeit independent (36).
Lp(a) levels for definition of CVD risk
Different statistical approaches were used to evaluate the association of Lp(a) levels with CVD, with different studies reporting risk ratios on the basis of different cutoff levels, comparison of mean values, or comparison of the top and bottom tertiles or quartiles of baseline Lp(a). However, in clinical practice, specific cut points are needed to identify individuals at risk for CVD who may require intervention. More than 30 years ago, a report on Lp(a) and MI (42) suggested an Lp(a) cutoff value of 30 mg/dl to reflect an increased risk for MI. This cutoff value is still widely used by clinicians to define patients at risk. At a workshop on Lp(a) and CVD organized by the National Heart, Lung, and Blood Institute, the consensus recommendation, valid for methods reporting Lp(a) in nanomoles per liter, was to consider individuals to be at increased risk if their Lp(a) values exceed the 75 percentile of the Framingham population (25). This percentile corresponds to an Lp(a) value ∼75 nM. However, the consensus recommendation cautioned that more studies using standardized methods to measure Lp(a) are required to define the clinical utility of this cut point for white individuals and to establish appropriate cut points for populations of different ethnicities. The 2010 European Atherosclerosis Society Consensus Panel recommended using an Lp(a) cutoff value of 50 mg/dl, which approximates the 80th percentile of the Danish general population (43).
The CVD risk associated with Lp(a) may also vary by ethnicity as Lp(a) levels and other CVD risk factors can be substantially different depending on the ethnic group (44). However, the vast majority of studies on Lp(a) have been performed in white populations, which limits the applicability of the conclusions to other ethnic or racial groups. One report from the Atherosclerosis Risk in Communities Study suggested that Lp(a) was a less significant predictor of risk in African Americans than Caucasians (45), whereas a later study reported that Lp(a) levels were positively associated with the risk of incident CVD to the same extent in African Americans and Caucasians (46). In contrast, a recent report from the Multi-Ethic Study of Atherosclerosis suggested that Lp(a) levels are a better predictor of CVD events in African Americans than Caucasians (47). The authors also suggested that an Lp(a) cutoff value of 30 mg/dl is suitable for blacks, whereas higher cutoff values should be used for whites and Hispanic individuals.
It is evident that common cut points and valid comparisons of Lp(a) levels among laboratories is not feasible at this point in time because of substantial differences in Lp(a) methods and calibrations and differences among reference populations. Therefore, for an Lp(a) cut point value to be meaningful for clinicians and patients, the method and calibration used to measure Lp(a) in patient samples should be the same as that used to establish the cut point. Furthermore, the patient should be from the same race/ethnicity as the one used to establish the cut point.
Using our ELISA method, we have determined Lp(a) level distribution for three different racial groups, Caucasian Americans from the fifth examination of the Framingham Offspring Study (17), African Americans from the fourth examination of the Coronary Artery Risk Development Study (26), and Japanese Americans from the Honolulu Heart Study (48). The Lp(a) percentiles in the three different populations are presented in Table 1. It is evident that Lp(a) distribution varies greatly among these three racial cohorts with a value of 75 nM approximating the 75th percentile for whites, the 50th percentile for blacks, and the 90th percentile for Japanese individuals. If a common cut point Lp(a) value such as 75 nM is used to define the risk for CVD, ∼25% of whites, 50% of blacks, and only 10% of Japanese individuals will be considered at increased risk. Based on clinical and epidemiological data, it may be reasonable that 25% of white individuals may be at increased CVD risk based on their elevated Lp(a) value. However, it is unlikely that 50% of black individuals and only 10% of Japanese individuals may be at increased CVD risk based on this common Lp(a) cut point.
TABLE 1.
Lp(a) percentiles
| n | Lp(a) Percentiles (nM) | ||||||||||||||||
| 10 | 20 | 30 | 40 | 50 | 60 | 70 | 75 | 80 | 85 | 90 | 95 | 98 | 99 | 99.5 | 99.9 | ||
| Caucasian Americans | 2,929 | 1 | 4 | 8 | 13 | 20 | 32 | 40 | 73 | 100 | 124 | 154 | 209 | 270 | 320 | 360 | 470 |
| African Americans | 1,899 | 16 | 30 | 44 | 58 | 74 | 91 | 114 | 130 | 148 | 166 | 199 | 234 | 305 | 368 | 407 | 531 |
| Japanese Americans | 1,379 | 3 | 6 | 11 | 14 | 19 | 25 | 34 | 40 | 49 | 60 | 75 | 103 | 150 | 194 | 237 | 348 |
Due to the lack of firmly established race-specific clinical cut points for Lp(a), we do not provide in our reports to clinicians any defined threshold Lp(a) value as an indication of increased risk for CVD. Instead, we have implemented a computerized approach to calculate for each patient’s Lp(a) value the corresponding percentile for each of the three racial groups, and this information is included as part of the clinical report. For white individuals, Lp(a) levels above the 75th or 80th percentile can be reasonably suggested to clinicians as an indicator of increased risk for CVD. However, for patients who are black, Japanese, or from other ethnic/racial groups, no such cut points have been established; therefore, clinicians need to exercise their best judgment for the risk evaluation of these individuals.
The availability of new therapeutic approaches to effectively and specifically lower Lp(a) levels (49) will result in a substantial increase in the request of Lp(a) measurements to screen for individuals with high Lp(a). Additionally, clinical trials will be performed to evaluate whether lowering Lp(a) will result in clinical benefits. Therefore, in addition to the necessity to validate the methods for their suitability to accurately measure Lp(a) and to define common units to report the results, specific recommendations should be made for rigorous sample collection, storage, and shipment conditions. Due to the large variability in sensitivity and linearity of different methods, Lp(a) levels that are below the lower calibrator value should not be extrapolated but reported as being below the specific assay limits. Conversely, and more importantly, Lp(a) levels that exceed the highest calibrator value should be reanalyzed after performing an appropriate dilution because extrapolation of the values can result in gross miscalculation of the true Lp(a) levels.
CONCLUSIONS
The structural heterogeneity of apo(a) has a profound impact on the measurement of Lp(a) obtained by different analytical methods resulting in inaccuracy of Lp(a) levels and in lack of comparability of results.
Major obstacles in the standardization of Lp(a) measurements are differences in approaches used to assign a target value to the assay calibrators, differences in units of expression of Lp(a) levels, and lack of commonly accepted guidelines for method validations.
Robust and precise analytical methods based on the use of Denka reagent are available from different manufacturers. Lp(a) concentrations in these assays are reported in nanomoles per liter, and the values are traceable to the WHO/IFCC reference material.
We wish to emphasize that the antibodies used in these assays are isoform dependent and that the reduced impact of apo(a) size variation is primarily due to the use of five different calibrators. To minimize the sensitivity to apo(a) size, each calibrator should be carefully selected in terms of Lp(a) values and apo(a) isoforms. Additionally, each lot of the five-point calibrator set needs to be validated on different analytical platforms using the described multistep standardization protocol to verify that Lp(a) values are accurately determined in patient samples with a minimum contribution of apo(a) isoform size variability. Upon request, manufacturers should provide the certificate of the evaluation of the calibrator and reagent lots with the relative expiration date.
Analytical methods based on the Denka reagent but with calibrator values assigned in milligrams per deciliter of Lp(a) mass with no traceability to a common reference material and no validation of the accuracy of results cannot be considered standardized, and no claim should be made by the manufacturers or by the users on their independence from apo(a) size variability.
No factor, independently of how established, should be used to convert Lp(a) levels from nanomoles per liter to milligrams per deciliter, or vice versa.
At present, a common Lp(a) cut point value to define individuals at high risk for CVD or for treatment assignment cannot be proposed.
The numerous problems plaguing the accurate measurements of Lp(a), as discussed in this review, cannot be ignored. The different issues need to be carefully evaluated and discussed by an international group of experts with the objective to propose possible approaches to circumvent the problems and, most importantly, to make commonly accepted decisions.
In light of the introduction of novel therapeutic approaches to lower Lp(a), the availability of well-standardized assays that provide comparability of results obtained by different laboratories is indispensable for the selection and classification of individuals at high risk.
Introduction of new therapeutic approaches will require performance of clinical trials to evaluate the clinical usefulness of Lp(a) lowering. In addition to the use of suitable methods, major efforts should be made in the standardization of sample collection and storage conditions.
Footnotes
Abbreviations:
- CV
- coefficient of variation
- IFCC
- International Federation of Clinical Chemistry
- K4
- kringle 4
- LDL-C
- LDL cholesterol
- Lp(a)
- lipoprotein (a)
- MAb
- monoclonal antibody
- MI
- myocardial infarction
- PRM
- proposed reference material
- WHO
- World Health Organization
REFERENCES
- 1.Brunner C., Kraft H. G., Utermann G., and Muller H. J.. 1993. Cys4057 of apolipoprotein(a) is essential for lipoprotein(a) assembly. Proc. Natl. Acad. Sci. USA. 90: 11643–11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koschinsky M. L., Côté G. P., Gabel B., and van der Hoek Y. Y.. 1993. Identification of the cysteine residue in apolipoprotein(a) that mediates extracellular coupling with apolipoprotein B-100. J. Biol. Chem. 268: 19819–19825. [PubMed] [Google Scholar]
- 3.van der Hoek Y. Y., Wittekoek M. E., Beisiegel U., Kastelein J. J., and Koschinsky M. L.. 1993. The apolipoprotein(a) kringle IV repeats which differ from the major repeat kringle are present in variably-sized isoforms. Hum. Mol. Genet. 2: 361–366. [DOI] [PubMed] [Google Scholar]
- 4.Gaubatz J. W., Hoogeveen R. C., Hoffman A. S., Ghazzaly K. G., Pownall H. J., Guevara J. Jr., Koschinsky M. L., and Morrisett J. D.. 2001. Isolation, quantitation, and characterization of a stable complex formed by Lp(a) binding to triglyceride-rich lipoproteins. J. Lipid Res. 42: 2058–2068. [PubMed] [Google Scholar]
- 5.McLean J. W., Tomlinson J. E., Kuang W. J., Eaton D. L., Chen E. Y., Fless G. M., Scanu A. M., and Lawn R. M.. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 330: 132–137. [DOI] [PubMed] [Google Scholar]
- 6.Guevara J. Jr., Knapp R. D., Honda S., Northup S. R., and Morrisett J. D.. 1992. A structural assessment of the apo(a) protein of human lipoprotein(a). Proteins. 12: 188–199. [DOI] [PubMed] [Google Scholar]
- 7.Lackner C., Cohen J. C., and Hobbs H. H.. 1993. Molecular definition of the extreme size polymorphism in apolipoprotein(a). Hum. Mol. Genet. 2: 933–940. [DOI] [PubMed] [Google Scholar]
- 8.Scanu A. M., and Edelstein C.. 1997. Learning about the structure and biology of human lipoprotein(a) through dissection by enzymes of the elastase family: facts and speculations. J. Lipid Res. 38: 2193–2206. [PubMed] [Google Scholar]
- 9.Albers J. J., and Hazzard W. R.. 1974. Immunochemical quantification of human plasma Lp(a) lipoprotein. Lipids. 9: 15–26. [DOI] [PubMed] [Google Scholar]
- 10.Albers J. J., Adolphson J. L., and Hazzard W. R.. 1977. Radioimmunoassay of human plasma Lp(a) lipoprotein. J. Lipid Res. 18: 331–338. [PubMed] [Google Scholar]
- 11.Fless G. M., and Santiago J. Y.. 1997. Molecular weight determination of lipoprotein(a) in solutions containing either NaBr or D20: relevance to the number of apolipoprotein(a) subunits in Lp(a). Biochemistry. 36: 233–238. [DOI] [PubMed] [Google Scholar]
- 12.Marcovina S. M., Albers J. J., Gabel B., Koschinsky M. L., and Gaur V. P.. 1995. Effect of the number of apolipoprotein(a) kringle 4 domains on immunochemical measurements of lipoprotein(a). Clin. Chem. 41: 246–255. [PubMed] [Google Scholar]
- 13.Albers J. J., Kennedy H., and Marcovina S. M.. 1996. Evidence that Lp(a) contains one molecule of apo(a) and one molecule of apoB: evaluation of amino acid analysis data. J. Lipid Res. 37: 192–196. [PubMed] [Google Scholar]
- 14.Marcovina S. M., Zhang Z. U., Gaur V. P., and Albers J. J.. 1993. Identification of 34 apolipoprotein(a) isoforms: differential expression of apolipoprotein(a) alleles between American blacks and whites. Biochem. Biophys. Res. Commun. 191: 1192–1196. [DOI] [PubMed] [Google Scholar]
- 15.Marcovina S. M., Hobbs H. H., and Albers J. J.. 1996. Relationship between number of apolipoprotein(a) kringle 4 repeats and mobility of isoforms in agarose gel: basis for a standardized nomenclature. Clin. Chem. 42: 436–439. [PubMed] [Google Scholar]
- 16.Lassman M. E., McLaughlin T. M., Zhou H., Pan Y., Marcovina S. M., Laterza O., and Roddy T. P.. 2014. Simultaneous quantitation and size characterization of apolipoprotein(a) by ultra-performance liquid chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 28: 1101–1106. [DOI] [PubMed] [Google Scholar]
- 17.Lamon-Fava S., Marcovina S. M., Albers J. J., Kennedy H., Deluca C., White C. C., Cupples A., McNamara J. R., Seman L. J., Bongard V., et al. 2011. Lipoprotein(a) levels, apo(a) isoform size, and coronary heart disease risk in the Framingham Offspring Study. J. Lipid Res. 52: 1181–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rifai N., Ma J., Sacks F. M., Ridker P. M., Hernandez W. J., Stampfer M. J., and Marcovina S. M.. 2004. Apolipoprotein(a) size and lipoprotein(a) concentration and future risk of angina pectoris with evidence of severe coronary atherosclerosis in men: the Physicians’ Health Study. Clin. Chem. 50: 1364–1371. [DOI] [PubMed] [Google Scholar]
- 19.Ridker P. M., Hennekens C. H., and Stampfer M. J.. 1993. A prospective study of lipoprotein(a) and the risk of myocardial infarction. J. Am. Med. Assoc. 270: 2195–2199. [PubMed] [Google Scholar]
- 20.Ridker P. M., Stampfer M. J., and Hennekens C. H.. 1995. Plasma concentration of lipoprotein(a) and the risk of future stroke. J. Am. Med. Assoc. 273: 1269–1273. [PubMed] [Google Scholar]
- 21.Ridker P. M., Stampfer M. J., and Rifai N.. 2001. Novel risk factors for systemic atherosclerosis: a comparison of C-reactive protein, fibrinogen, homocysteine, lipoprotein(a), and standard cholesterol screening as predictors of peripheral arterial disease. J. Am. Med. Assoc. 285: 2481–2485. [DOI] [PubMed] [Google Scholar]
- 22.Tate J. R., Berg K., Couderc R., Dati F., Kostner G. M., Marcovina S. M., Rifai N., Sakurabayashi I., and Steinmetz A.. 1999. International Federation of Clinical Chemistry and Laboratory Medicine (IFCC) Standardization Project for the measurement of lipoprotein(a). Phase 2: selection and properties of a proposed secondary reference material for lipoprotein(a). Clin. Chem. Lab. Med. 37: 949–958. [DOI] [PubMed] [Google Scholar]
- 23.Marcovina S. M., Albers J. J., Scanu A. M., Kennedy H., Giaculli F., Berg K., Couderc R., Dati F., Rifai N., Sakurabayashi I., et al. 2000. Use of a reference material proposed by the International Federation of Clinical Chemistry and Laboratory Medicine to evaluate analytical methods for the determination of plasma lipoprotein(a). Clin. Chem. 46: 1956–1967. [PubMed] [Google Scholar]
- 24.Dati F., Tate J. R., Marcovina S. M., and Steinmetz A.. 2004. First WHO/IFCC international reference reagent for lipoprotein(a) for immunoassay – Lp(a) SRM 2B. Clin. Chem. Lab. Med. 42: 670–676. [DOI] [PubMed] [Google Scholar]
- 25.Marcovina S. M., Koschinsky M. L., Albers J. J., and Skarlatos S.. 2003. Report of the National Heart, Lung, and Blood Institute Workshop on lipoprotein(a) and cardiovascular disease: recent advances and future directions. Clin. Chem. 49: 1785–1796. [DOI] [PubMed] [Google Scholar]
- 26.Marcovina S. M., Albers J. J., Wijsman E., Zhang Z., Chapman N. H., and Kennedy H.. 1996. Differences in Lp(a) concentrations and apo(a) polymorphs between black and white Americans. J. Lipid Res. 37: 2569–2585. [PubMed] [Google Scholar]
- 27.Brown W. V., Ballantyne C. M., Jones P. H., and Marcovina S.. 2010. Management of Lp(a). J. Clin. Lipidol. 4: 240–247. [DOI] [PubMed] [Google Scholar]
- 28.Kostner K. M., März W., and Kostner G. M.. 2013. When should we measure lipoprotein(a)? Eur. Heart J. 34: 3268–3276. [DOI] [PubMed] [Google Scholar]
- 29.McConnell J. P., Guadagno P. A., Dayspring T. D., Hoefner D. M., Thiselton D. L., Warnick G. R., and Harris W. S.. 2014. Lipoprotein(a) mass: a massively misunderstood metric. J. Clin. Lipidol. 8: 550–553. [DOI] [PubMed] [Google Scholar]
- 30.Marcovina S. M., and Koschinsky M. L.. 1998. Lipoprotein(a) as a risk factor for coronary artery disease. Am. J. Cardiol. 82: 57U–66U. [DOI] [PubMed] [Google Scholar]
- 31.Kamstrup P. R., Benn M., Tybjaerg-Hansen A., and Nordestgaard B. G.. 2008. Extreme lipoprotein(a) levels and risk of myocardial infarction in the general population: the Copenhagen City Heart Study. Circulation. 117: 176–184. [DOI] [PubMed] [Google Scholar]
- 32.Kamstrup P. R., Tybjaeeg-Hansen A., and Nordestgaard B. G.. 2014. Elevated Lipoprotein (a) and risk of aortic valve stenosis in the general population. J. Am. Coll. Cardiol. 63: 470–477. [DOI] [PubMed] [Google Scholar]
- 33.O’Donoghue M. L., Morrow D. A., Tsimikas S., Sloan S., Ren A. F., Hoffman E. B., Desai N. R., Solomon S. D., Domanski M., Arai K., et al. 2014. Lipoprotein(a) for risk assessment in patients with established coronary heart disease. J. Am. Coll. Cardiol. 63: 520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig W. Y., Neveux L. M., Palomaki G. E., Cleveland M. M., and Haddow J. E.. 1998. Lipoprotein(a) as a risk factor for ischemic heart disease: meta-analysis of prospective studies. Clin. Chem. 44: 2301–2306. [PubMed] [Google Scholar]
- 35.Danesh J., Collins R., and Peto R.. 2000. Lipoprotein (a) and coronary heart disease. Meta-analysis of prospective studies. Circulation. 102: 1082–1085. [DOI] [PubMed] [Google Scholar]
- 36.Erqou S., Kaptoge S., Perry P. L., Di Angelantonio E., Thompson A., White I. R., Marcovina S. M., Collins R., Thompson S. G., and Danesh J.; Emerging Risk Factors Collaboration. 2009. Lipoprotein(a) concentrations and the risk of coronary heart disease, stroke, and nonvascular mortality. J. Am. Med. Assoc. 302: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamstrup P. R., Tybjaerg-Hansen A., Steffensen R., and Nordestgaard B. G.. 2009. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. J. Am. Med. Assoc. 301: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 38.Kronenberg F., Trenkwalder E., Dieplinger H., and Utermann G.. 1996. Lipoprotein(a) in stored plasma samples and the ravages of time. Why epidemiological studies might fail. Arterioscler. Thromb. Vasc. Biol. 16: 1568–1572. [DOI] [PubMed] [Google Scholar]
- 39.Maher V. M., Brown B. G., Marcovina S. M., Hillger L. A., Zhao X. Q., and Albers J. J.. 1995. Effects of lowering elevated LDL cholesterol on cardiovascular risk of lipoprotein(a). J. Am. Med. Assoc. 274: 1771–1774. [PubMed] [Google Scholar]
- 40.Berg K., Dahlen G., Christophersen B., Cook T., Kjekshus J., and Petersen T.. 1997. Lp(a) lipoprotein level predicts survival and major coronary events in the Scandinavian Simvastatin Study. Clin. Genet. 52: 254–261. [DOI] [PubMed] [Google Scholar]
- 41.von Eckardstein A., Schulte H., Cullen P., and Assmann G.. 2001. Lipoprotein(a) further increases the risk of coronary events in men with high global cardiovascular risk. J. Am. Coll. Cardiol. 37: 434–439. [DOI] [PubMed] [Google Scholar]
- 42.Kostner G. M., Avogaro P., Cazzolato G., Marth E., Bittolo-Bon G., and Qunici G. B.. 1981. Lipoprotein Lp(a) and the risk for myocardial infarction. Atherosclerosis. 38: 51–61. [DOI] [PubMed] [Google Scholar]
- 43.Nordestgaard B. G., Chapman M. J., Ray K., Borén J., Andreotti F., Watts G. F., Ginsberg H., Amarenco P., Catapano A., Descamps O. S., et al. ; European Atherosclerosis Society Consensus Panel. 2010. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur. Heart J. 31: 2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandholzer C., Hallman D. M., Saha N., Sigurdsson G., Lackner C., Császár A., Boerwinkle E., and Utermann G.. 1991. Effects of the apolipoprotein(a) size polymorphism on the lipoprotein(a) concentration in 7 ethnic groups. Hum. Genet. 86: 607–614. [DOI] [PubMed] [Google Scholar]
- 45.Sharrett A. R., Ballantyne C. M., Coady S. A., Heiss G., Sorlie P. D., Catellier D., and Patsch W.; Atherosclerosis Risk in Communities Study Group. 2001. Coronary heart disease prediction from lipoprotein cholesterol levels, triglycerides, lipoprotein(a), apolipoproteins A-I and B, and HDL density subfractions: the Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 104: 1108–1113. [DOI] [PubMed] [Google Scholar]
- 46.Virani S. S., Brautbar A., Davis B. C., Nambi V., Hoogeveen R. C., Sharrett A. R., Coresh J., Mosley T. H., Morrisett J. D., Catellier D. J., et al. 2012. Associations between lipoprotein(a) levels and cardiovascular outcomes in black and white subjects: the Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 125: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan W., Cao J., Steffen B. T., Post W. S., Stein J. H., Tattersall M. C., Kaufman J. D., McConnell J. P., Hoefner D. M., Warnick R., et al. 2015. Race is a key variable in assigning lipoprotein(a) cutoff values for coronary heart disease risk assessment: the Multi-Ethnic Study of Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 35: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Austin M. A., Rodriguez B. L., McKnight B., McNeely M. J., Edwards K. L., Curb J. D., and Sharp D. S.. 2000. Low-density lipoprotein particle size, triglycerides, and high-density lipoprotein cholesterol as risk factors for coronary heart disease in older Japanese-American men. Am. J. Cardiol. 86: 412–416. [DOI] [PubMed] [Google Scholar]
- 49.Tsimikas S., Viney N., Hughes S., Singleton W., Graham M., Baker B., Burkey J., Yang Q., Marcovina S. M., Geary R., et al. 2015. Antisense therapy targeting apolipoprotein(a): a randomized, double-blind, placebo-controlled phase 1 study. Lancet. 386: 1472–1483. [DOI] [PubMed] [Google Scholar]