Abstract
We have examined the posttranslational modification of the human chromatin protein DEK and found that DEK is phosphorylated by the protein kinase CK2 in vitro and in vivo. Phosphorylation sites were mapped by quadrupole ion trap mass spectrometry and found to be clustered in the C-terminal region of the DEK protein. Phosphorylation fluctuates during the cell cycle with a moderate peak during G1 phase. Filter binding assays, as well as Southwestern analysis, demonstrate that phosphorylation weakens the binding of DEK to DNA. In vivo, however, phosphorylated DEK remains on chromatin. We present evidence that phosphorylated DEK is tethered to chromatin throughout the cell cycle by the un- or underphosphorylated form of DEK.
The DEK protein was initially identified as a fusion protein with CAN nucleoporin in a subtype of acute myeloid leukemia involving the t(6;9) chromosomal translocation (34). Subsequently, DEK was found to be the target of autoantibodies in several diseases, including systemic lupus erythematosus (9, 10, 38), juvenile rheumatoid arthritis (10, 32), and sarcoidosis (9, 10). Interestingly, DEK has also been linked to ataxia-telangiectasia, because a fragment of DEK cDNA reverses the mutagen-sensitive phenotype of cells from ATM patients (28). Despite the number of clinical observations, the biological function of DEK remains unclear (33).
Several reports suggest an involvement of DEK in transcriptional regulation. By using cofractionation and coimmunoprecipitation, it was demonstrated that the transcriptional corepressor hDaxx associates with DEK (21). However, the exact function of DEK in hDaxx-mediated repression is not clear. DEK has also been found to be associated with the latency-associated nuclear antigen, which is constitutively expressed in Kaposi's sarcoma-associated herpesvirus latent infection (25). The data indicate that the latency-associated nuclear antigen is tethered to chromatin through its interaction with the DEK protein and the methyl CpG binding protein MeCP2. In addition, DEK interacts with cell type-specific transcription factor AP-2α in vitro and stimulates transactivation activity of AP-2α over the APOE promoter (8). Thus, DEK could be involved in linking several different proteins to chromatin. It has been reported that DEK binds to DNA and specifically recognizes the peri-ets sites in the human immunodeficiency virus type 2 enhancer (11, 14, 15) and to class II major histocompatibility complex Y-box sequences (1). However, our experiments had shown that DEK recognizes DNA structures rather than DNA sequences and preferentially binds supercoiled and four-way junction DNAs (35). These data suggest that DEK functions as an architectural protein in chromatin. Indeed, DEK is a constituent of oligonucleosomes, generated by micrococcal nuclease digestion of chromatin in isolated nuclei (22) and associates with metaphase chromosomes (13). Purified DEK changes the topology of DNA in viral minichromosomes and reduces the accessibility of chromatin to DNA binding factors including components of the replication machinery (4). The DEK-induced change in topology is due to the introduction of positive supercoils into the DNA (36).
DEK does not belong to a known family of proteins. Apart from four stretches of acidic amino acids, the only recognizable feature is a homology to a DNA binding motif, the SAF-box (scaffold attachment factor) (17, 24), also termed the SAP (SAF-A/B acinus and pias) domain (6), between amino acids 149 and 187 of the DEK protein. Analysis of a series of deletion mutant forms of DEK revealed that the SAF-box is indeed responsible for DEK-DNA interaction. In addition to the SAF-box, a second DNA binding domain between amino acids 270 and 350 appears to be involved in DEK-DNA interaction. This domain is also involved in DEK-DEK multimerization (23).
DEK is known to be a phosphoprotein, and it is possible that DEK activity is regulated by phosphorylation (13). However, it is not known whether phosphorylation of DEK changes within the cell cycle, which kinases are responsible, and whether phosphorylation influences the activities of DEK.
We report here that DEK is phosphorylated by CK2 (formerly termed casein kinase 2) in vitro and in vivo. Phosphorylation peaks in the G1 phase of the cell cycle and influences DEK-DNA interactions. In addition, our data suggest that a mediator protein, most likely DEK in its dephosphorylated form, is involved in the binding of phosphorylated DEK to chromatin in vivo.
MATERIALS AND METHODS
Cell culture, cell cycle synchronization, in vivo labeling with 32Pi, cell fractionation, immunoprecipitation, and fluorescence-activated cell sorter (FACS) analysis.
HeLa-S3 cells were cultivated and synchronized by a double-thymidine block as described previously (22). Labeling of cell cultures with 32Pi was performed by washing cells two times with SSC buffer (0.15 M NaCl, 15 mM sodium citrate) and once with phosphate-free medium (Sigma) and then cultivating them in 5 ml of phosphate-free medium supplemented with 200 μCi of 32Pi (carrier free; ICN) per 94-mm-diameter dish.
Cell fractionation was carried out as described previously, with minor modifications (22). Nuclei were subjected to a one-step extraction with 450 mM NaCl after disruption of the nuclear envelope with 0.5% NP-40. Immunoprecipitations from the 450 mM NaCl extracts were performed with a total of 3 μg of monospecific antibodies per sample. After 1 h of incubation on ice, 30 μl of a 50% protein A-Sepharose solution (Pharmacia) was added and the mixture was rolled at 4°C for another hour. The immunocomplexes were washed three times with 1 ml of nuclear extraction buffer (450 mM NaCl, 20 mM HEPES [pH 7.4], 0.5 mM MgCl2). Immunocomplexes were removed from protein A beads with 2% sodium dodecyl sulfate (SDS) containing 5% β-mercaptoethanol at 37°C for 1 h following three wash steps with the same buffer. The supernatants were pooled, and the proteins were concentrated as previously described (37).
For FACS analysis, HeLa cells were washed three times with phosphate-buffered saline (NaCl, Pi) supplemented with 5 mM EDTA. For DNA staining, 4 × 105 cells were permeabilized with 0.5% Triton X-100 and stained with a total of 18 μg of propidium iodide (Sigma). Fluorescence was measured with a FACScalibur flow cytometer (BD Bioscience), and results were visualized and evaluated with the Cell Quest software (BD Bioscience).
Recombinant protein expression, dephosphorylation, preparation of antibodies, and immunoblotting.
The open reading frame of human DEK was cloned into the multiple cloning site of pBlueBacHis2A (Invitrogen). SF-9 cells (Invitrogen) were cotransfected with linearized Autographa californica nuclear polyhedrosis virus DNA with the Bac-N-Blue DNA transfection kit (Invitrogen) in accordance with the manufacturer's protocol. Five days after cotransfection, the supernatant was harvested and subjected to plaque assays. Single plaques were picked, and a high-titer virus stock was raised. Three days postinfection with a high-titer virus stock, HighFive cells were harvested and washed three times with phosphate-buffered saline prior to lysis with 2 ml of lysis buffer per 175-cm2 flask (100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1% NP-40, 5 mM imidazole). To disrupt DNA-protein and protein-protein interactions, the lysed cells were further incubated in the presence of 1.3 M NaCl for 20 min at room temperature. The lysate was cleared (100,000 × g, 10 min), adjusted to 10% glycerol, diluted with lysis buffer to a final concentration of 700 mM NaCl, and incubated with 10 μl of equilibrated 50% Ni-nitrilotriacetic acid (NTA)-agarose (QIAGEN) per 2 ml of lysate. After binding for 1 h at 4°C, the beads were washed three times with 10 volumes of buffer 1 (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 50 mM imidazole), three times with 10 volumes of buffer 2 (50 mM Tris-Cl [pH 7.5], 300 mM NaCl, and 50 mM imidazole), and again with 10 volumes of buffer 1. Elution was performed with 50 mM Tris-Cl [pH 7.5]-150 mM NaCl-500 mM imidazole. Aliquots of recombinant protein were stored at −70°C in elution buffer. Prior to use, His-DEK was dephosphorylated with 400 U of λ-phosphatase per 10 pmol of DEK in accordance with the manufacturer's protocol (New England Biolabs) for 90 min at 30°C. Recombinant DEK protein was used to raise antibodies in rabbits. Monospecific antibodies were purified from serum by affinity chromatography with Sulfo-Link (Pierce) coupled with recombinant DEK protein. Immunoblotting was carried out as previously described (22).
In vitro phosphorylation.
S-20 extracts were prepared from HeLa cells at the indicated time points after release from the double-thymidine block. Cell cycle stages were confirmed in parallel by FACS analysis. Cells were resuspended in hypotonic buffer (10 mM HEPES [pH 7.5], 5 mM KCl, 1.5 mM MgCl2, 0.1 mM dithiothreitol [DTT]) supplemented with 250 mM sucrose. The cell pellet was kept on ice for 30 min and Dounce homogenized to prepare the soluble cytoplasmic and nucleoplasmic proteins. S-20 extract (150 μg of total protein) was incubated with dephosphorylated recombinant human His-tagged DEK or human His-tagged geminin in the presence of 1 mM Na-vanadate, 20 mM NaF, a protease inhibitor mixture (EDTA-free Complete; Roche), and 30 μCi of [γ-32P]ATP (specific radioactivity, >4,000 Ci/mmol; ICN) at 30°C. Treatment with 4,5,6,7-tetrabromobenzotriazole (TBB) (29) was performed by incubation of the S-20 extracts for 10 min on ice prior to the phosphorylation reaction. Reactions were stopped on ice by addition of 500 mM NaCl, 0.5% NP-40, and 40 μM ATP. DEK was purified with Ni-NTA beads (QIAGEN), and geminin, used as a control protein, was immunoprecipitated as previously described (26). The purified proteins were investigated by immunoblotting and autoradiography.
In-gel kinase assay.
Five, 10, and 20 μg of total protein from an S-20 extract, obtained by asynchronously growing HeLa cells, were subjected to electrophoresis in an SDS-10% polyacrylamide gel electrophoresis (PAGE) gel that had been polymerized in the presence of 40 μg of baculovirus-expressed, His-tagged DEK (after dephosphorylation) per ml of gel matrix. Following electrophoresis, the gel was washed twice for 30 min with 100 ml of 50 mM Tris-HCl (pH 8)-20% isopropanol, twice for 30 min with 100 ml of buffer B (50 mM Tris-HCl [pH 7.5], 5 mM β-mercaptoethanol), and once for 1 h with 200 ml of buffer B containing 6 M guanidinium-HCl. Proteins were then renatured in the gel overnight at 4°C in 200 ml of buffer B containing 0.05% Tween 20, with five buffer changes. The gel was then incubated in kinase buffer (25 mM HEPES [pH 7.8], 10 mM MgCl2, 2 mM CaCl2, 25 μM ATP) containing 10 μCi of [γ-32P]ATP per ml at 25°C for 1 h. Incubation was followed by five washes, each with 200 ml of 5% trichloroacetic acid-1% sodium pyrophosphate at room temperature. Gels were stained with Coomassie brilliant blue, dried, and subjected to autoradiography.
Filter binding assay with purified kinases and radiolabeled DNA.
Recombinant His-tagged DEK (5 pmol, dephosphorylated) was incubated in kinase buffer (20 mM HEPES [pH 7.8], 10 mM MgCl2, 10 mM CaCl2, 20 mM NaF, 1 mM vanadate) in the presence of 1 μCi of [γ-32P]ATP and purified CK2 (native holoenzyme purified to homogeneity from rat liver). Reactions were stopped at indicated time points by adding 5 ml of 5% sodium pyrophosphate. To assay protein kinase C (PKC; Promega), we used a buffer containing 20 mM HEPES (pH 7.4), 2 mM CaCl2, 1 mM DTT, and 10 mM MgCl2. The buffer for the glycogen synthetase kinase 3 (GSK-3) assay contained 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 5 mM DTT. All reactions were performed with 10 U of each of the kinases. The samples were then transferred to nitrocellulose filters (Schleicher & Schuell), washed three times with 5% sodium pyrophosphate, dried, and counted in a liquid scintillation counter.
SV40 DNA labeling for filter binding was carried out as described in the section on Southwestern analysis. Reactions were set up in a total volume of 30 μl, either in nE 50 (20 mM HEPES [pH 7.6], 50 mM NaCl, 10 mM sodium bisulfite, 1 mM EDTA) or in nE 100 (100 mM NaCl). Ten nanograms of labeled DNA (105 cpm) was incubated with increasing amounts of either dephosphorylated or CK2-rephosphorylated His-DEK for 1 h at 37°C. The samples were transferred to nitrocellulose filters and washed three times with nE50 or nE100 (1 ml of each). Filters were dried and counted as described above.
Southwestern analysis.
For Southwestern analysis, identical amounts of dephosphorylated and CK2-rephosphorylated DEK were blotted onto a nitrocellulose membrane and blocked with 5% nonfat milk powder in 50 mM Tris-HCl (pH 7.5)-50 mM NaCl-1 mM DTT-1 mM EDTA for 1 h at room temperature. As a probe, HinfI-digested and end-labeled (Klenow polymerase; New England Biolabs) SV40 DNA was used (106 cpm/ml). After 1 h of incubation, the membrane was washed five times with 10 mM Tris-HCl, pH 7.5-50 mM NaCl-1 mM DTT-1 mM EDTA and subjected to autoradiography or immunoblotting.
Sucrose gradients with salt-treated minichromosomes.
Dephosphorylated His-DEK was incubated with salt-treated SV40 minichromosomes at a molar ratio of 60 (DEK/DNA) for 1 h in nE 50 in the presence of 10 U of topoisomerase I (wheat germ; Promega), followed by addition of 2 mM Na-vanadate, 30 mM NaF, 50 μM ATP, and 60 μCi of [γ-32P]ATP. To start the in situ phosphorylation, 300 U of purified CK2 was added to the DEK-supplemented SV40 minichromosomes and the mixture was incubated for 1 h at 37°C. Samples were loaded on 5 to 30% sucrose gradients (in nE 70 supplemented with 2 mM Na-vanadate and 30 mM NaF) and centrifuged for 3 h at 36,000 rpm in an SW-40 rotor (Beckman). Gradients were fractionated into 20 fractions and adjusted to 2% SDS following incubation for 30 min at 55°C. One-half of each fraction was used for DNA extraction (proteinase K digestion and ethanol precipitation) and visualization (agarose gel electrophoresis and ethidium bromide staining). The other half of each fraction was used for protein precipitation (37) and SDS-PAGE, followed by blotting to nitrocellulose membrane, autoradiography, and immunoblotting with DEK-specific antibodies.
Micrococcal nuclease digestion.
Treatment of isolated nuclei with micrococcal nuclease was carried out as previously described (22). Briefly, nuclei were prepared from asynchronously growing HeLa S3 cells by cell fractionation. Nuclei were washed three times with extraction buffer 50 (20 mM HEPES [pH 7.8], 0.5 mM MgCl2, 50 mM NaCl) after disruption of the nuclear envelope with 0.5% NP-40. The resulting pellet was adjusted to 500 μg of DNA per ml and 2 mM CaCl2, before 10 U of micrococcal nuclease (Sigma Aldrich) per 50 μg of DNA was added, and incubated for 10 min at 14°C. The reaction was stopped by addition of 8 mM EDTA and incubation on ice for 10 min. Mono- to oligonucleosomal fragments were separated from insoluble material by centrifugation and adjusted to 2 mM Na-vanadate-30 mM NaF and supplemented with a protease inhibitor cocktail (EDTA-free Complete; Roche). Either 1.5 μg of unphosphorylated or CK2-phosphorylated His-DEK was added to the nucleosomal fragments, and the mixture was incubated for 1 h at 37°C. The samples were loaded onto 5 to 40% sucrose gradients in elution buffer 100 (20 mM HEPES [pH 7.5], 0.5 mM MgCl2, 100 mM NaCl, 1 mM Na-vanadate, 30 mM NaF) and centrifuged for 14 h at 36,000 rpm in an SW-40 rotor (Beckman). Gradients were fractionated and treated as described above.
In-gel digestion.
For in-gel-digestion and subsequent mass spectrometry (MS) analysis of dephosphorylated and CK2-phosphorylated His-DEK, the bands were cut out and destained in a 60% acetonitrile-water mixture for 20 min at 25°C. After lyophilization of the gel piece, a solution of 50 mM NH4HCO3 was added for rehydration and the mixture was incubated for 20 min at 25°C. This procedure was repeated two times, and the final rehydration was performed with the protease solution (12.5 ng of trypsin [modified trypsin; Promega] per μl in 50 mM NH4HCO3) at 4°C for 45 min. The gel spots were incubated afterwards for 12 h at 37°C, followed by elution of protein fragments for 3 to 4 h in a 60% acetonitrile-water mixture. The eluates were lyophilized to dryness and desalted prior to analysis with ZipTipC18 (as recommended by Millipore) and eluted with a GELoader tip (Eppendorf) into a nanospray needle with 5 μl of 50% methanol-0.1% CH3COOH.
Triple-quadrupole ion trap MS.
The Q TRAP mass spectrometer (Applied Biosystems MDS SCIEX Q TRAP LC/MS/MS system) was used to determine the phosphoacceptor sites. We took advantage of the fact that phosphopeptides tend to lose the phosphor moiety upon low-energy collisional activation in a tandem MS experiment. The advanced-scan modes of neutral loss and precursor ion scans were used for scanning peptides that lose PO3 with m/z 79 Da in the negative-ion mode or 98 Da in the positive-ion mode, corresponding to either H3PO4 or HPO32− and H2O (b elimination, neutral loss). So we used the quadrupole ion trap method to isolate the phosphopeptides, perform MS/MS, and characteristically to show loss of the phosphate group, which serves as a signature for a phosphopeptide or a phosphate-containing peptide.
Database searching and enumeration of phosphopeptides.
All MS/MS data were directly used for a database search procedure with Analyst software (Applied Biosystems) in combination with the MASCOT MS/MS Ion Search Engine (www.matrixscience.com). The enzyme used for cleavage was modified trypsin (Promega), and the modification introduced into the search was phosphorylation and oxidation of methionine. For all of the sequences reported, spectra were manually validated and contained sufficient information to assign not only the sequence but also the site of phosphorylation.
RESULTS
DEK is differentially phosphorylated during the cell cycle.
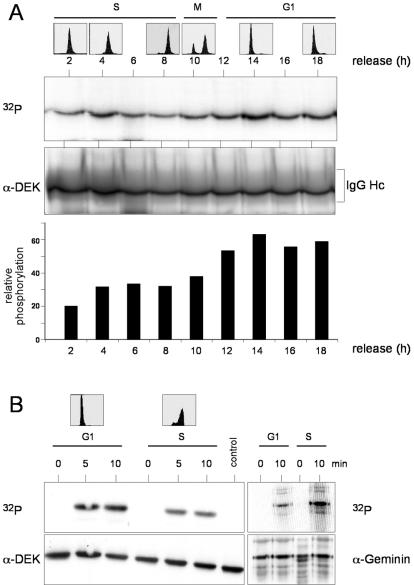
Transiently transfected DEK is known to be phosphorylated in COS cells (13). In order to determine when endogenous DEK is phosphorylated during the cell cycle, we used synchronized HeLa cells and performed in vivo labeling experiments with 32Pi for 2-h periods after release from the double-thymidine block. Cell cycle stages were confirmed by FACS analysis (Fig. 1A, top). Protein extracts were prepared from the chromatin-bound fraction and immunoprecipitated with DEK-specific antibodies. We found significant incorporation of 32Pi into DEK. Comparison of the autoradiograph (Fig. 1A, 32P) and the immunoblot (Fig. 1A, α-DEK) revealed a moderate increase in phosphorylation during the G1 phase (Fig. 1A, bottom).
FIG. 1.
Phosphorylation of DEK in vivo and in vitro. (A) Phosphorylation in vivo. HeLa cells were synchronized by a double-thymidine block and radiolabeled with 32Pi for 2 h at the indicated times after release. Cells were fractionated, and the 450 mM NaCl eluate was treated with DEK antibodies. After electrophoresis, the immunocomplexes were analyzed by autoradiography (32P) and Western blotting (α-DEK). For FACS analysis, cells were treated in parallel and harvested at the indicated time points (top). For densitometric analysis, bands were scanned and analyzed by NIH Imager and the ratio of the autoradiograph to the Western blot was plotted against the time of release (bottom). IgG (immunoglobulin G) Hc indicates the position of the antibody heavy chain. (B) Phosphorylation in vitro. Recombinant, dephosphorylated, His-tagged DEK protein (600 ng) was incubated with G1 phase (15.5 h after release) and S phase (6 h after release) extracts in the presence of [γ-32P]ATP (control lane, without extract). FACS analysis was taken from cells treated in the same way (top). Reactions were stopped at the indicated times, and DEK was purified from the samples with Ni-NTA-agarose. The eluates were precipitated and analyzed by autoradiography (32P) and Western blotting (α-DEK). As a control, human geminin was incubated in the same extracts, immunoprecipitated, and processed as described above.
To confirm the differential phosphorylation of DEK during the cell cycle, we used an independent approach and measured the activity of phosphorylating enzymes in protein extracts prepared at different cell cycle stages (Fig. 1B, top). Protein extracts prepared at 6 h (S phase extract) and at 15.5 h (G1 phase extract) after release from a double-thymidine block were incubated with recombinant His-tagged DEK as the substrate. Since recombinant His-tagged DEK from insect cells is highly phosphorylated, it had to be treated with λ-phosphatase before use. After incubation for 5 to 10 min under phosphorylation conditions, DEK was purified by Ni-NTA-agarose and processed for immunoblotting and autoradiography. In accordance with the in vivo data, G1 phase extracts are around twofold more active in phosphorylating DEK protein than are S phase extracts (Fig. 1B, 32P). Recombinant human geminin served as a control in these assays and was shown to be highly phosphorylated in S phase extracts and much less in G1 phase extracts (26).
DEK is phosphorylated by CK2 in vivo and in vitro.
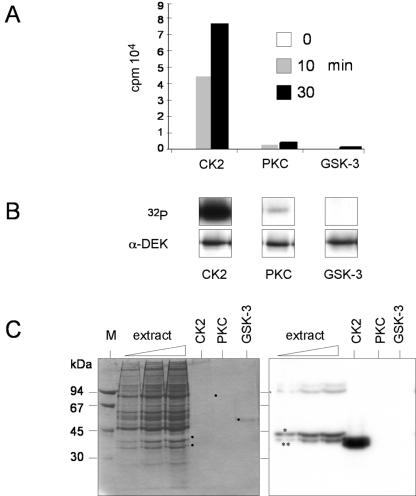
To identify the kinase(s) involved in DEK phosphorylation, we analyzed the DEK sequence for potential phosphorylation sites with the Scansite program (http://scansite.mit.edu). Phosphorylation sites were detected for CK2, for GSK-3, and for PKC (see Fig. 4, predicted sites). We therefore compared the activities for DEK as a substrate for CK2, PKC, and GSK-3. We found that CK2 very efficiently phosphorylated His-DEK in vitro, whereas PKC and GSK-3 were far less active in standard filter binding assays (Fig. 2A) and in autoradiography after gel electrophoresis (Fig. 2B). In addition, we performed in-gel kinase assays to identify the kinase(s) involved in DEK phosphorylation in unfractionated extracts (Fig. 1B) (16). To this end, dephosphorylated DEK was polymerized within the gel matrix and S-20 extracts from unsynchronized cells were separated on the gel. The proteins were renatured in the gel and incubated with [γ-32P]ATP. Labeled bands were identified in comparison with the positions of the purified CK2, PKC, and GSK-3 kinases run in parallel on the gel (Fig. 2 C, dots). We found that CK2 is the main kinase phosphorylating DEK in crude cell extracts (Fig. 2C, right side). The differences in electrophoretic mobility between CK2 in the cell extract and purified CK2 are most likely due to limited proteolysis of the α CK2 subunit in purified CK2. This increases the catalytic activity and leads to a mobility similar to that of the α′ CK2 subunit, as described previously (30). The minor band of phosphorylating activity observed in the presence of increasing amounts of extracts could be due to PKC.
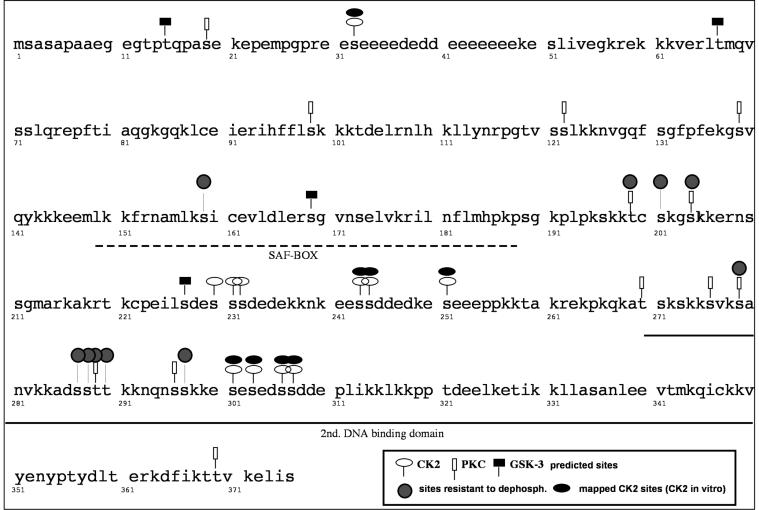
FIG. 4.
Phosphorylation sites mapped by in vitro phosphorylation with purified CK2. The DEK protein sequence is shown. The broken line indicates the position of the SAF-box DNA binding motif, and the solid line indicates the position of the second DNA binding domain.
FIG. 2.
DEK is phosphorylated by CK2. (A) Filter binding assay. After dephosphorylation with λ-phosphatase, recombinant His-DEK (5 pmol) was incubated in the presence of [γ-32P]ATP and purified protein kinases (10 U). Reactions were stopped at the indicated time points, transferred to nitrocellulose filters, washed, and then counted. Incorporated radioactivity for the individual kinases is given in counts per minute (cpm, 104). (B) Products from the filter binding assay were analyzed by SDS-PAGE, autoradiography (32P), and Western blotting (α-DEK). (C) In-gel kinase assay. Increasing amounts of HeLa S-20 extract, together with purified CK2, PKC, and GSK-3, were subjected to electrophoresis in a 10% polyacrylamide gel that was polymerized in the presence of 40 μg of His-DEK per ml. The proteins were renatured, and the gel was incubated with [γ-32P]ATP. Shown are the Coomassie-stained gel (left) and the autoradiograph (right). Dots indicate the positions of purified kinases. The asterisk and double asterisk indicate the positions of the CK2 α and α′ subunits, respectively.
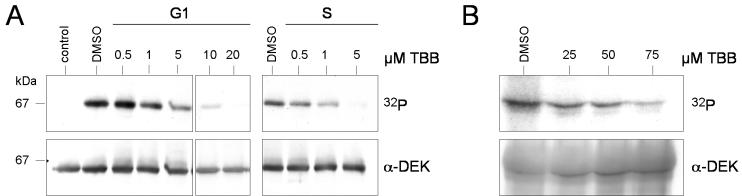
Next we tested whether phosphorylation of DEK could be inhibited in vitro and in vivo by a highly specific inhibitor of CK2, TBB (31). We found that 5 μM TBB was sufficient to inhibit phosphorylation of DEK in S phase extracts, whereas 20 μM TBB was necessary to completely inhibit DEK phosphorylation in G1 phase extracts (Fig. 3A). This could be explained by modified proportions of isoforms in the CK2 holoenzyme (e.g., holoenzyme versus free catalytic subunits or holoenzyme with different α and α′ subunit compositions) present in the extracts, whose susceptibility to TBB could vary (30). However, the possibility cannot be excluded that another kinase that is sensitive to TBB at high concentrations contributes to DEK phosphorylation in G1 phase extracts.
FIG. 3.
Inhibition of DEK phosphorylation by TBB. (A) In vitro phosphorylation. Recombinant His-DEK (600 ng) was dephosphorylated with λ-phosphatase and incubated with S-20 extracts from HeLa cells (150 μg; G1 and S phases), [γ-32P]ATP, and increasing amounts of TBB. Dimethyl sulfoxide (DMSO), at the highest concentration, was used as a control (control lane, without extract). Reactions were stopped after 10 min, and DEK was purified with Ni-NTA-agarose. The eluates were subjected to SDS-PAGE and analyzed by autoradiography (32P) and Western blotting (α-DEK). The molecular masses of markers are indicated in kilodaltons. (B) Inhibition of DEK phosphorylation in vivo. Asynchronously growing HeLa cells were incubated with the indicated amounts of TBB for 1 h in phosphate-free medium. Cells were washed and incubated with TBB and 200 μCi of 32Pi for another 2 h. DEK was immunoprecipitated from the 450 mM NaCl eluate. The autoradiograph (32P) and the immunoblot (α-DEK) are shown.
The influence of TBB on DEK phosphorylation was also tested in vivo. For that purpose, asynchronously growing HeLa cells were labeled with 32Pi and treated for 3 h with increasing amounts of TBB. Cells were fractionated, and DEK was immunoprecipitated from the 450 mM NaCl eluate. Comparison of the immunoblot and the autoradiograph revealed that phosphorylation of DEK was inhibited by TBB up to 80% in vivo (Fig. 3B). In summary, these data demonstrate that CK2 is the main kinase phosphorylating the DEK protein in vivo.
In order to determine the sites phosphorylated by CK2, we used quadrupole ion trap MS, which offers enhanced resolution and mass accuracy combined with superior full-scan sensitivity compared to conventional three-dimensional ion trap MS and standard MS. As a substrate we used recombinant His-DEK purified from infected insect cells, which was first dephosphorylated with λ-phosphatase and then rephosphorylated with purified CK2. Samples were digested in gel with trypsin and then subjected to nano-ESI-MS analysis. Thus, we determined that Ser32, Ser243, Ser244, Ser251, Ser301, Ser303, Ser306, and Ser307 are phosphorylated by CK2 (Fig. 4). Sequence coverage and identified phosphorylated peptides are shown in Fig. 5. Except for Ser32, all of the other phosphorylation sites are located in the C terminus of DEK protein (Fig. 4). Interestingly, a cluster of phosphorylation sites from Ser301 to Ser307 is located in the second DNA binding domain of DEK as recently identified and characterized (23).
FIG. 5.
Coverage and identified peptides. (A) Sequence coverage of DEK protein phosphorylated with CK2 identified after nano-ESI-Q-TRAP measurements. Phosphorylated peptides are in the black box. Unphosphorylated peptides are in boldface and cover 33% of the whole protein. Taking phosphorylation and oxidation of methionine into account, the coverage increased to 99%. (B) Sequence coverage of DEK protein dephosphorylated with λ-phosphatase identified after nano-ESI-Q-TRAP measurements. The dephosphorylated peptides that were formerly phosphorylated are in the broken-line box. Taking into consideration the newly dephosphorylated peptides, the coverage of the protein increased to 45%. The phosphorylated peptides resistant to dephosphorylation are shown in the black box. Taking phosphorylation and oxidation of methionine into account, the coverage increased to 70%. (C) Phosphorylated peptides identified by nano-ESI-Q-TRAP.
We also found various phosphorylated sites within the dephosphorylated His-DEK protein that are resistant to λ-phosphatase treatment. This is not surprising, because it is known that recombinant proteins are highly phosphorylated in baculovirus-infected insect cells. We note, however, that DEK, bearing phosphatase-resistant phosphate groups, behaves exactly like bacterially expressed (unphosphorylated) glutathione S-transferase-DEK in DNA binding and supercoiling assays. On the other hand, untreated His-DEK from insect cells is not able to bind DNA and introduce supercoils (data not shown).
Phosphorylation of DEK changes protein-DNA interactions.
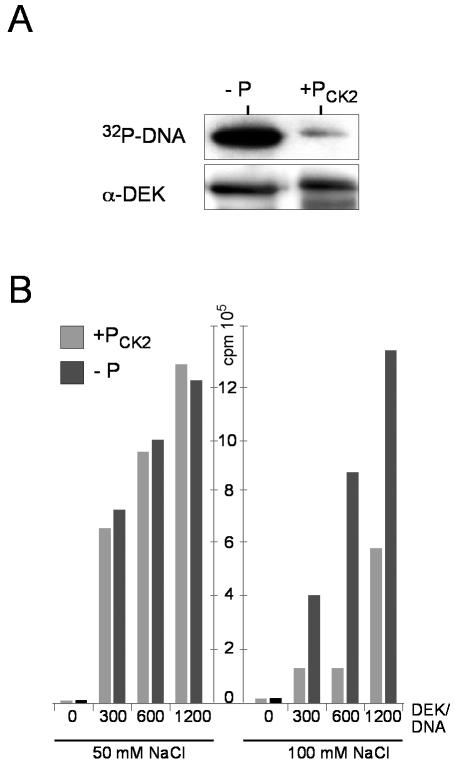
We have investigated whether DEK phosphorylation influences DEK-DNA interactions. We first compared the DNA binding properties of phosphorylated and dephosphorylated DEK by Southwestern analysis. Equal amounts of phosphorylated and dephosphorylated DEK were transferred from polyacrylamide gels onto nitrocellulose membranes and incubated with radioactively labeled, HinfI-digested SV40 DNA. Dephosphorylated DEK bound strongly to DNA, while phosphorylated DEK bound rather weakly (Fig. 6A). For further analysis, we performed filter binding assays at different ionic strengths. At 50 mM salt, similar amounts of DNA bound dephosphorylated and phosphorylated DEK (Fig. 6B). However, at 100 mM salt, the amount of DNA bound to phosphorylated DEK was clearly lower than the amount of DNA bound to dephosphorylated DEK (Fig. 6B). In fact, the residual binding observed with the preparation of phosphorylated DEK could be due to a fraction of DEK molecules that may be underphosphorylated (compare Fig. 7). The conclusion is that phosphorylation decreases the affinity of DEK for DNA.
FIG. 6.
Influence of DEK phosphorylation on DNA binding. (A) Southwestern analysis. DEK was transferred to a nitrocellulose membrane and incubated with radioactively labeled, HinfI-digested SV40 DNA. The autoradiograph (32P-DNA) and the Western blot (α-DEK) are shown (−P, dephosphorylated DEK; +Pck2, DEK phosphorylated by CK2 in vitro). (B) Filter binding assay. 32P-labeled, HinfI-digested SV40 DNA was incubated in the absence (0) or presence of increasing amounts of either dephosphorylated (−P) or phosphorylated (+Pck2) DEK. The assay was performed with either 50 or 100 mM NaCl. DEK/DNA molar ratios are indicated.
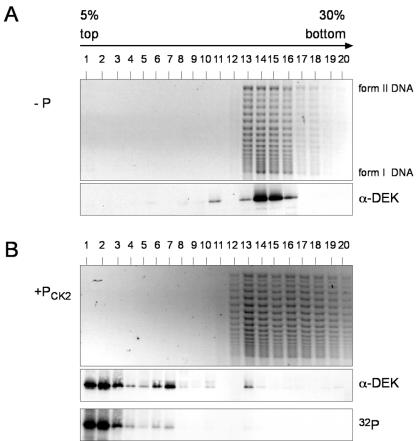
FIG. 7.
Interaction of dephosphorylated and phosphorylated DEK with salt-treated SV40 minichromosomes. (A) Salt-treated SV40 minichromosomes were incubated with dephosphorylated DEK (−P) (at a ratio of 60 mol of DEK/mol of DNA) in the presence of topoisomerase I and separated on 5 to 30% sucrose gradients for 3 h. Individual fractions were analyzed by agarose gel electrophoresis (top) and Western blotting (α-DEK). I, supercoiled DNA; II, relaxed, closed circular, and nicked DNA. (B) Dephosphorylated His-DEK was incubated with salt-treated SV40 minichromosomes as in panel A, followed by in situ phosphorylation with CK2 in the presence of [γ-32P]ATP, and processed as described above (+Pck2). 32P indicates the autoradiograph.
The question arises of whether the interaction of DEK with chromatin is also influenced by phosphorylation. To address this point, we used salt-treated SV40 minichromosomes (18) that were incubated with dephosphorylated DEK in the presence of topoisomerase I. As known from previous work, treatment with DEK in the presence of topoisomerase I causes a change in the superhelical density of the SV40 minichromosomes (4, 36) (Fig. 7A and B, top). One part of the minichromosome preparation, carrying DEK, was phosphorylated with CK2 in the presence of [γ-32P]ATP, and the other part served as a control. Minichromosomes with dephosphorylated and phosphorylated DEK were separated on 5 to 30% sucrose gradients with 100 mM salt. Individual gradient fractions were analyzed by Western blotting with DEK-specific antibodies. In addition, fractions were deproteinized and analyzed by agarose gel electrophoresis (Fig. 7A and B). In the control sample, we found essentially all of the DEK protein in association with the minichromosomes (Fig. 7A, α-DEK). However, phosphorylation by CK2 caused a release of DEK that appeared at the top of the gradient (Fig. 7B). Identical results were obtained by gradient analysis of protein-free DNA incubated with dephosphorylated and phosphorylated DEK; again, only the dephosphorylated form of DEK was found in association with DNA (data not shown).
What keeps phosphorylated DEK on chromatin during the cell cycle?
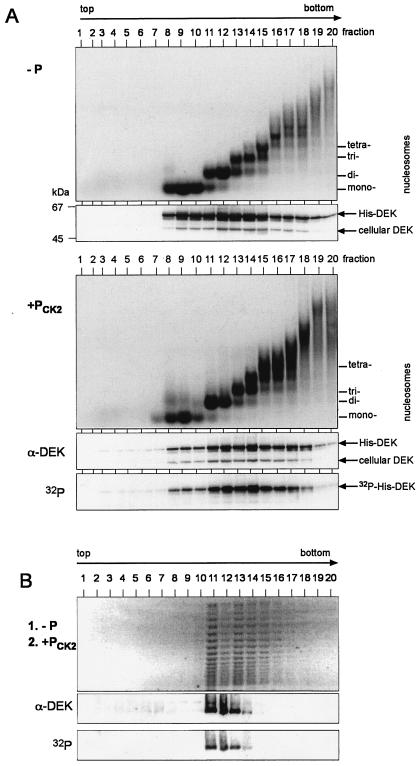
As just shown, DEK after in situ phosphorylation no longer binds to salt-treated SV40 minichromosomes. This observation stands in contrast to those of earlier experiments, which had shown that the amount and localization of DEK do not change during the cell cycle (22). As salt-treated minichromosomes were prepared with 500 mM salt, they mainly contained the core histones while all other chromatin-associated proteins were removed (18). Since untreated native minichromosomes are difficult to investigate because they are densely packed and tend to aggregate, we used oligonucleosomes generated by limited micrococcal nuclease digestion from cellular chromatin to determine whether other chromatin proteins are required to tether phosphorylated DEK to chromatin (22). The oligonucleosomal fragments were incubated with dephosphorylated and phosphorylated His-DEK. The products were separated on sucrose gradients, and individual fractions were analyzed by agarose gel electrophoresis and Western blotting (Fig. 8A). We found that both dephosphorylated (Fig. 8A, −P) and phosphorylated (Fig. 8A, +Pck2) DEK cosedimented with chromatin fragments, indicating that phosphorylated DEK did bind native chromatin. Thus, in contrast to salt-treated minichromosomes, we found a binding of DEK to native oligonucleosomes, suggesting that a chromatin-associated factor may be necessary to mediate the interaction of phosphorylated DEK with chromatin. We have shown before that DEK is able to multimerize upon phosphorylation (23). Therefore, the possibility arises that phosphorylated DEK and dephosphorylated DEK reside together in a complex. To investigate this possibility, we incubated salt-treated SV40 minichromosomes first with dephosphorylated DEK and topoisomerase I (Fig. 7A) and then added in vitro CK2-phosphorylated DEK (Fig. 8B). The minichromosomes were analyzed on a 5 to 30% sucrose gradient with 100 mM salt. In contrast to Fig. 7B, we found in this experiment that all of the phosphorylated DEK protein comigrated with minichromosomes (Fig. 8B; compare with Fig. 7B), suggesting that bound, dephosphorylated DEK can link phosphorylated DEK to chromatin.
FIG. 8.
Association of dephosphorylated and phosphorylated DEK with native oligonucleosomes. (A) HeLa cell nuclei were digested for 10 min at 14°C with micrococcal nuclease. Released oligonucleosomes were incubated with dephosphorylated (−P) and CK2-phosphorylated (+PCK2) DEK and separated on 5 to 40% sucrose gradients for 14 h. Purified DNA from the individual fractions was analyzed by agarose gel electrophoresis and ethidium bromide staining (top). Positions of DNA fragments corresponding to mono-, di-, tri-, and tetranucleosomes are indicated. Proteins were separated by SDS-PAGE and analyzed by Western blotting with DEK-specific antibodies (α-DEK). 32P-labeled DEK was detected by autoradiography (32P). Positions of endogenous DEK, cellular DEK, and exogenously added DEK (His-DEK) are indicated. Marker molecular masses are indicated in kilodaltons. (B) Salt-treated SV40 minichromosomes were incubated with dephosphorylated DEK (at a ratio of 30 mol of DEK/mol of DNA) in the presence of topoisomerase I for 1 h at 37°C (1. −P). In vitro CK2-phosphorylated DEK was added (at a ratio of 30 mol of DEK/mol of DNA), the mixture was incubated for another hour at 37°C (2. +PCK2), and samples were processed as described for Fig. 7. Shown are the agarose gel (top), the Western blot (α-DEK), and the autoradiograph (32P).
DISCUSSION
The purpose of this study was to obtain more information about the ubiquitous chromatin protein DEK. More specifically, we asked whether DEK is phosphorylated and, if so, whether phosphorylation occurs at specific stages of the cell cycle.
To address these questions, we first performed pulse-labeling experiments in which we added radioactive phosphate to HeLa cells that proliferate in synchrony after release from a block in early S phase. We found that phosphorylation of DEK occurs throughout the cell cycle, with a maximum in late G1 phase. Our evidence suggests that DEK is mainly phosphorylated by the protein kinase CK2. This conclusion is supported by three lines of evidence. First, mass spectroscopic analysis of phosphopeptides reveals that most phosphorylated peptides contain amino acid motifs known to form consensus sites for CK2 kinase (Fig. 4). Second, the highly specific CK2 kinase inhibitor TBB (31) inhibits the in vivo phosphorylation of DEK (Fig. 3B). Third, a DEK-phosphorylating activity in protein extracts from cells has the electrophoretic properties of CK2 kinase (as shown by an in-gel phosphorylation assay [16]) (Fig. 3C). In fact, the in vitro phosphorylating activity could be inhibited by TBB, just as it inhibited the in vivo phosphorylation of DEK (Fig. 3A). This does not imply that CK2 kinase is the only activity responsible for DEK phosphorylation. In fact, data suggest that PKC could contribute to DEK phosphorylation, but its contribution to overall DEK phosphorylation is minor compared to that of CK2 kinase. It may be surprising that CK2 kinase has such an important function in DEK phosphorylation given the fact that the enzyme is ubiquitous and constitutively expressed and active (19). Indeed, more than 300 protein targets of CK2 kinase have been described, and these targets include important regulatory proteins like Myc, Jun, Fos, and others (27). Furthermore, CK2 kinase has been shown to regulate several basic cellular processes like DNA replication, transcription, and chromatin remodeling and appears to be involved in the regulation of cell growth, probably by its participation in signal transduction (3, 5, 12, 19, 20, 7). It is therefore not surprising that CK2 kinase could have oncogenic effects in certain cell lines when ectopically expressed (20). Thus, even though much has to be learned yet about the regulation of CK2 kinase activity, it seems to be safe to conclude that its role in DEK phosphorylation must have consequences of physiological importance. We have investigated this possibility and found that CK2 phosphorylation modifies the interaction of DEK with DNA and with chromatin. Indeed, DNA binding experiments in Southwestern assays revealed that the binding of phosphorylated DEK to DNA is less stable than the binding of unmodified DEK (Fig. 6A), and classic filter binding assays showed that complexes between phosphorylated DEK and DNA are disrupted at 100 mM NaCl whereas complexes between unmodified DEK and DNA remain stable (Fig. 6B).
Interestingly, the major DNA binding domain of DEK, the SAF-box, is quite removed from the major CK2 phosphorylation sites, which cluster in the carboxy-terminal region. This region is involved in DEK-DEK interactions (di- or multimerization) but also includes a second DNA binding domain (23). Therefore, the effects of phosphorylation on DNA binding could be due to an effect on DEK-DEK interactions or an impairment of this second DNA binding domain. We will discuss this point below but will first point out an apparent paradox. This is that phosphorylation of DEK changes during the cell cycle, yet the amount of DEK on chromatin remains essentially constant at all phases of the cell cycle (22). This appears to be in contrast to the biochemical evidence showing that phosphorylation decreases the DNA binding activity of DEK. To investigate this point, we studied the association of DEK with chromatin. We demonstrated that phosphorylated DEK does not bind salt-treated SV40 minichromosomes (Fig. 7). As shown before (18), treatment of SV40 minichromosomes with 0.5 M NaCl causes the removal of essentially all nonhistone chromatin proteins, including enzymes and HMG (high-mobility group) proteins, but does not affect the nature and arrangement of the 25 to 28 nucleosomes on SV40 DNA. Unmodified DEK binds to this structure (Fig. 7A), while phosphorylated DEK does not (Fig. 7B), mirroring the behavior of DEK toward DNA and suggesting that DEK contacts DNA sites in the minichromosome. Since untreated native minichromosomes are difficult to investigate because they are densely packed and tend to aggregate, we used oligonucleosomes generated by limited micrococcal nuclease digestion from cellular chromatin for further experiments. As previously shown, the oligonucleosomal fragments carry endogenous DEK protein (22). We show now that unmodified DEK, as well as phosphorylated DEK, does bind native chromatin fragments, suggesting that untreated chromatin carries a constituent that tethers phosphorylated DEK to chromatin. Our evidence suggests that this binding partner could be endogenous, probably unmodified DEK. This conclusion is corroborated by studies in this laboratory showing that DEK is able to multimerize in vitro and in yeast two-hybrid screens and that DEK-DEK interactions are affected by phosphorylation (23).
What does this mean for the function of DEK in chromatin? Work in this and other laboratories has provided evidence that DEK functions as an architectural protein comparable to the better-known HMG proteins (2). Thus, DEK is ubiquitous and abundant, with >106 copies per nucleus (22) in all of the metazoan cells examined. It binds DNA in a sequence-unspecific but structure-specific manner and prefers four-way junction and bent DNAs over classic B-form DNA (35). DEK introduces supercoils into topologically closed DNA forms (4, 36). This and the observation that DEK proteins are believed to interact with transcriptional activator proteins (AP-2 α) (8) and with transcriptional repressors (hDaxx) (21) probably indicate that it influences the expression of genes just as HMG proteins do in enhanceosomes (2). The possible regulatory functions most likely depend on DEK's ability to bind DNA, and phosphorylation may be one way of regulating DEK's role in gene expression by introducing structural changes into DNA and chromatin.
Acknowledgments
We are grateful to Tanja Waldmann, Anita Krebs, and Eric Carstens for stimulating discussions and critical reading of the manuscript.
This work was supported by grants from DFG to C.G.
REFERENCES
- 1.Adams, B. S., H. C. Cha, J. Cleary, T. Haiying, H. Wang, K. Sitwala, and D. M. Markovitz. 2003. DEK binding to class II MHC Y-box sequences is gene- and allele-specific. Arthritis Res. Ther. 5:R226-R233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agresti, A., and M. E. Bianchi. 2003. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13:170-178. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, K., D. A. Gerber, and C. Cochet. 2002. Joining the cell survival squad: an emerging role for protein kinase CK2. Trends Cell Biol. 12:226-230. [DOI] [PubMed] [Google Scholar]
- 4.Alexiadis, V., T. Waldmann, J. Andersen, M. Mann, R. Knippers, and C. Gruss. 2000. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the replication efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 14:1308-1312. [PMC free article] [PubMed] [Google Scholar]
- 5.Allende, J. E., and C. C. Allende. 1995. Protein kinases. 4. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. [DOI] [PubMed] [Google Scholar]
- 6.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 7.Barz, T., K. Ackermann, G. Dubois, R. Eils, and W. Pyerin. 2003. Genome-wide expression screens indicate a global role for protein kinase CK2 in chromatin remodeling. J. Cell Sci. 166:1563-1577. [DOI] [PubMed] [Google Scholar]
- 8.Campillos, M., M. A. García, F. Valdivieso, and J. Vázquez. 2003. Transcriptional activation by AP-2α is modulated by the oncogene DEK. Nucleic Acids Res. 31:1571-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, X., M. A. Michelis, J. Wang, R. Bose, T. DeLange, and W. H. Reeves. 1998. Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum. 41:1505-1510. [DOI] [PubMed] [Google Scholar]
- 10.Dong, X., J. Wang, F. N. Kabir, M. Shaw, A. M. Reed, L. Stein, L. E. Andrade, V. F. Trevisani, M. L. Miller, T. Fujii, M. Akizuki, L. M. Pachman, M. Satoh, and W. H. Reeves. 2000. Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum. 43:85-93. [DOI] [PubMed] [Google Scholar]
- 11.Faulkner, N. E., J. M. Hilfinger, and D. M. Markovitz. 2001. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J. Biol. Chem. 276:25804-25812. [DOI] [PubMed] [Google Scholar]
- 12.Faust, M., and M. Montenarh. 2000. Subcellular localization of protein kinase CK2. A key to its function? Cell Tissue Res. 301:329-340. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod, M., J. Boer, S. van Baal, M. Jaegle, M. von Lindern, K. G. Murti, D. Davis, J. Bonten, A. Buijs, and G. Grosveld. 1995. Relocation of the carboxy-terminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene 10:1739-1748. [PubMed] [Google Scholar]
- 14.Fu, G. K., G. Grosveld, and D. M. Markovitz. 1997. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc. Natl. Acad. Sci. USA 94:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, G. K., and D. M. Markovitz. 1996. Purification of the pets factor, a nuclear protein that binds to the inducible TG-rich element of the human immunodeficiency virus type 2 enhancer. J. Biol. Chem. 271:19599-19605. [DOI] [PubMed] [Google Scholar]
- 16.Geahlen, R. L., M. Anostario, Jr., P. S. Low, and M. L. Harrison. 1986. Detection of protein kinase activity in sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 153:151-158. [DOI] [PubMed] [Google Scholar]
- 17.Göhring, F., B. L. Schwab, P. Nicotera, M. Leist, and F. O. Fackelmayer. 1997. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 16:7361-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruss, C., and R. Knippers. 1995. The SV40 minichromosome, p. 101-113. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 7. Academic Press, Orlando, Fla. [Google Scholar]
- 19.Guerra, B., B. Boldyreff, S. Sarno, L. Cesaro, O. G. Issinger, and L. A. Pinna. 1999. CK2: a protein kinase in need of control. Pharmacol. Ther. 82:303-313. [DOI] [PubMed] [Google Scholar]
- 20.Guerra, B., and O. G. Issinger. 1999. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis 20:391-408. [DOI] [PubMed] [Google Scholar]
- 21.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 115:3319-3330. [DOI] [PubMed] [Google Scholar]
- 22.Kappes, F., K. Burger, M. Baack, F. O. Fackelmayer, and C. Gruss. 2001. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 276:26217-26323. [DOI] [PubMed] [Google Scholar]
- 23.Kappes, F., I. Scholten, N. Richter, C. Gruss, and T. Waldmann. 2004. Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 24:6000-6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kipp, M., B. L. Schwab, M. Przybylski, P. Nicotera, and F. O. Fackelmayer. 2000. Apoptotic cleavage of scaffold attachment factor A (SAF-A) by caspase-3 occurs at a noncanonical cleavage site. J. Biol. Chem. 275:5031-5036. [DOI] [PubMed] [Google Scholar]
- 25.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulartz, M., S. Kreitz, E. Hiller, E. C. Damoc, M. Przybylski, and R. Knippers. 2003. Expression and phosphorylation of the replication regulator protein geminin. Biochem. Biophys. Res. Commun. 305:412-420. [DOI] [PubMed] [Google Scholar]
- 27.Meggio, F., and L. A. Pinna. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349-368. [DOI] [PubMed] [Google Scholar]
- 28.Meyn, M. S., J. M. Lu-Kuo, and L. B. K. Herzing. 1993. Expression cloning of multiple human cDNAs that complement the phenotypic defects of ataxia-telangiectasia group D fibroblasts. Am. J. Hum. Genet. 53:1206-1216. [PMC free article] [PubMed] [Google Scholar]
- 29.Ruzzene, M., D. Penzo, and L. A. Pinna. 2002. Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells. Biochem J. 364:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarno, S., S. Moro, F. Meggio, G. Zagotto, D. Dal Ben, P. Ghisellini, R. Battistutta, G. Zanotti, and L. A. Pinna. 2002. Toward the rational design of protein kinase casein kinase-2 inhibitors. Pharmacol. Ther. 93:159-168. [DOI] [PubMed] [Google Scholar]
- 31.Sarno, S., H. Reddy, F. Meggio, M. Ruzzene, S. P. Davies, A. Donella-Deana, D. Shugar, and L. A. Pinna. 2001. Selectivity of 4,5,6,7-tetrabromobenzotriazole, an ATP site-directed inhibitor of protein kinase CK2 (′casein kinase-2′). FEBS Lett. 496:44-48. [DOI] [PubMed] [Google Scholar]
- 32.Sierakowska, H., K. R. Williams, I. S. Szer, and W. Szer. 1993. The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis. Clin. Exp. Immunol. 94:435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sitwala, K. V., N. Mor-Vaknin, and D. M. Markovitz. 2003. Minireview: DEK and gene regulation, oncogenesis and AIDS. Anticancer Res. 23:2155-2158. [PubMed] [Google Scholar]
- 34.von Lindern, M., M. Fornerod, S. van Baal, M. Jaegle, T. de Wit, A. Buijs, and G. Grosveld. 1992. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol. 12:1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldmann, T., M. Baack, N. Richter, and C. Gruss. 2003. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 31:7003-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldmann, T., C. Eckerich, M. Baack, and C. Gruss. 2002. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J. Biol. Chem. 277:24988-24994. [DOI] [PubMed] [Google Scholar]
- 37.Wessel, D., and U. I. Flügge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141-143. [DOI] [PubMed] [Google Scholar]
- 38.Wichmann, I., N. Respaldiza, J. R. Garcia-Lozano, M. Montes, J. Sanchez-Roman, and A. Nunez-Roldan. 2000. Autoantibodies to DEK oncoprotein in systemic lupus erythematosus (SLE). Clin. Exp. Immunol. 119:530-532. [DOI] [PMC free article] [PubMed] [Google Scholar]