Abstract
DEK was originally described as a proto-oncogene protein and is now known to be a major component of metazoan chromatin. DEK is able to modify the structure of DNA by introducing supercoils. In order to find interaction partners and functional domains of DEK, we performed yeast two-hybrid screens and mutational analyses. Two-hybrid screening yielded C-terminal fragments of DEK, suggesting that DEK is able to multimerize. We could localize the domain to amino acids 270 to 350 and show that multimerization is dependent on phosphorylation by CK2 kinase in vitro. We also found two DNA binding domains of DEK, one on a fragment including amino acids 87 to 187 and containing the SAF-box DNA binding motif, which is located between amino acids 149 and 187. This region is sufficient to introduce supercoils into DNA. The second DNA binding domain is located between amino acids 270 and 350 and thus overlaps the multimerization domain. We show that the two DNA-interacting domains differ in their binding properties and in their abilities to respond to CK2 phosphorylation.
The proto-oncogene protein DEK was first identified as a fusion protein with the nucleoporin CAN in patients suffering from a subtype of acute myeloid leukemia (AML) (30). A recent study showed that more than two-thirds of AML patients expressing the DEK-CAN fusion also have a specific internal tandem duplication in the FMS-like tyrosine kinase (FLT3) gene (23, 28), resulting in a constitutively active kinase. In contrast to AML with the DEK-CAN rearrangement, the incidence of the activating FLT3 internal tandem duplication in all AML patients is only around 20%. This suggests that DEK-CAN might contribute to the generation of FLT3 internal tandem repeats as a possible mechanism leading to carcinogenesis (4, 19, 28).
In addition to its involvement in leukemia, DEK has been shown to be a major antigen in several autoimmune diseases like systemic lupus erythematosus (6, 7, 33), juvenile rheumatoid arthritis (7, 25), and sarcoidosis (6, 7). Despite these associations with several human disorders, little is known about how DEK could functionally be involved in these diseases.
In search of the cellular function of DEK, several groups are trying to find interaction partners of DEK. DEK has been reported to be involved in transcriptional repression by interaction with the corepressor Daxx (13), as well as in transcriptional activation stimulating the binding of activator protein AP-2α to its target DNA sequences (5). Furthermore, DEK has been reported to bind to the pets site, a regulatory element of the human immunodeficiency virus type 2 enhancer, in a sequence-specific manner (8, 10). It has also been reported that DEK recognizes the Y box of the class II major histocompatibility complex gene promoter, a known transcription factor binding site with sequence similarities to the pets site (1, 26). In contrast to these findings, we could not detect sequence-specific binding of DEK to DNA (31). Instead, we observed structure-specific binding of DEK to DNA crossovers, as found in supercoiled or four-way junction DNAs (31). Moreover, DEK binding causes a change in the superhelical density of closed circular plasmid DNA and simian virus 40 (SV40) minichromosomes in vitro (2, 32).
DEK is a nuclear phosphoprotein (9, 25) that remains associated with chromatin during the entire cell cycle (14). While the amount of DEK on chromatin does not change during the cell cycle, we detected a moderate increase in DEK phosphorylation during the G1 phase (15). It appears that DEK is mainly phosphorylated by CK2 kinase in vitro and in vivo. Moreover, phosphorylation reduces the DNA binding activity of DEK. This is surprising, because most of the phosphorylation sites were mapped within the C-terminal region of the protein (15, 31, 32), whereas a potential DNA binding motif, the SAF-box (scaffold attachment factor) (11, 18), also known as the SAP-box (SAF-A/B acinus and pias) (3), is located between amino acids 149 and 187. The SAF-box is the only recognizable feature besides some highly acidic regions of the protein. The SAF-box was identified as a novel DNA binding domain that binds specifically to AT-rich SAR/MAR (scaffold/matrix attachment region) DNA (11, 18). SAF-box-containing proteins are involved in different processes, such as DNA repair (Ku70, PARP), pre-mRNA processing (SAF-A, SAF-B, SPRY), transcriptional elongation (Tho1), and nuclear architecture (SAF-A, SAF-B) (3).
To investigate whether the SAF-box is the DNA binding domain of DEK, we expressed several deletion mutant forms of DEK and tested them for DNA binding in electrophoretic mobility shift assays (EMSAs) and for the ability to change the supercoils of closed circular DNA. We show here that a DEK fragment containing the SAF-box is able to bind DNA and appears to be responsible for supercoiling activity. Moreover, the SAF-box-containing fragment induces intermolecular interactions between two or more DNA molecules. In addition, we have identified a second functional domain within the C-terminal region of DEK. This domain is able to bind DNA and induce DEK-DEK interactions. These two functional activities are regulated by phosphorylation.
MATERIALS AND METHODS
Cloning.
Full-length DEK cDNA was subcloned from pRSET-A-dek into pBlueBacHis2-A (Invitrogen) via the BamHI and EcoRI restriction sites. DEK fragments with 5′ EcoRI and 3′ XhoI cutting sites were generated by PCR. A stop codon was introduced before the XhoI site. The PCR products were digested and ligated into pBlueBacHis2-B (Invitrogen) that was cut with EcoRI and SalI, creating a hybrid SalI/XhoI site at the 3′ end of the insert. All constructs were checked by sequencing.
Expression and purification of his-DEK fragments.
The different pBlueBacHis2 constructs of DEK were cotransfected with linearized Autographa californica nuclear polyhedrosis virus DNA (Bac-N-Blue DNA) into SF9 cells as described in the manufacturer's (Invitrogen) protocol. Passage 3 virus stocks were used to infect HighFive cells for protein expression. Cells at 3 days postinfection were washed twice with phosphate-buffered saline and then lysed with 2 ml of lysis buffer per 175-mm2 flask (100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1% NP-40, 5 mM imidazole). To disrupt DNA-protein and protein-protein interactions, the lysed cells were further incubated in the presence of 1.3 M NaCl for 20 min at room temperature. The lysate was cleared by centrifugation at 100,000 × g. The supernatant was adjusted to 10% glycerol, diluted with lysis buffer to a final concentration of 700 mM NaCl, and incubated with 8 μl of 50% Ni-nitrilotriacetic acid (NTA)-agarose beads (QIAGEN). After 1 h of incubation at 4°C, the beads were washed twice with WB-150 (100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 50 mM imidazole) and transferred into a 10-ml Bio-Rad column. The beads were washed twice with WB-300 (100 mM Tris-Cl [pH 7.5], 300 mM NaCl, 50 mM imidazole) and again with WB-150. The His-tagged proteins were then eluted 10 times with 20 μl of elution buffer (100 mM Tris-Cl [pH 7.5], 150 mM NaCl, 500 mM imidazole) per 175-cm2 flask. All DEK-containing fractions were pooled and stored at −70°C.
Far Western analysis.
A protein kinase A (PKA) phosphorylation site (amino acid motif RRASV) was cloned into pBlueBacHis2-A-DEK with BamHI and the annealed oligonucleotides PKA forward (5′ GATCC CGT CGT GCA TCT GTT G) and PKA reverse (5′ GATCC AAC AGA TGC ACG ACG G). Correct orientation of the insert was checked by sequencing. After expression and purification, 1 μg of PKA-His-tagged DEK was diluted 1/10 with wash buffer WB-150 (see description of expression and purification) and rebound to 100 μl of settled Ni-NTA-agarose for 2 h at 4°C. The resin was washed twice with 1× PKA phosphorylation buffer (New England Biolabs) by centrifuging the batch for 15 s at maximum speed. [γ-32P]ATP labeling was then carried out on column, with 1 μl of PKA (New England Biolabs) in a total volume of 400 μl for 30 min at 37°C. After being washed three times with 1 ml of wash buffer, labeled DEK was eluted from the Ni-NTA-agarose with 3× 300 μl of elution buffer. The eluates were analyzed by Western blotting and autoradiography; radioactively labeled DEK peaked in the second eluate.
Two-hybrid screening.
For two-hybrid screening, we used the “interaction hunt” protocol of Golemis et al. (12). Full-length DEK and a truncated DEK cDNA coding for amino acids 1 to 350 were cloned into the bait vector pEG202 with EcoRI and XhoI, generated by PCR. We used two libraries, an oligo(dT)-primed HeLa cell library (Origene) and a random-primed human kidney library (Invitrogen), for multiple screenings, both with full-length and truncated DEK as bait.
Sedimentation analysis.
For sedimentation analysis, 2 μg of recombinant His-DEK was used either dephosphorylated or phosphorylated. Dephosphorylation was carried out for 90 min with λ-phosphatase (New England Biolabs) at 30°C in accordance with the manufacturer's protocol (for 10 pmol of DEK, 400 U of λ-phosphatase was used). Phosphorylation with CK2 was performed with CK2 buffer (20 mM HEPES [pH 7.8], 10 mM MgCl2, 10 mM CaCl2, 20 mM NaF, and 1 mM vanadate supplemented with 100 μM ATP) for 90 min at 37°C (for 10 pmol of DEK, 40 U of CK2 was used). After incubation for 60 min at 37°C in nE100 (20 mM HEPES [pH 7.6], 100 mM NaCl, 10 mM sodium bisulfite, 1 mM EDTA), samples were loaded onto 5 to 30% sucrose gradients in nE100 or nE800 (800 mM NaCl) and centrifuged for 14 h at 36,000 rpm in an SW40 rotor (Beckman). Gradients were fractionated, and proteins were concentrated and then immunoblotted with DEK-specific antibodies. The pellet fraction was dissolved in 2% sodium dodecyl sulfate (SDS) and treated in the same way.
Topology assay.
Recombinant DEK fragments were dephosphorylated with λ-phosphatase (New England Biolabs) as described above. After incubation at 30°C for 90 min, the samples were dialyzed on Whatman filters (type VS; pore size, 0.025 μm) against nE300 buffer (20 mM HEPES [pH 7.6], 300 mM NaCl, 10 mM sodium bisulfite, 1 mM EDTA) in the presence of 1 μg of bovine serum albumin (New England Biolabs) per microliter for 90 min at 4°C. Supercoiled SV40 DNA (0.0028 pmol) was incubated with increasing amounts of the different DEK fragments in the presence of 1 U of topoisomerase I (wheat germ; Promega). The amounts of protein are indicated in the figure legends. The reactions were performed in buffer L0 (20 mM HEPES [pH 7.6], 10 mM dithiothreitol, 25 μg of bovine serum albumin per ml) in a total volume of 90 μl. After proteinase K (Roche) digestion, DNA was precipitated and analyzed on 0.8% agarose gels in 0.5× TBE (45 mM Tris-borate, 1 mM EDTA) at 2 V/cm for 16 h. The gels were stained with SybrGold (MobiTec).
EMSA.
Purified DEK fragments were treated as described for the topology assay. Linearized SV40 DNA (0.05 pmol) was incubated with increasing amounts of DEK as indicated in the figure legends. The reactions were performed in a total volume of 35 μl, and mixtures were loaded directly onto a 0.6% agarose gel in 0.5× TBE (50 mM Tris-borate, 1 mM EDTA [adjusted to pH 7.8 with boric acid]) and run at 2 V/cm overnight.
Ligase assay.
A 123-bp DNA ladder (Pharmacia) was digested with AvaI. Twenty-four nanograms (0.3 pmol) of the 123-bp fragment was incubated with 1.2 pmol of the different DEK fragments for 1 h at 37°C in ligase buffer (4 mM Tris [pH 7.8], 1 mM dithiothreitol, 0.5 mM ATP, 5 mM MgCl2) in a final volume of 20 μl. T4 DNA ligase (1 U) was added for 10 min at 37°C; the ligase was then inactivated for 15 min at 65°C. To distinguish between linear and circular products, the samples were treated with 50 U of exonuclease III. Samples were digested with proteinase K and analyzed on 8% polyacrylamide gels for 3 h at 10 V/cm. The reaction products were then analyzed by SybrGold (MobiTec) staining.
RESULTS
Multimerization of DEK protein.
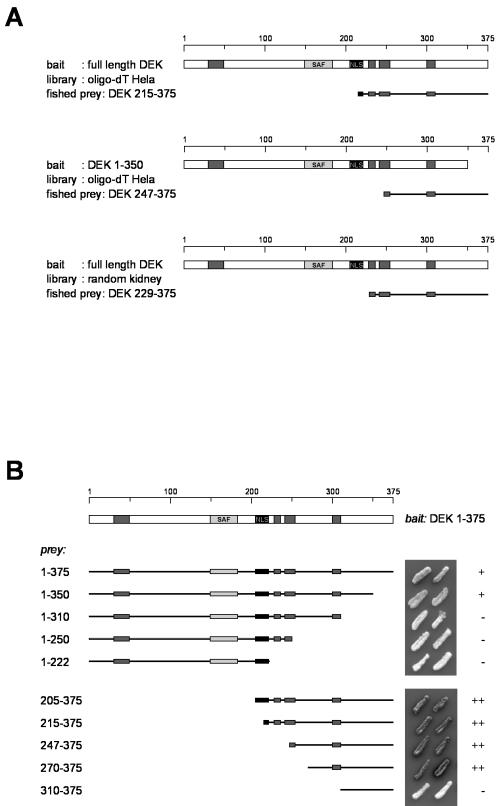
To identify DEK interaction partners, we used the yeast two-hybrid system. Two different libraries were screened with full-length DEK or DEK 1 -350 as bait. By using full-length DEK as bait, we recovered sequences containing amino acids 215 to 375 of DEK from an oligo(dT)-primed HeLa library. With DEK 1-350 as bait, we isolated DEK 247-375 from the same cDNA library. To avoid the bias for carboxy-terminal regions intrinsic to oligo(dT)-primed libraries, we also used a random primed kidney cDNA library and full-length DEK as bait. Again, a cDNA encoding a C-terminal fragment of DEK (amino acids 229 to 375) was found (Fig. 1A). These data indicate that DEK′s preferred interaction partner is DEK itself and that amino acids 247 to 350 of DEK contain a di- or multimerization domain.
FIG. 1.
Two-hybrid screening. (A) Results of three different two-hybrid interaction hunts. Full-length DEK (top and bottom) and DEK 1-350 (middle) were used as bait. We screened against two different human libraries and obtained prey that codes for carboxy-terminal fragments of DEK. (B) Localization of the DEK-DEK multimerization domain. We tested different deletion mutant forms of DEK (indicated on the left side) against full-length DEK in the two-hybrid system. LacZ activation is shown on the right. Dark grey boxes, highly acidic regions; light grey box, SAF-box, a potential DNA binding domain; black box, nuclear localization sequence (NLS).
To confirm this finding and localize the multimerization domain more precisely, we tested several fragments of DEK versus the full-length protein in the two-hybrid system (Fig. 1B). All fragments containing amino acids 270 to 350 were able to interact with DEK, showing that the amino-terminal region up to residue 270 is not involved in di- or multimerization.
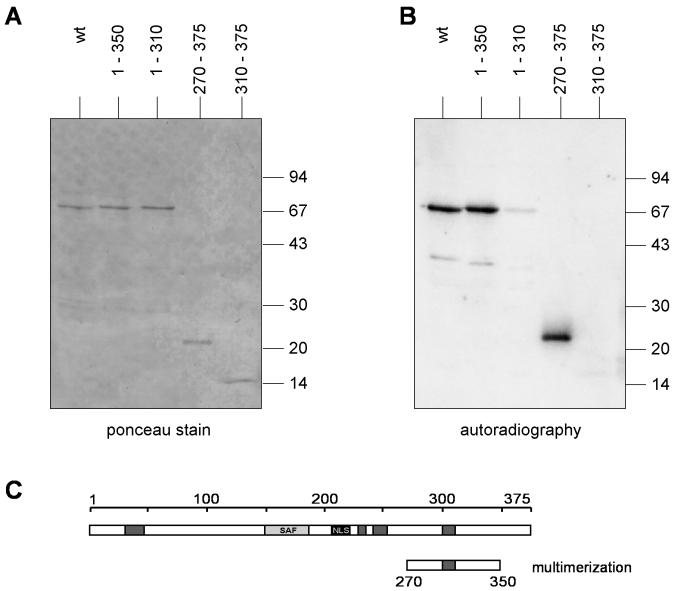
We obtained independent support for this conclusion by far Western blotting. A PKA consensus sequence (amino acid motif RRASV) was linked 5′ to the amino terminus of full-length DEK. The protein was expressed in the baculovirus system and labeled in vitro with [γ-32P]ATP by PKA. Equal amounts of deletion mutant forms and full-length DEK were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. After incubation with labeled full-length DEK and extensive washing, the blot was analyzed by autoradiography (Fig. 2B). Full-length DEK and fragments with amino acids 1 to 350 and 270 to 375 gave positive autoradiographic signals, whereas amino acids 1 to 310 yielded only a faint autoradiographic band (Fig. 2B). Thus, these data confirmed that a protein-protein interaction domain is located between amino acids 270 and 350 of the DEK protein (Fig. 2C). Here it is important to mention that recombinant His-DEK as purified from the baculovirus system is phosphorylated (15). Therefore we tested phosphorylated fragments of DEK and also used phosphorylated PKA-DEK as a probe.
FIG. 2.
Far Western analysis. Twenty nanomoles of full-length DEK (wt, lane 1) and different DEK deletion mutant forms were separated on a 10 to 18% polyacrylamide gel and blotted onto a nitrocellulose membrane. The membrane was then incubated with 32P-labeled full-length DEK. A, Ponceau staining; B, autoradiography. Molecular sizes of marker proteins are given in kilodaltons. (C) DEK protein with the multimerization domain. Dark grey boxes, highly acidic regions; light grey box, SAF-box, a potential DNA binding domain; black box, nuclear localization sequence (NLS). wt, wild type.
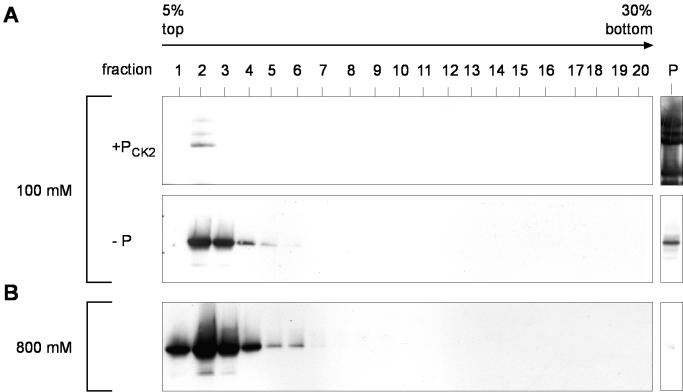
Since DEK-DNA interaction is affected by phosphorylation (15), we tested whether phosphorylation also influences DEK multimerization. We compared the sedimentation behavior of phosphorylated and dephosphorylated full-length DEK on sucrose gradients. We used recombinant full-length DEK that was purified from the baculovirus system in a phosphorylated form. This substrate was first dephosphorylated by λ-phosphatase (Fig. 3A, −P) to remove endogenous phosphates and then rephosphorylated by CK2 in vitro (Fig. 3A, +PCK2). CK2 is the main kinase responsible for the majority of DEK phosphorylation in vitro and in vivo (15).
FIG. 3.
Sedimentation analysis of phosphorylated and dephosphorylated DEK. Recombinant full-length DEK was dephosphorylated with λ-phosphatase (−P), rephosphorylated with CK2 (+PCK2), analyzed on 5 to 30% sucrose gradients, and then centrifuged for 14 h at 36,000 rpm. The gradients were fractionated and analyzed by immunoblotting with DEK-specific antibodies. (A) Sedimentation analysis of dephosphorylated DEK (−P) and phosphorylated DEK (+PCK2) in the presence of 100 mM NaCl. (B) Sedimentation analysis of phosphorylated DEK in the presence of 800 mM NaCl. P corresponds to the pellet fraction.
When sucrose gradients were run under nearly physiological salt conditions (100 mM NaCl), phosphorylated DEK formed large complexes that were found in the pellet of the gradient (Fig. 3A, +PCK2). However, dephosphorylated DEK sedimented at physiological salt concentrations and DEK sedimented mainly as monomers close to the top of the gradient (Fig. 3A, −P). At 800 mM salt (Fig. 3B), phosphorylated DEK had sedimentation properties like those of dephosphorylated DEK at 100 mM NaCl, showing that a high salt concentration disrupts the interaction of phosphorylated DEK (compare Fig. 3A, −P, and B). Thus, phosphorylation of DEK induces DEK-DEK interactions at moderate salt concentrations, whereas the dephosphorylated protein mainly exists in the monomeric form under these conditions.
Similar results were obtained by native gel electrophoresis: phosphorylated DEK remained close to the gel slots as a high-molecular-weight complex, while unphosphorylated DEK migrated as monomers (data not shown). A conclusion from sucrose gradient and native gel electrophoretic analyses is that phosphorylated DEK tends to aggregate in vitro instead of forming discrete multimers.
Characterization of the DNA binding properties of the SAF-box-containing fragment.
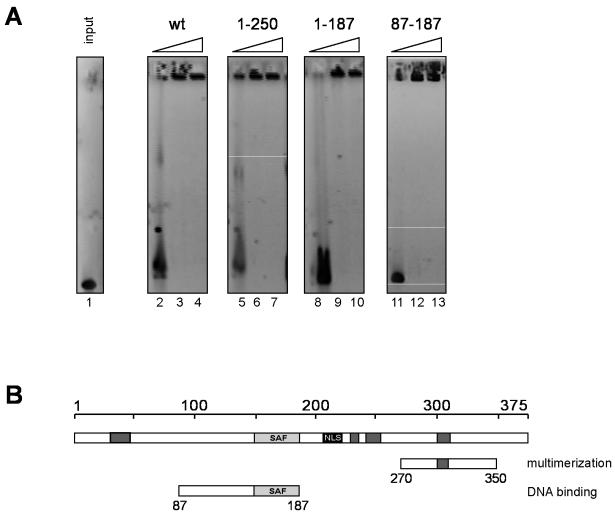
The region of DEK between amino acids 149 and 187 has similarity to a DNA binding motif that is known as the SAF-box (11) and that is also termed the SAP-box (3). To test if the SAF-box is indeed the DNA binding domain of the DEK protein, we used DEK deletion mutant forms with or without the SAF-box in EMSAs. As we have shown before (31, 32), the binding of full-length DEK results in large DNA protein complexes that cannot enter the gel (Fig. 4A, lanes 2 to 4). To investigate if the multimerization domain (amino acids 270 to 350, Fig. 1 and 2) was responsible for this binding mode, we successively removed the C-terminal part of DEK and tested these fragments by EMSA (Fig. 4A, lanes 5 to 13). Surprisingly, we found that even the smallest fragment (amino acids 87 to 187) that contains the SAF-box at its C-terminal region (amino acids 149 to 187) was able to induce this high-molecular-weight complex (Fig. 4A, lanes 11 to 13). Therefore, the previously identified protein-protein interaction domain (amino acids 270 to 350, Fig. 1 and 2) seems to have no influence on the DNA binding properties of DEK. This could indicate that another multimerization domain may exist in the region of amino acids 87 to 187, probably dependent on DNA binding, which induces intermolecular interactions between two or more DNA molecules. Interestingly, the SAF-box-containing protein SAF-A also multimerizes upon DNA binding (18).
FIG. 4.
EMSAs with different dephosphorylated SAF-box fragments. (A) One hundred seventy-five nanograms (0.05 pmol) of linearized SV40 DNA was incubated with 9, 28, and 46 pmol of dephosphorylated DEK fragments (lanes 2 to 4, 5 to 7, 8 to 10, 11 to 13, and 14 to 16, respectively) or without protein (input, lane 1) for 1 h at 37°C. Nucleoprotein complexes were analyzed on 0.6% agarose gels in 0.5× TBE (pH 7.8; see Materials and Methods) and visualized by ethidium bromide staining. The fragments used are indicated above the blots. (B) Map showing the identified DNA binding domain of DEK. Dark grey boxes, highly acidic regions; light grey box, SAF-box, a potential DNA binding domain; black box, nuclear localization sequence (NLS). wt, wild type.
The SAF-box-containing fragment induces intermolecular interactions and is sufficient for the introduction of supercoils.
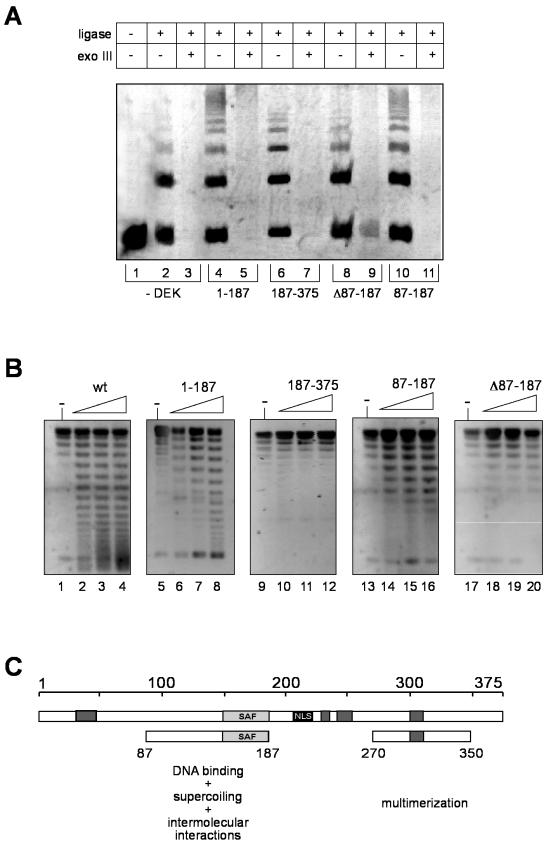
We have recently shown that full-length DEK (glutathione S-transferase and His tagged) promotes the linkage of DNA fragments via a ligase-mediated end-to-end joining assay (31). To determine whether this reaction depends on the SAF-box-containing domain (amino acids 87 to 187), we incubated various deletion mutant DEK proteins with 123-bp DNA fragments in the presence of T4 ligase. Our data show (Fig. 5A) that all DEK constructs containing the amino acid 87 to 187 region stimulated the formation of linear multimers (Fig. 5A, lanes 4 and 10), whereas DEK constructs without the amino acid 87 to 187 region could not (Fig. 5A, lanes 6 and 8). However, all ligation products were sensitive to exonuclease III (Fig. 5A, lanes 5, 7, 9, and 11), showing that none of the DEK fragments were able to bend DNA, as shown before for full-length DEK (31). These data indicate that the region between amino acids 87 and 187 is responsible for the stimulation of intermolecular interactions and thus might include a DNA-dependent multimerization domain.
FIG. 5.
Ligase and topology assay. (A) Ligase assay with a 123-bp fragment. Twenty-four nanograms (0.3 pmol) of a 123-bp fragment with AvaI sticky ends was incubated in the absence (−DEK, lanes 1 to 3) or presence of 1.2 pmol of each DEK fragment (lanes 4 to 13). T4 DNA ligase was added to the reaction mixture as indicated, and the mixture was incubated for 10 min at 37°C. Samples in lanes 3, 5, 7, 9, 11, and 13 were treated with exonuclease III to remove linear products. The reaction products were analyzed by an 8% polyacrylamide gel and SybrGold staining. The positions of the 123-bp multimers are indicated. (B) Topology assays with different DEK fragments. Ten nanograms of circular SV40 DNA (0.003 pmol) was incubated in the absence (−, lanes 1, 5, 9, 13, and 17) or presence of increasing amounts of the indicated DEK fragments (lanes 2 to 4, 6 to 8, 10 to 12, 14 to 16, and 18 to 20; 1.8, 2.4, and 3 pmol, respectively) and 1 U of topoisomerase I. The samples were deproteinized with proteinase K, precipitated with ethanol, and analyzed by a 0.8% agarose gel and SybrGold staining. The positions of forms I (supercoiled) and II (relaxed) are indicated. (C) DEK protein with the newly identified domains. Dark grey boxes, highly acidic regions; light grey box, SAF-box, a potential DNA binding domain; black box, nuclear localization sequence (NLS). wt, wild type.
Recently, we have shown that DEK causes supercoiling of circular DNA in the presence of topoisomerase I (32). In order to determine which domain is responsible for this activity, we tested recombinant DEK fragments in the topology assay (32). We used dephosphorylated DEK fragments and incubated them with circular SV40 DNA in the presence of topoisomerase I. Again, only the fragments containing the amino acid 87 to 187 region were able to change the superhelicity of the DNA (Fig. 5B, lanes 1 to 4, 5 to 8, and 13 to 16).
Thus, we have shown that the region between amino acids 87 and 187, including the SAF-box (amino acids 149 to 187), binds to DNA, induces intermolecular interactions, probably owing to a DNA-dependent multimerization, and is responsible for the introduction of supercoils into circular DNA.
Identification of a second DNA binding domain in the C-terminal region of DEK.
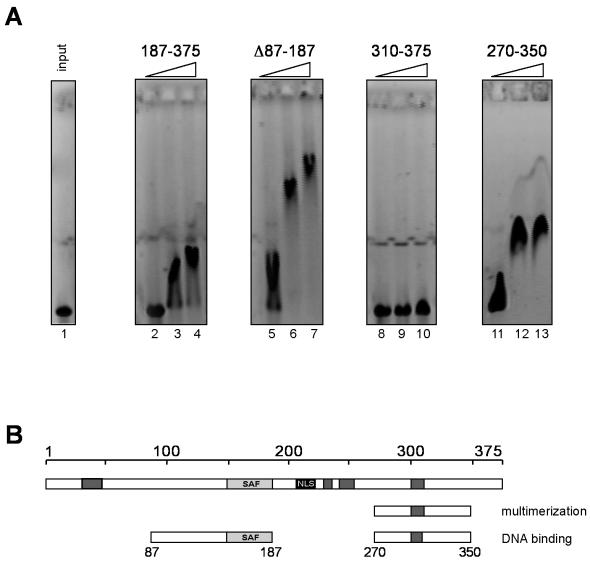
During our investigations of different SAF-box fragments, we also tested the N-terminally truncated deletion mutant forms of DEK by EMSA. Surprisingly, we found that these fragments were able to bind DNA (Fig. 6A, lanes 2 to 4), but we detected no large DNA-protein complexes, even when we used very large amounts of protein (data not shown). We then performed EMSAs with shorter C-terminal fragments (Fig. 6A) and found that the amino acid 310 to 375 fragment showed very weak, if any, binding, even with the largest amount of protein (Fig. 6A, lane 10), whereas a fragment consisting of amino acids 270 to 350 induced a complete shift at low protein concentrations (Fig. 6A, lanes 11 to 13). Large immobile complexes, like those involving amino acids 87 to 187, were never observed. We have thus identified a second DNA binding domain within amino acids 270 to 350 that does not multimerize upon DNA binding like the SAF-box-containing fragments (Fig. 4A).
FIG. 6.
EMSAs with C-terminal fragments of DEK. (A) One hundred seventy-five nanograms (0.05 pmol) of linearized SV40 DNA was incubated with 9, 28, and 46 pmol of dephosphorylated DEK fragments (lanes 2 to 4, 5 to 7, 8 to 10, 11 to 13, and 14 to 16, respectively) or without protein (input, lane 1) for 1 h at 37°C. Nucleoprotein complexes were analyzed on 0.6% agarose gels in 0.5× TBE (pH 7.8, see Materials and Methods) and visualized by ethidium bromide staining. The fragments used are indicated above the blots. (B) Map showing the identified DNA binding domain of DEK. Dark grey boxes, highly acidic regions; light grey box, SAF-box, a potential DNA binding domain; black box, nuclear localization sequence (NLS). wt, wild type.
It appears that the domain between amino acids 270 and 350 has several functions. As just shown, it includes a DNA binding activity (Fig. 6), but in addition it contains a multimerization domain (Fig. 2) and also most of the phosphorylation sites. Above we have shown that phosphorylation affects the interaction between DEK proteins (Fig. 3), and therefore we ask now whether phosphorylation also influences DNA binding.
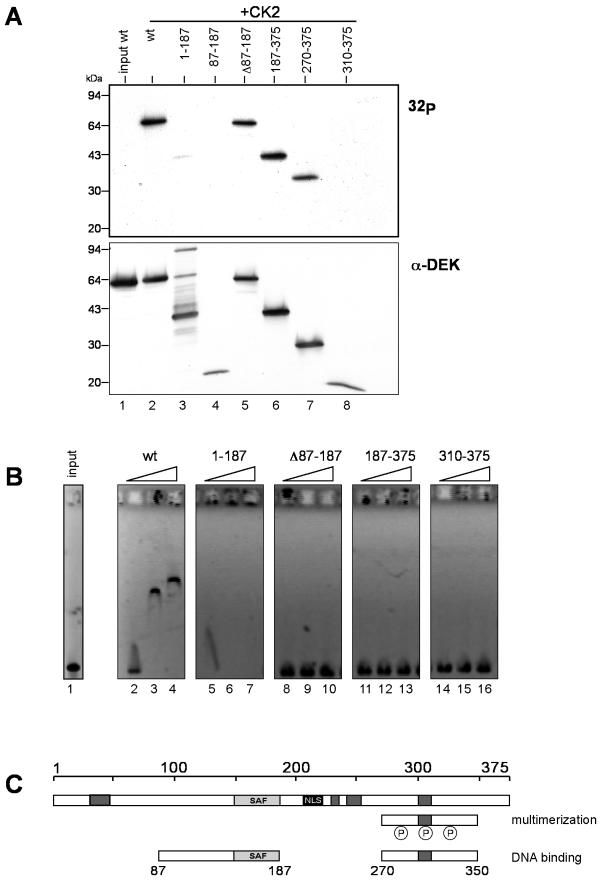
We showed that the sequence between amino acids 187 and 310 can be phosphorylated in vitro (Fig. 7A). This was expected, as most of the in vitro phosphorylation sites mapped to this region (15). We then investigated whether phosphorylation affects DNA binding and found that the in vitro-phosphorylated C-terminal fragments had completely lost the ability to bind DNA (Fig. 7B; compare with Fig. 6). A control was fragment 1 to 187, which still bound DNA after CK2 phosphorylation (Fig. 7B, lanes 5 to 7). Another control was full-length DEK, whose DNA binding activity was reduced after phosphorylation (Fig. 7B, lanes 2 to 4; compare with Fig. 4). This indicates that phosphorylation has an overall effect on DEK′s ability to bind DNA.
FIG. 7.
EMSAs with phosphorylated DEK fragments. (A) Nine, 28, and 46 pmol of the different DEK fragments were phosphorylated with CK2 in vitro as described before. The samples were separated by SDS-10 to 18% polyacrylamide gel electrophoresis and analyzed by autoradiography (32P) and Western blotting with DEK-specific antibodies (α-DEK). The positions of the molecular weight markers are indicated. (B) One hundred seventy-five nanograms (0.05 pmol) of linearized SV40 DNA was incubated with 9, 28, and 46 pmol of phosphorylated DEK fragments (lanes 2 to 4, 5 to 7, 8 to 10, 11 to 13, and 14 to 16, respectively) or without protein (input, lane 1) for 1 h at 37°C. The nucleoprotein complexes were analyzed on 0.6% agarose gels in 0.5× TBE (pH 7.8, see Materials and Methods) and visualized by ethidium bromide staining. (C) DEK protein with newly identified regions. Dark grey boxes, highly acidic regions; light grey box, SAF-box, a potential DNA binding domain; black box, nuclear localization sequence (NLS). wt, wild type.
DISCUSSION
We used two-hybrid screening and mutational analysis to identify interaction partners and functional domains of the proto-oncogene protein DEK. Two-hybrid screening yielded C-terminal fragments of DEK, suggesting that DEK is able to multimerize. We localized the multimerization domain to amino acids 270 to 350 and showed furthermore that multimerization is dependent on phosphorylation by CK2 in vitro. We also found two DNA binding domains of DEK, one on a fragment consisting of amino acids 87 to 187 that contains a known DNA binding motif, the SAF-box (amino acids 149 to 187) (11). This region is sufficient to introduce supercoils into DNA and to induce intermolecular interactions, as shown by ligase-mediated end-to-end joining. A second DNA binding domain is located between amino acids 270 and 350 and thus overlaps the multimerization domain. The two DNA-interacting domains differ in their binding modes and in their abilities to be regulated by CK2 phosphorylation.
The SAF-box-containing fragment (amino acids 87 to 187) introduces supercoils and induces intermolecular interactions.
The SAF-box has been described as a DNA binding motif that consists of two predicted amphipathic helices separated by a region that includes an invariant glycine. SAF-box modules are found throughout all eukaryotes (from yeast to humans) but not in prokaryotes (18). Most of the SAF-box proteins have additional functional modules that contribute to their specific function in the cell. It is thought that the SAF module may target these proteins to the sites of active transcription and therefore link nuclear processes like transcription, pre-mRNA processing, and DNA repair by binding to SAR/MAR regions in the nucleus (3). DEK, however, has no known additional module, which may suggest its participation in a specific nuclear process. In fact, no specific binding to SAR/MAR DNA has been detected.
However, DEK has some features in common with other SAR/MAR-binding proteins. It binds preferentially to DNA crossovers (31) like histone H1, and like SAF-A, DEK multimerizes upon DNA binding (Fig. 5A) (31) and is partially localized at the nuclear matrix (Kappes, unpublished data). In addition, DEK has some properties in common with the known nuclear architectural HMG proteins. Thus, DEK induces intra- and intermolecular interactions (31, 32) and changes the topology of circular DNA substrates (24, 27, 29). Taken together, these data suggest a role for DEK in nuclear architecture. This is also thought to be one function of the SAF-A and SAF-B proteins, the founding members of the SAF-box family of proteins (18, 22).
We were able to show here that the SAF-box-containing region (amino acids 87 to 187) of DEK is sufficient to introduce supercoils and to induce intermolecular interactions between two or more DNA molecules (Fig. 5). The ligase assay suggests that the SAF-box fragment induces intermolecular interaction of DEK-DNA complexes. This multimerization must be dependent on DNA binding, because it was not detectable by far Western analysis in the absence of DNA (Fig. 2B).
These results may suggest a mechanism by which DEK introduces supercoils into closed circular DNA. DEK-DEK interaction upon DNA binding would change the twist of DNA, resulting in torsional stress of the DNA molecule. This tension is then compensated by changing the writhing number and the introduction of compensating supercoils. While topoisomerase I is able to relax these compensating supercoils, the change in twist is constrained when DEK is bound (32). Such a mechanism has been discussed for 13S condensin (16, 17) and for bacterial HMf proteins (21), which both introduce positive supercoils into circular DNA.
The region of amino acids 270 to 350 is responsible for multimerization and DNA binding.
We have searched for proteins that interact with DEK. However, in several independent yeast two-hybrid screens, we identified only one interaction partner, namely, DEK itself. Testing different fragments of DEK against the full-length protein showed that the putative di- or multimerization domain is located in the C-terminal region of DEK, between amino acids 270 and 350 (Fig. 1B). These results were confirmed by far Western analysis (Fig. 3). Why were other interacting proteins not detected given that DEK-binding partners such as histone H2A/H2B, AP-2α, SRm160, or Daxx and HDAC have been described in the literature (2, 5, 13, 20)?
One possibility is that putative interaction partners are misfolded or not properly posttranslationally modified in yeast. It is also possible that the prey libraries do not contain sequences encoding the respective proteins. Notably, the intrinsic restriction of the two-hybrid system for testing one-to-one interactions could make it difficult to detect larger protein complexes such as the complex formed by Daxx, HDAC, histones, and DEK (13).
We showed by EMSA that there exists a second DNA binding domain in the C-terminal part of DEK (Fig. 6A). We could map this second DNA binding domain to the region between amino acids 270 and 350 (Fig. 6A), which is also the region of protein-protein interaction (Fig. 1 and 2). The nucleoprotein complexes formed by the second DNA binding domain migrated into the gel, suggesting that no large DNA-protein complexes are induced by this domain (Fig. 6A). Consistent with this, the second DNA binding domain does not induce intermolecular interactions upon DNA binding (Fig. 5A, lanes 8 and 10) and fails to create supercoils into circular DNA (Fig. 5B, lanes 9 to 12 and 17 to 20).
Interestingly, the region of amino acids 270 to 350, including the multimerization domain and the second DNA binding domain, also contains most of the mapped CK2 phosphorylation sites (15). Indeed, fragments containing the C-terminal region of DEK are highly phosphorylated by CK2 in vitro (Fig. 7A, lanes 2, 5, 6, and 7). Furthermore, phosphorylation of amino acids 270 to 350 disrupts the DNA binding of these fragments (Fig. 7B, lanes 5 to 16) but triggers DEK-DEK interaction (Fig. 3A). Therefore, the functional activity of this domain appears to be changed upon phosphorylation: it binds to DNA when dephosphorylated but dissociates and multimerizes upon phosphorylation. Furthermore, phosphorylation of the full-length protein seems to negatively regulate the DNA binding activity of the SAF-box (Fig. 7B, lanes 2 to 4; compare with Fig. 4A, lanes 2 to 4). This could be due to conformational changes in the whole protein or to a release of the phosphorylated second DNA binding domain (amino acids 270 to 350) from the DNA. DEK-DEK interactions could then weaken the binding of the SAF-box domain (amino acids 87 to 187).
In summary, these data could provide an explanation for the diverse and sometimes conflicting data about DEK. Thus, DEK may be involved in nuclear architecture via the SAF-box region (amino acids 87 to 187). In fact, just like SAF-A, DEK preferentially binds to DNA crossovers (31) and induces inter- and intramolecular interactions (31, 32).
Published data suggest that DEK may be involved in transcriptional regulation of chromatin. This could be connected to the fact that DEK is released from DNA upon phosphorylation, thereby making target sequences accessible for transcriptional activators or repressors. This implies that DEK on chromatin is dynamic. However, further experiments are needed to support this assumption.
Acknowledgments
We are grateful to Rolf Knippers, Martina Baack, and Anita Krebs for stimulating discussions and critical reading of the manuscript and Lorenzo Pinna for providing CK2.
This work was supported by grants from DFG to C.G. I.S. was supported by BIF (Boehringer Ingelheim Fonds).
REFERENCES
- 1.Adams, B. S., H. C. Cha, J. Cleary, T. Haiying, H. Wang, K. Sitwala, and D. M. Markovitz. 2003. DEK binding to class II MHC Y-box sequences is gene- and allele-specific. Arthritis Res. Ther. 5:R226-R233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexiadis, V., T. Waldmann, J. Andersen, M. Mann, R. Knippers, and C. Gruss. 2000. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 14:1308-1312. [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 4.Birg, F., M. Courcoul, O. Rosnet, F. Bardin, M. J. Pebusque, S. Marchetto, A. Tabilio, P. Mannoni, and D. Birnbaum. 1992. Expression of the FMS/KIT-like gene FLT3 in human acute leukemias of the myeloid and lymphoid lineages. Blood 80:2584-2593. [PubMed] [Google Scholar]
- 5.Campillos, M., M. A. Garcia, F. Valdivieso, and J. Vázquez. 2003. Transcriptional activation by AP-2α is modulated by the oncogene DEK. Nucleic Acids Res. 31:1571-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong, X., M. A. Michelis, J. Wang, R. Bose, T. DeLange, and W. H. Reeves. 1998. Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum. 41:1505-1510. [DOI] [PubMed] [Google Scholar]
- 7.Dong, X., J. Wang, F. N. Kabir, M. Shaw, A. M. Reed, L. Stein, L. E. Andrade, V. F. Trevisani, M. L. Miller, T. Fujii, M. Akizuki, L. M. Pachman, M. Satoh, and W. H. Reeves. 2000. Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum. 43:85-93. [DOI] [PubMed] [Google Scholar]
- 8.Faulkner, N. E., J. M. Hilfinger, and D. M. Markovitz. 2001. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J. Biol. Chem. 276:25804-25812. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod, M., J. Boer, S. van Baal, M. Jaegle, M. von Lindern, K. G. Murti, D. Davis, J. Bonten, A. Buijs, and G. Grosveld. 1995. Relocation of the carboxy-terminal part of CAN from the nuclear envelope to the nucleus as a result of leukemia-specific chromosome rearrangements. Oncogene 10:1739-1748. [PubMed] [Google Scholar]
- 10.Fu, G. K., G. Grosveld, and D. M. Markovitz. 1997. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc. Natl. Acad. Sci. USA 94:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Göhring, F., and F. O. Fackelmayer. 1997. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry 36:8276-8283. [DOI] [PubMed] [Google Scholar]
- 12.Golemis, E. A., et al. 1999. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 13.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 115:3319-3330. [DOI] [PubMed] [Google Scholar]
- 14.Kappes, F., K. Burger, M. Baack, F. O. Fackelmayer, and C. Gruss. 2001. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 276:26317-26323. [DOI] [PubMed] [Google Scholar]
- 15.Kappes, F., C. Damoc, R. Knippers, M. Przybylski, L. A. Pinna, and C. Gruss. 2004. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol. Cell. Biol. 24:6011-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimura, K., and T. Hirano. 1997. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90:625-634. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, K., V. V. Rybenkov, N. J. Crisona, T. Hirano, and N. R. Cozzarelli. 1999. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell 98:239-248. [DOI] [PubMed] [Google Scholar]
- 18.Kipp, M., F. Gohring, T. Ostendorp, C. M. van Drunen, R. van Driel, M. Przybylski, and F. O. Fackelmayer. 2000. SAF-box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 20:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libura, M., V. Asnafi, A. Tu, E. Delabesse, I. Tigaud, F. Cymbalista, A. Bennaceur-Griscelli, P. Villarese, G. Solbu, A. Hagemeijer, K. Beldjord, O. Hermine, and E. Macintyre. 2003. FLT3 and MLL intragenic abnormalities in AML reflect a common category of genotoxic stress. Blood 102:2198-2204. [DOI] [PubMed] [Google Scholar]
- 20.McGarvey, T., E. Rosonina, S. McCracken, Q. Li, R. Arnaout, E. Mientjes, J. A. Nickerson, D. Awrey, J. Greenblatt, G. Grosveld, and B. J. Blencowe. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell Biol. 150:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musgrave, D. R., K. M. Sandman, and J. N. Reeve. 1991. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc. Natl. Acad. Sci. USA 88:10397-10401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romig, H., F. O. Fackelmayer, A. Renz, U. Ramsperger, and A. Richter. 1992. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11:3431-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosnet, O., M. G. Mattei, S. Marchetto, and D. Birnbaum. 1991. Isolation and chromosomal localization of a novel FMS-like tyrosine kinase gene. Genomics 9:380-385. [DOI] [PubMed] [Google Scholar]
- 24.Sheflin, L. G., and S. W. Spaulding. 1989. High mobility group protein 1 preferentially conserves torsion in negatively supercoiled DNA. Biochemistry 28:5658-5664. [DOI] [PubMed] [Google Scholar]
- 25.Sierakowska, H., K. R. Williams, I. S. Szer, and W. Szer. 1993. The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis. Clin. Exp. Immunol. 94:435-439. [Erratum, 96:177, 1994.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitwala, K. V., K. Adams, and D. M. Markovitz. 2002. YY1 and NF-Y binding sites regulate the transcriptional activity of the dek and dek-can promoter. Oncogene 21:8862-8870. [DOI] [PubMed] [Google Scholar]
- 27.Stros, M., J. Stokrova, and J. O. Thomas. 1994. DNA looping by the HMG-box domains of HMG1 and modulation of DNA binding by the acidic C-terminal domain. Nucleic Acids Res. 22:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiede, C., C. Steudel, B. Mohr, M. Schaich, U. Schakel, U. Platzbecker, M. Wermke, M. Bornhauser, M. Ritter, A. Neubauer, G. Ehninger, and T. Illmer. 2002. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood 99:4326-4335. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, J. O. 2001. HMG1 and 2: architectural DNA binding proteins. Biochem. Soc. Trans. 29:395-401. [DOI] [PubMed] [Google Scholar]
- 30.von Lindern, M., M. Fornerod, S. van Baal, M. Jaegle, T. de Wit, A. Buijs, and G. Grosveld. 1992. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol. 12:1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldmann, T., M. Baack, N. Richter, and C. Gruss. 2003. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 31:7003-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldmann, T., C. Eckerich, M. Baack, and C. Gruss. 2002. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J. Biol. Chem. 277:24988-24994. [DOI] [PubMed] [Google Scholar]
- 33.Wichmann, I., N. Respaldiza, J. R. Garcia-Lozano, M. Montes, J. Sanchez-Roman, and A. Nunez-Roldan. 2000. Autoantibodies to DEK oncoprotein in systemic lupus erythematosus (SLE). Clin. Exp. Immunol. 119:530-532. [DOI] [PMC free article] [PubMed] [Google Scholar]