Abstract
SUMO (also called Sentrin) is a ubiquitin-like protein that plays an important role in regulating protein function and localization. It is known that several nuclear receptors are modified by SUMO; however, the effect of desumoylation in regulating nuclear receptor function has not been elucidated. Here we show that androgen receptor (AR)-mediated transcription is markedly enhanced by SENP1, a member of SUMO-specific protease family. SENP1's ability to enhance AR-dependent transcription is not mediated through desumoylation of AR, but rather through its ability to deconjugate histone deacetylase 1 (HDAC1), thereby reducing its deacetylase activity. HDAC1's repressive effect on AR-dependent transcription could be reversed by SENP1 and by deletion of its sumoylation sites. RNA interference depletion of endogenous HDAC1 also reduced SENP1's effect. Thus, SENP1 could regulate AR-dependent transcription through desumoylation of HDAC1. These studies provide insights on the potential role of desumoylation in the regulation of nuclear receptor activity.
SUMO is a ubiquitin-like protein that can be covalently attached to a large number of proteins through the formation of isopeptide bonds with specific lysine residues of target proteins (23, 40, 42, 50, 53). Sumoylation requires a specific activating-enzyme complex (Uba2/Aos1), conjugation enzyme (Ubc9), and ligases (PIAS, RanBP2, and Pc2) (17-19, 26-28, 46, 47). A large number of sumoylated proteins, including RanGAP1, PML, IκBα, p53, c-Jun, Sp3, Elk-1, p300, and many nuclear receptors, have been identified (8, 9, 15, 20, 29, 33, 38, 41, 48, 49). Sumoylation has emerged as an important regulatory mechanism for protein function and localization (23, 40, 42, 53).
Sumoylation is a dynamic process that is mediated by activating, conjugating, and ligating enzymes and that is readily reversed by a family of SUMO-specific proteases (36, 53). In Saccharomyces cerevisiae, there are two SUMO-specific proteases, Ulp1 and Ulp2/Smt4 (1, 36, 37). Ulp1 is essential for the G2/M transition of the cell cycle (36), whereas Ulp2/Smt4 is not essential for viability (37). In the mammalian system, four SUMO-specific proteases have been reported (2, 19, 22, 31, 44, 45, 53). SENP1 is a nuclear protease that appears to deconjugate a large number of sumoylated proteins (19). SENP2 is a nuclear-envelope-associated protease that appears to have activity similar to that of SENP1 when overexpressed (19, 22, 55). The mouse SENP2 was named SMT3IP2/Axam2 (44). There is a spliced isoform of mouse SENP2, called SuPr1, which could alter the distribution of nuclear PML oncogenic domain-associated proteins, such as CBP and Daxx, and which converted Sp3 to a strong activator with diffuse nuclear localization (2, 49). Two additional SUMO-specific proteases (SENP3/SMT3IP1 and SENP6/SUSP1) have also been reported (31, 53). SENP3/SMT3IP1 is a nucleolar protein (E. T. H. Yeh, unpublished data), whereas SENP6/SUSP1 is located in the cytosol (31). However, very little is known about the biological activities of SENP6/SUSP1 or SENP3/SMT3IP1. Although the ability of SENPs to reverse sumoylation is established, it remains to define the specificity of each SENP and to analyze the difference in each regulatory pathway mediated by these SENPs.
The androgen receptor (AR) is a ligand-regulated transcription factors belonging to the nuclear receptor superfamily (13, 39). It mediates the effects of androgen on the regulation of cell growth, differentiation, and maintenance of male reproductive functions (13, 39). AR can be subdivided into distinct functional domains: the N-terminal transactivation domain, the central DNA-binding domain, and the C-terminal ligand-binding domain (13, 24, 39). In the absence of ligands, AR locates primarily in cytoplasm and associates with heat shock proteins in an inactive state (13). Upon binding to ligand, AR undergoes a series of changes, including conformational change, translocation from the cytoplasm to the nucleus, and binding to a specific DNA sequence called the androgen response element (ARE) in the promoter regions of target genes to regulate transcription of these genes (13).
Like those of other nuclear receptors, the actions of AR are subjected to modulation by a large number of coregulators including coactivators and corepressors (24, 39, 51). These regulatory proteins are recognized by different functional domains of the AR and mediate transactivation (by coactivators) and transrepression (by corepressors) functions of AR (24). Coactivators function either as molecular bridges to enhance recruitment of the basal transcription machinery to the promoters of target genes or as factors that overcome the repressive effect of chromatin structure on transcription (16). The coactivators of AR include SRC-1, SRC-2/GRIP1, ACTR/AIB1/RAC3/pCIP, CBP, p300, and pCAF (16, 39). These coactivators possess histone acetyltransferase activity, which plays a role in chromatin remodeling to allow for active transcription of DNA (39). Conversely, corepressors including SMRT and N-CoR can attenuate AR-dependent transcription by recruiting histone deacetylases (HDACs) such as HDAC1 and HDAC2 in a repression complex (39, 43, 51). It has been shown that expression levels of some coregulators of AR were altered in the genesis and progression of prostate cancer, suggesting that they may be involved in the promotion or progression of prostate cancer through regulating AR activity (4, 35).
AR activity also can be regulated by posttranslational modification, such as phosphorylation and sumoylation (34, 48). AR is sumoylated in vivo at lysine residues 386 and 520 (48). Mutation of these residues increases the transactivation ability of AR, suggesting that sumoylation is involved in the regulation of AR activity (48). Interestingly, four AR coregulators, SRC-1, SRC-2/GRIP1, p300, and HDAC1, have also been found to be sumoylated (3, 5, 15, 32). SRC-1 has five sumoylation sites, and two major sites were localized in the NR box situated in nuclear receptor interacting region 1 (3). It is observed that sumoylation can increase the interaction of SRC-1 with the progesterone receptor. Two residues located in the nuclear receptor interacting region of SRC-2/GRIP1 were found to be sumoylated (32). Substitution at these two sumoylation sites could attenuate the activity of SRC-2/GRIP1 on AR-dependent transcription. In p300, two sumoylated sites located in the CRD1 domain are required for its transcriptional-repression function. Mutations that reduce SUMO modification increase p300-mediated transcriptional activity (15). HDAC1 was also found to be sumoylated (5). Mutation of two sumoylation sites of HDAC1 greatly reduced HDAC1-mediated transcriptional repression (5). It is unknown, however, whether desumoylation of these AR coregulators may be involved into the regulation of AR-mediated transcription.
Here, we showed that one of the SUMO-specific proteases, SENP1, profoundly enhances AR-dependent transcription. Both AR and HDAC1 were targets of SENP1, but the effect of SENP1 on AR-dependent transcription was mediated mostly through desumoylation of HDAC1. SENP1 could overcome the HDAC1 repressive function and reduce HDAC1 deacetylase activity. Thus, our data strongly support a role for SENP1 as a novel activator of AR-dependent transcription through desumoylation of HDAC1.
MATERIALS AND METHODS
Plasmids and antibodies.
Flag-AR, ARE-luciferase, Gal4-luciferase, Gal4-DBD, and hemagglutinin (HA)-SUMO-1 plasmids have been described previously (5, 30, 54). PB (−426/+28)-Luciferase, Flag-SENP1, Flag-SENP1 mutant (R630L, K631M), Flag-SENP2, His-SENP3, Flag-HDAC1, Flag-HDAC1 DM (K444, 476R), Gal4-DBD-HDAC1, Gal4-DBD-HDAC1 DM (K444, 476R), and Flag-AR DM (K386R, K520E) were prepared by standard cloning and PCR-based mutagenesis. Details of constructions are available upon request. Flag-SRC-1 was a gift from Ming-Jer Tsai (Baylor College of Medicine). The p300 expression plasmid was a gift from Yongzhong Wu (Virigina Commonwealth University). We used antibodies against Flag (M2; Sigma), HA (HA-7; Sigma), HDAC1 (2E10; Upstate Biotechnology), and actin (I-19; Santa. Cruz Biotechnology). An anti-AR antibody was raised against AR from a rabbit immunized with a bacterial recombinant AR N-terminal (amino acids 1 to 322) peptide.
Cell transfection and luciferase assays.
PC-3 and LNCaP cells were grown in phenol red-free RPMI 1640 (GIBCO-BRL) supplemented with 5% charcoal-dextran-stripped fetal bovine serum. COS-7 and HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. After 24 h of cultivation, these cells were transiently transfected with expression plasmids by Lipofectamine (Invitrogen) according to the manufacturer's instructions. Luciferase was assayed as described previously (52). β-Galactosidase activity was used as an internal control.
Immunoprecipitations and glutathione-Sepharose pull-down.
Cells (1 × 106) were lysed in 400 μl of lysis buffer (50 mM Tris [pH 7.5], 300 mM NaCl, 10 mM MgCl2, 0.5% NP-40, 0.3% Triton X-100, and protease inhibitors). The supernatant was cleared by centrifugation and immunoprecipitated by a specific antibody and protein A-Sepharose or pulled-down by glutathione-Sepharose.
Western blotting.
Western blotting was carried out as described in our previous publication (30).
PSA ELISA assay.
A PSA enzyme-linked immunosorbent assay (ELISA) kit was purchased from MP Biomedicals (Orangeburg, N.Y.). The cultural media of LNCaP cells were collected as samples for PSA ELISA according the manufacturer's instructions. The data were normalized to the total protein concentrations of samples.
Deacetylase assay.
An HDAC assay kit was purchased from Upstate Biotechnology and used according the manufacturer's instructions. Briefly, biotinylated histone H4 peptide, active PCAF, and [3H]acetyl-coenzyme A (CoA) in histone acetyltransferase assay buffer were incubated for 4 h at 30°C to label histone H4 with [3H]acetyl-CoA. Streptavidin-agarose was used to capture the labeled biotinylated histone H4 peptide. Flag-tagged HDAC and HDAC1 DM were immunoprecipitated with an anti-Flag M2 antibody from transfected HeLa nuclear extracts. After being washed with HDAC assay buffer, the beads were mixed with 50,000 cpm of captured streptavidin-agarose-[3H]acetyl-CoA-labeled histone H4 peptide in HDAC assay buffer and incubated for 5 h at 37°C. The counts of released [3H]acetate per minute in the supernatant were determined by liquid scintillation counting.
RNAi.
An HDAC1 small interfering RNA (siRNA) assay kit was purchased from Upstate Biotechnology for the RNA interference (RNAi) assay. The kit includes four pooled SMART-selected HDAC1 siRNA duplexes and nonspecific siRNA duplexes. For the SENP1 siRNA system, a 21-nucleotide SENP1 siRNA (GTGAACCACAACTCCGTATTC) was synthesized (Dharmacon). The same sequence in the inverted orientation was used as a nonspecific siRNA control. The SENP1 and nonspecific siRNA oligonucleotides were inserted into the pSuppressorNeo vector (IMGENEX Corporation) according to the manufacturer's instructions. PC-3 and LNCaP cells were grown in a 24-well plate. Cells were transfected with the oligonucleotides (40 pmol/well) or siRNA plasmid (200 ng) once (for PC-3) or three times within 12-h intervals (for LNCaP) with Lipofectamine 2000 (Invitrogen). PSA ELISA or a luciferase assay was performed.
RESULTS
SENP1 markedly enhances AR-dependent transcription.
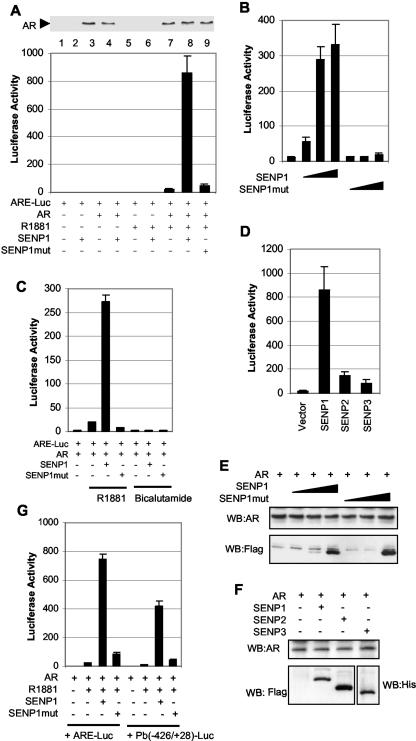
Since both AR and four of its coregulators are sumoylated, we speculated that one of the SUMO-specific proteases might play an important role in the regulation of AR-dependent transcription. For this purpose, we carried out a luciferase reporter gene assay to examine whether SENP1 could affect AR-dependent transcription. AR and ARE-luciferase reporter plasmids (ARE-Luc) were cotransfected into PC-3 cells with plasmids encoding either SENP1 or a catalytically inactive mutant SENP1. As shown in Fig. 1A, SENP1 dramatically enhanced AR transcriptional activity by 45-fold. This effect is dependent on the presence of AR ligand R1881. SENP1's catalytic activity is required for this effect, as the catalytically inactive mutant SENP1 (R630L, K631M) has a minor effect on AR-dependent transcription (Fig. 1A, bottom, lane 8 versus 9). Titration of SENP1 showed a dose-dependent effect of SENP1 on AR-dependent transcription (Fig. 1B). Even at very low levels of cotransfected DNA, SENP1 still could induce AR transactivation. In contrast, increasing levels of the mutant SENP1 did not alter significantly AR-dependent transcription (Fig. 1B), further validating the need for SENP1's enzymatic activity in the transcriptional regulation of AR. As the antagonists of androgen can also bind to the AR, we tested whether SENP1 could act on antagonist-bound AR. As shown in Fig. 1C, no effect of SENP1 on AR-dependent transcription was noted in the presence of bicalutamide. The enhancement of AR transcriptional activity by SENP1 is specific, as SENP2 and SENP3, the other members of the SENP family, only modestly enhanced AR-dependent transcription (Fig. 1D). Western analysis demonstrated that AR protein levels were not affected by exogenous expression of SENP1, mutant SENP1 (Fig. 1A and E), SENP2, or SENP3 (Fig. 1F). SENP1 and mutant SENP1 were expressed at similar levels (Fig. 1E), and SENP1 and SENP2 were also expressed similarly (Fig. 1F).
FIG. 1.
SENP1 markedly enhances AR-dependent transcription. (A) Enhancement of AR-dependent transcription by SENP1, but not by mutant SENP1. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase (50 ng) reporter plasmids in the absence or presence of wild-type or mutant SENP1 plasmids (150 ng). After 12 h of transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured. Transfection efficiency was normalized with a β-galactosidase expression construct, and the results are presented as activation over that for an empty vector. The expression level of AR was analyzed by Western blotting with the anti-AR antibody. (B) Dose response of SENP1 action. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase (50 ng) reporter plasmids in the absence or presence of increasing amounts of wild-type or mutant SENP1 plasmids (10, 50, and 150 ng). After 12 h of transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured as described for panel A. (C) SENP1 could not activate antagonist-bound AR. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase (50 ng) reporter plasmids in the absence or presence of wild-type or mutant SENP1 plasmids (150 ng). Twelve hours after transfection, cells were treated with 10 nM R1881 or 5 μM bicalutamide for 24 h, and the luciferase activity was measured as described for panel A. (D) SENP1, but not SENP2 or SENP3, markedly activates AR transactivation. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase (50 ng) reporter plasmids in the absence or presence of SENP1, SENP2, or SENP3 plasmids (150 ng). Twelvehours after transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured as described in for panel A. (E and F) Western blots of cell extracts from panels B (E) and D (F). (G) SENP1 enhances probasin promoter activity. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase or PB(−426/+28)-luciferase (50 ng) reporter plasmids in the absence or presence of wild-type or mutant SENP1 plasmids (150 ng). After 12 h of transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured as described for panel A.
We also used the promoter of rat probasin, another AR target gene-driven luciferase, to test the effect of SENP1 on AR transactivation. SENP1 could also increase AR-dependent transcription (Fig. 1G). Thus, the effect of SENP1 can be generalized to at least two AR-dependent promoter systems.
We also performed an ARE-luciferase reporter assay on AR-positive cell line LNCaP cells. In the presence of a ligand, expression of SENP1 induced endogenous AR activity by sevenfold (Fig. 2A). This effect was also dependent on SENP1 catalytic activity, as mutant SENP1 has a minor effect (Fig. 2A). We further examined the effects of SENP1 on endogenous androgen-responsive PSA gene expression in LNCaP cells. As PSA is a secreted protein, we used ELISA to examine PSA secretion in the cultural medium. As shown in Fig. 2B, the concentration of secreted PSA protein in the cultural medium was increased by R1881. The increase was further enhanced in the SENP1-transfected cell, but not in the mutant-SENP1-transfected cell (Fig. 2B). To further confirm the effect of SENP1 in regulation of AR-dependent transcription, we used siRNA to silence endogenous SENP1 in LNCaP cells. As expected, PSA expression was decreased in SENP1 siRNA-transfected cells (Fig. 2C). The level of endogenous SENP1 mRNA in LNCaP cells transfected with SENP1 siRNA plasmids was decreased by 53% (Fig. 2D). Collectively, these data strongly indicate that SENP1 acts as strong activator for AR-dependent transcription and that the catalytic activity of SENP1 is required for this action.
FIG. 2.
SENP1 increases PSA expression. (A) SENP1 induces endogenous AR-dependent transcription in LNCaP cells. LNCaP cells were transfected with ARE-luciferase reporter plasmids (50 ng) in the absence or presence of increasing amounts of wild-type or mutant SENP1 plasmids (150 ng). After 12 h of transfection, cells were treated with or without R1881 (10 nM) for 24 h, and the luciferase activity was measured as described for Fig. 1A. (B) SENP1 increases PSA expression in LNCaP cells. LNCaP cells were transfected with empty vector, SENP1, or mutant SENP1 expression plasmids and treated with or without R1881 (20 nM) for 24 or 48 h. The cultural media were collected as samples. (C) Silencing endogenous SENP1 decreases PSA expression. LNCaP cells were transfected with nonspecific siRNA (NS-siRNA) and SENP1 siRNA expression plasmids and treated with or without R1881 for 24 h. PSA released into the media was measured by ELISA. (D) Endogenous SENP1 is knocked down by RNAi. SENP1 levels in LNCaP cells transfected with either nonspecific siRNA or specific SENP1 siRNA plasmids were measured by real-time PCR, and each sample was run in triplicate. SENP1 mRNA expression was normalized to β-actin expression.
SENP1 induction of AR transcriptional activity occurs independent of AR desumoylation.
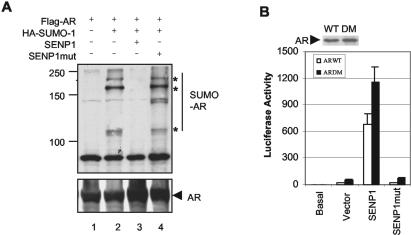
Since AR could be conjugated by SUMO (48), we examined whether AR could be desumoylated by SENP1 in vivo. Plasmids encoding Flag-tagged AR and HA-tagged SUMO-1 were transfected into COS-7 cells with plasmids encoding SENP1 or a catalytically inactive mutant SENP1. Immunoprecipitation with the anti-AR antibody followed by blotting with the anti-HA antibody showed that AR was sumoylated (Fig. 3A, top, lane 2) and that SUMO-1-AR conjugates were depleted in cells coexpressing SENP1 (lane 3), but not in cells coexpressing mutant SENP1 (Fig. 3A, top, lane 4). Immunoblotting of the same filter with the anti-AR antibody showed that AR was immunoprecipitated equally in all lanes (Fig. 3A, bottom). These results indicate that AR is a target protein of SENP1.
FIG. 3.
SENP1 desumoylates AR, but sumoylation of AR is not required for SENP1 activation. (A) SENP1 deconjugates SUMO-1 from AR in vivo. Whole-cell lysates from COS-7 cells transfected with the indicated plasmids were immunoprecipitated with the anti-AR antibody and analyzed by Western blotting with the anti-HA antibody (top) or anti-AR antibody (bottom). Asterisks, sumoylated proteins. (B) Sumoylation of AR is not required for SENP1 activation. PC-3 cells were transfected with either wild type AR (AR WT) or mutant AR (AR DM) plasmids (10 ng) and ARE-luciferase reporter plasmids (50 ng) in the absence or presence of either wild-type or mutant SENP1 plasmids. After 12 h of transfection, cells were treated with or without (basal) R1881 (10 nM) for 24 h, and the luciferase activity was measured as described for Fig. 1A. Expression levels of AR WT and AR DM were analyzed by Western blotting with the anti-AR antibody.
Since SENP1 can desumoylate conjugated AR, we speculated that SENP1 could regulate AR-dependent transcription by direct desumoylation of AR. We therefore generated an AR sumoylation mutant protein AR DM (K386R, K520E). The mutant AR was then compared to the wild-type AR in the ARE-luciferase reporter system. Consistent with a previous study (48), the mutant AR exhibited higher transcriptional activity than wild-type AR (2.8-fold; Fig. 3B). However, coexpression of SENP1 markedly enhanced the mutant AR's transcriptional activity by 24-fold compared to that for the vector control. SENP1's effect on the mutant AR was similar to that of the wild-type AR and was also dependent on its catalytic activity (Fig. 3B). These data suggest that most of SENP1's enhancement of AR-dependent transcription is independent of the sumoylation status of AR.
SRC-1 and p300 are not major targets for SENP1 action on AR-dependent transcription.
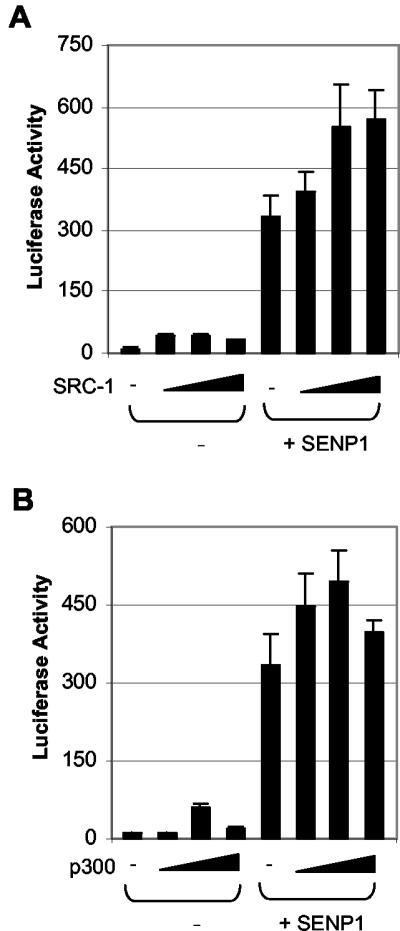
The transcriptional activity of AR could be modulated by coregulatory proteins. Because some AR coregulators, specifically SRC-1, SRC-2/GRIP1, p300, and HDAC1, are sumoylated (3, 5, 15, 24, 32, 39, 51), these coregulators would be the target for the SENP1 effect on AR-dependent transcription. SRC-2/GRIP1 is unlikely to account for SENP1's enhancement of AR-dependent transcription as the SRC-2/GRIP1(K239, 731,788R) mutation could attenuate the effect of SRC-2/GRIP1 on AR-dependent transcription (32); therefore, we focused on SRC-1, p300, and HDAC1.
We first examined whether SRC-1 or p300 could be involved in SENP1's enhancement of AR-dependent transcription by a cotransfection assay. As shown in Fig. 4A, SRC-1 alone could enhance AR-dependent transcriptional activity by ∼3-fold and SENP1 alone could enhance AR-dependent transcriptional activity by 40-fold. However, coexpression of SRC-1 with SENP1 could not significantly enhance SENP1's activity, suggesting that SRC-1 was not the major target in SENP1's enhancement of AR-dependent transcription. Similarly, coexpression of p300 had little effect on SENP1's activity, suggesting that p300 also did not play a significant role in SENP1's enhancement of AR-dependent transcription (Fig. 4B).
FIG. 4.
SRC-1 and p300 are not major targets for SENP1 action on AR-dependent transcription. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase (50 ng) plus SENP1 (150 ng) plasmids in the absence or presence of increased amounts of SRC-1 or p300 plasmids (10, 50, and 150 ng). After 12 h of transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured as described for Fig. 1A.
HDAC1 can be desumoylated by SENP1 in vivo.
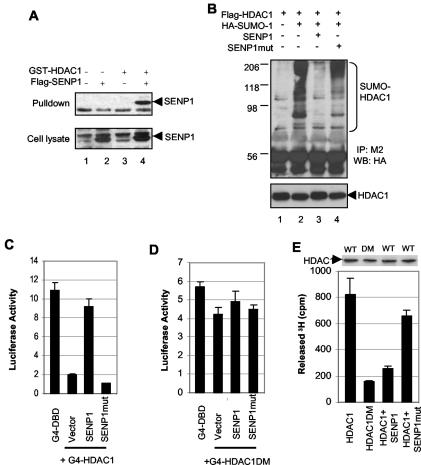
We next examined whether HDACs are responsible for SENP1 enhancement of AR-dependent transcription. There are at least 10 HDACs in the mammalian genome (7). Both HDAC1 and HDAC2 are involved in the formation of the repression complex for the AR (51), but only HDAC1 could be conjugated by SUMO (5). Thus, HDAC1 could be a potential mediator in SENP1's enhancement of AR-dependent transcription. To test this hypothesis, we first determined whether HDAC1 could associate with SENP1 in vivo. HDAC1 coprecipitated with SENP1 in cell extracts (Fig. 5A, lane 4). We further examined whether HDAC1 could be desumoylated by SENP1. We found that HDAC1 was sumoylated and that sumoylated HDAC1 could be deconjugated by SENP1 (Fig. 5B, top). This was dependent on SENP1's catalytic activity because mutant SENP1 could not deconjugate sumoylated HDAC1 (Fig. 5B, top). HDAC1 levels were also evaluated to ensure that immunoprecipitates are equal in all lanes (Fig. 5B, bottom).
FIG. 5.
SENP1 desumoylates HDAC1 and inhibits HDAC1 repressive activity. (A) SENP1 physically interacts with HDAC1 in vivo. Precipitates with glutathione-Sepharose (top) or whole-cell lysates (bottom) from COS-7 cells transfected with the indicated expression plasmid were analyzed by Western blotting with anti-Flag M2. (B) SENP1 deconjugates SUMO-1 from HDAC1 in vivo. Whole-cell lysates from COS-7 cells transfected with the indicated expression plasmids were immunoprecipitated with the anti-Flag M2 antibody and analyzed by Western blotting with the anti-HA antibody (top) or anti-HDAC1 antibody (bottom). (C and D) SENP1 overcomes HDAC1's repressive activity. PC-3 cells were transfected with Gal4-luciferase (100 ng) and either Gal4-DBD, Gal4-DBD-HDAC1, or Gal4-DBD-HDAC1 DM (100 ng) in the absence or presence of wild-type SENP1 or mutant SENP1 (SENP1mut) plasmids (150 ng). The luciferase activity was measured. (E) SENP1 reduces the deacetylase activity of HDAC1. HDAC1 or HDAC1 DM was immunoprecipitated by the anti-Flag M2 antibody from the nuclear extracts of HeLa cells transfected with Flag-HDAC1, Flag-HDAC1 DM, or Flag-HDAC1 plus either SENP1 or mutant SENP1 plasmids and assayed for HDAC activity. The immunoprecipitates were quantified by Western blotting with the anti-HDAC1 antibody.
SENP1 inhibits HDAC1's transcriptional repression.
We next examined whether SENP1 could affect HDAC1's transcriptional repression activity. Two approaches were used for this purpose. First we used a Gal4-DBD reporter system in PC-3 cells. HDAC1 tethered to DNA via fusion to Gal4-DBD was shown to repress the reporter gene activity driven by a minimal promoter harboring five Gal4 binding sites (5) (Fig. 5C). Coexpression of SENP1 strongly overcame the HDAC1-mediated transcriptional repression, whereas mutant SENP1 could not overcome this effect (Fig. 5C). To further determine whether SENP1 action is through the desumoylation of HDAC1, we generated a Gal4-DBD-fused mutant HDAC1 (HDAC1 DM), where Lys at two major sumoylation sites, 444 and 476, was replaced by Arg, and tested the activity using the same assay. The sumoylation site mutant HDAC has minimal repression activity (Fig. 5D). Coexpression of SENP1 had no effect on the repression of HDAC1 DM (Fig. 5D), suggesting that the SENP1 action is mainly mediated through the direct desumoylation of HDAC1. The second approach was to examine the effect of SENP1 on HDAC1 deacetylase activity, which is believed to be the major mechanism for repression of transcription. In the presence of SENP1, the deacetylase activity of HDAC1 precipitates had a 70% reduction (Fig. 5E). In contrast, expression of mutant SENP1 did not affect HDAC1's deacetylase activity (Fig. 5E). We also examine the deacetylase activity of HDAC1 DM and showed that this mutant HDAC had a markedly reduced HDAC activity (Fig. 5E). Collectively, these data indicated that SENP1 could inhibit the repressive effect of HDAC1 through desumoylation of HDAC1.
HDAC1 mediates SENP1 action on AR-dependent transcription.
The above results indicated that the effect of SENP1 on HDAC1 is dependent on SENP1's desumoylation activity. To further confirm that desumoylation of HDAC1 is required for SENP1 to overcome HDAC1's repressive effect on AR-dependent transcription, we compared the repressive effect of HDAC1 DM on AR-dependent transcription to that of wild-type HDAC1. As shown in Fig. 6A, while wild-type HDAC1 repressed the AR activity up to 90%, the mutant HDAC1 repressed AR transactivation less than 50%. These data indicate that SENP1's ability to inhibit HDAC1's repressive effect is mediated in part through desumoylation of HDAC1.
FIG. 6.
HDAC1 is required for SENP1's effect on AR transactivation. (A) Mutation of K444 and K476 relieves repression of HDAC1 on AR transactivation. PC-3 cells were transfected with AR (10 ng) and ARE-luciferase (50 ng) reporter plasmids in the absence or presence of Flag-HDAC1 or Flag-HDAC1 DM plasmids (50 ng). After 12 h of transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured as described for Fig. 1A. Expression levels of HDAC1 and HDAC1 DM were analyzed by Western blotting with the anti-Flag M2 antibody. (B) Endogenous HDAC1 is knocked down by RNAi. Nuclear extracts from PC-3 cells untransfected or transfected with either HDAC-1 specific RNAi duplexes or a nonspecific RNAi control were analyzed by Western blotting with the anti-HDAC-1 antibody (top) or antiactin antibody (bottom). (C) HDAC1 siRNA increases PSA expression. LNCaP cells were transfected with either HDAC-1 specific RNAi duplexes (HDAC1-siRNA) or nonspeciic RNAi control (NS-siRNA) and treated with or without R1881 (20 nM) for 24 h. PSA released into the media was assayed by ELISA. (D) Enhancement of AR-dependent transcription by SENP1 requires HDAC1. Empty vector, wild-type SENP1, or mutant SENP1 plasmids were transfected into PC-3 cells with AR and ARE-luciferase in the presence of either HDAC1-specific RNAi duplexes or a nonspecific RNAi control. After 24 h of transfection, cells were treated with 10 nM R1881 for 24 h, and the luciferase activity was measured as described for Fig. 1A.
To confirm that HDAC1 is required for the enhancement of AR-dependent transcription by SENP1, we determined the effect of SENP1 on AR-dependent transcription with HDAC1 siRNA duplexes to knock down the expression of endogenous HDAC1. The expression of endogenous HDAC1 was markedly decreased by transfection of HDAC1 siRNA oligonucleotides (Fig. 6B, lane 2) but not the nonspecific siRNA. We analyzed endogenous PSA expression in LNCaP cells after transfection with HDAC1 siRNA. As expected, PSA expression was enhanced by transfection of HDAC1 siRNA (Fig. 6C). When SENP1 was cotransfected with HDAC1 siRNA or nonspecific control siRNA, we observed only 9.5-fold enhancement of AR-dependent transcription by SENP1 in HDAC1-silenced cells (Fig. 6D). In contrast, nonspecific siRNA oligonucleotides did not interfere with the SENP1's enhancement of AR-dependent transcription (50-fold) (Fig. 5D). Furthermore, mutant SENP1 had no effect on AR-dependent transcription in both types of siRNA oligonucleotide-transfected cells. These data clearly demonstrated that SENP1 targets mainly HDAC1 to enhance AR-dependent transcription.
DISCUSSION
The SUMO pathway of posttranslational protein modification has been shown to be a major regulator of transcription (10, 14, 23, 25, 53). Most of the reports have focused on the effect of SUMO modification through the action of the conjugation enzyme Ubc9 or E3 ligases. However, very little is known about the SUMO deconjugating systems for the regulation of gene transcription. In mammalian cells, there are at least four different SUMO-specific proteases that have been identified (53). SENP1 was the first identified SUMO-specific protease; it is localized in the nucleus and can deconjugate sumoylated PML, but not RanGAP1 (19). SENP2 is localized in the nuclear envelope and also has activity against sumoylated PML, but not RanGAP1 (19, 22). Moreover, a truncated form of SENP2, called SuPr1, was shown to regulate Sp3 activity and alter PML distribution (2, 49). The biological activities of SENP3 and SENP6 are not known. They were classified as SENP family members because of the conserved catalytic domain. Both SENP1 and SENP2 are expressed at very low level in different cell lines surveyed (Yeh, unpublished data). It is of interest to study transcription systems that could be regulated by the SENPs in order to further understand the role of sumoylation in transcription regulation. Here, we use AR and its coregulators to demonstrate a potent regulatory activity of SENP1.
The AR-dependent transcription system provides an attractive model to study the regulatory function of desumoylation because, in this system, both AR and four of its coregulators, SRC-1, SRC-2, p300, and HDAC1, are conjugated by SUMO (3, 5, 15, 32, 48). Sumoylation, in general, has a suppressive effect on AR-dependent transcription (48). For example, a mutant AR that cannot be sumoylated has a threefold increase in its transcriptional activity compared to wild-type AR. However, overexpression of SENP1 could enhance AR-dependent transcription up to 45-fold. Thus, SENP1 most likely regulates the AR-dependent transcription pathway either at multiple steps or at a key step. First, we demonstrated that sumoylation of AR could not account for the SENP1 effect because the sumoylation-deficient mutant AR still can be activated by SENP1. Furthermore, we showed that SRC-1 and p300 were not likely to account for the marked enhancement of AR-dependent transcription by SENP1. Instead, most of the SENP1's effect seems to be directed against sumoylated HDAC1. We demonstrated that SENP1 could remove SUMO from modified HDAC1 and reduces its deacetylase activity. The role of HDAC1 sumoylation in AR-dependent transcription is based on several results. First, sumoylation is essential for HDAC1 repression function, as indicated in previous studies (5). SENP1 can deconjugate SUMO-1 from sumoylated HDAC1, hence decreasing HDAC1 repression capabilities. Second, numerous reports suggest that HDAC1 can strongly repress AR-dependent transcription (11, 12, 51), and our data indicate that SENP1 can overcome such repression. Third, SENP1 can inhibit HDAC1 deacetylation activity, which is required for its transcriptional repression. Fourth, SENP1's ability to enhance AR transactivation was reduced when endogenous HDAC1 was knocked down by siRNA.
The effect of SENP1 on AR-dependent transcription was also demonstrated with the endogenous AR-regulated protein PSA. We showed that PSA production was enhanced when SENP1, but not mutant SENP1, was expressed in LNCaP cells. PSA levels are used as a diagnostic marker for prostate cancer screening; in prostate cancer, AR activity is increased and hence PSA expression is also increased (6, 13). The enhanced AR activity is essential for cancer cell growth, as prostate cancer in most cases will undergo regression in response to androgen removal therapy (4, 13). Even in the androgen-refractory prostate cancer, AR still plays a critical role in the growth of tumor cells (4, 6, 13). However, the mechanism that underlies the regulation of the activity of AR in the tumor cells is not elucidated. AR gene amplification and mutation and cross talk with growth factor-stimulated signal transduction pathways have been proposed as possible mechanisms to facilitate AR translocation and activity (4, 13, 21). Our studies have demonstrated that SENP1 functions as a strong activator of AR to markedly enhance AR-dependent transcription. In preliminary studies, we found that the SENP1 message is increased in prostatic intraepithelial neoplasm and prostate cancer cells, but not in normal prostate tissues (unpublished data). Further studies will be required to determine the direct connection between overexpression of SENP1 and pathogenesis of prostate cancer by using transgenic mice model.
Acknowledgments
We thank Ming-Jer Tsai for the SRC-1 plasmid and Yongzhong Wu for the p300 expression plasmid.
This work was supported in part by NIH grants RO1 DK065156 (Z.W.) and RO1 CA 80089 (E.T.H.Y) and Department of Defense grant DAMS17-01-0097 (Z.W.).
REFERENCES
- 1.Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S. J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9:1169-1182. [DOI] [PubMed] [Google Scholar]
- 2.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 3.Chauchereau, A., L. Amazit, M. Quesne, A. Guiochon-Mantel, and E. Milgrom. 2003. Sumoylation of the progesterone receptor and of the coactivator SRC-1. J. Biol. Chem. 278:12335-12343. [DOI] [PubMed] [Google Scholar]
- 4.Culig, Z., H. Klocker, G. Bartsch, H. Steiner, and A. Hobisch. 2003. Androgen receptors in prostate cancer. J. Urol. 170:1363-1369. [DOI] [PubMed] [Google Scholar]
- 5.David, G., M. A. Neptune, and R. A. DePinho. 2002. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277:23658-23663. [DOI] [PubMed] [Google Scholar]
- 6.Debes, J. D., and D. J. Tindall. 2002. The role of androgens and the androgen receptor in prostate cancer. Cancer Lett. 187:1-7. [DOI] [PubMed] [Google Scholar]
- 7.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 9.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 10.Freiman, R. N., and R. Tjian. 2003. Regulating the regulators: lysine modifications make their mark. Cell 112:11-17. [DOI] [PubMed] [Google Scholar]
- 11.Fu, M., C. Wang, J. Wang, X. Zhang, T. Sakamaki, Y. G. Yeung, C. Chang, T. Hopp, S. A. Fuqua, E. Jaffray, R. T. Hay, J. J. Palvimo, O. A. Janne, and R. G. Pestell. 2002. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 22:3373-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaughan, L., I. R. Logan, S. Cook, D. E. Neal, and C. N. Robson. 2002. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 277:25904-25913. [DOI] [PubMed] [Google Scholar]
- 13.Gelmann, E. P. 2002. Molecular biology of the androgen receptor. J. Clin. Oncol. 20:3001-3015. [DOI] [PubMed] [Google Scholar]
- 14.Gill, G. 2003. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13:108-113. [DOI] [PubMed] [Google Scholar]
- 15.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. p300 Transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 16.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 17.Gong, L., T. Kamitani, K. Fujise, L. S. Caskey, and E. T. Yeh. 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272:28198-28201. [DOI] [PubMed] [Google Scholar]
- 18.Gong, L., B. Li, S. Millas, and E. T. Yeh. 1999. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448:185-189. [DOI] [PubMed] [Google Scholar]
- 19.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 20.Gostissa, M., A. Hengstermann, V. Fogal, P. Sandy, S. E. Schwarz, M. Scheffner, and G. Del Sal. 1999. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18:6462-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grossmann, M. E., H. Huang, and D. J. Tindall. 2001. Androgen receptor signaling in androgen-refractory prostate cancer. J. Natl. Cancer Inst. 93:1687-1697. [DOI] [PubMed] [Google Scholar]
- 22.Hang, J., and M. Dasso. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961-19966. [DOI] [PubMed] [Google Scholar]
- 23.Hay, R. T. 2001. Protein modification by SUMO. Trends Biochem. Sci. 26:332-333. [DOI] [PubMed] [Google Scholar]
- 24.Heinlein, C. A., and C. Chang. 2002. Androgen receptor (AR) coregulators: an overview. Endocr. Rev. 23:175-200. [DOI] [PubMed] [Google Scholar]
- 25.Hochstrasser, M. 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107:5-8. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 27.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The Polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 28.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 29.Kamitani, T., K. Kito, H. P. Nguyen, H. Wada, T. Fukuda-Kamitani, and E. T. Yeh. 1998. Identification of three major sentrinization sites in PML. J. Biol. Chem. 273:26675-26682. [DOI] [PubMed] [Google Scholar]
- 30.Kamitani, T., H. P. Nguyen, and E. T. Yeh. 1997. Preferential modification of nuclear proteins by a novel ubiquitin-like molecule. J. Biol. Chem. 272:14001-14004. [DOI] [PubMed] [Google Scholar]
- 31.Kim, K. I., S. H. Baek, Y. J. Jeon, S. Nishimori, T. Suzuki, S. Uchida, N. Shimbara, H. Saitoh, K. Tanaka, and C. H. Chung. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102-14106. [DOI] [PubMed] [Google Scholar]
- 32.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J. Biol. Chem. 277:30283-31288. [DOI] [PubMed] [Google Scholar]
- 33.Le Drean, Y., N. Mincheneau, P. Le Goff, and D. Michel. 2002. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology 143:3482-3489. [DOI] [PubMed] [Google Scholar]
- 34.Lee, H. J., and C. Chang. 2003. Recent advances in androgen receptor action. Cell. Mol. Life Sci. 60:1613-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, P., X. Yu, K. Ge, J. Melamed, R. G. Roeder, and Z. Wang. 2002. Heterogeneous expression and functions of androgen receptor co-factors in primary prostate cancer. Am. J. Pathol. 161:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 37.Li, S.-J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 39.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 40.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 41.Muller, S., M. Berger, F. Lehembre, J. S. Seeler, Y. Haupt, and A. Dejean. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275:13321-13329. [DOI] [PubMed] [Google Scholar]
- 42.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 43.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 44.Nishida, T., F. Kaneko, M. Kitagawa, and H. Yasuda. 2001. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J. Biol. Chem. 276:39060-39066. [DOI] [PubMed] [Google Scholar]
- 45.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 46.Nishida, T., and H. Yasuda. 2002. PIAS1 and PIASxα function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J. Biol. Chem. 277:41311-41317. [DOI] [PubMed] [Google Scholar]
- 47.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 48.Poukka, H., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2000. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc. Natl. Acad. Sci. USA 97:14145-14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 50.Saitoh, H., R. T. Pu, and M. Dasso. 1997. SUMO-1: wrestling with a new ubiquitin-related modifier. Trends Biochem. Sci. 22:374-376. [DOI] [PubMed] [Google Scholar]
- 51.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 52.Su, B., E. Jacinto, M. Hibi, T. Kallunki, M. Karin, and Y. Ben-Neriah. 1994. JNK is involved in signal integration during costimulation of T lymphocytes. Cell 77:727-736. [DOI] [PubMed] [Google Scholar]
- 53.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]
- 54.Yu, X., P. Li, R. G. Roeder, and Z. Wang. 2001. Inhibition of androgen receptor-mediated transcription by amino-terminal enhancer of split. Mol. Cell. Biol. 21:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]