Abstract
Transcriptional repressors often employ multiple activities, but the molecular mechanisms and physiological relevance of this functional diversity remain obscure. The Drosophila melanogaster Knirps repressor uses CtBP corepressor-dependent and -independent pathways. To separately analyze the components of Knirps repression activity, we elucidated the specific repression properties of CtBP and of Knirps subdomains. Like Knirps, CtBP represses adjacent transcriptional activators; but unlike Knirps, CtBP is unable to repress basal promoter elements. We determined that the ability of CtBP to recapitulate only a subset of Knirps activities is due to a quantitative, rather than qualitative, deficiency in repression activity. The CtBP-dependent portion of Knirps synergizes with the CtBP-independent repression activity to potently repress promoter elements from enhancer- or promoter-proximal positions. This result indicates that multiple repression activities are combined to exceed critical thresholds on target genes. CtBP mutant proteins unable to bind NAD fail to interact with DNA-bound factors. We show that DNA-binding Gal4-CtBP fusion proteins also require NAD binding for activity, indicating that NAD plays a role in repression at a step subsequent to CtBP recruitment to the promoter.
Transcriptional repression is an essential feature of gene regulation widely utilized in development and other biological processes. Repression is effected by a variety of transcription factors and cofactors, which utilize different pathways to mediate their function. Repression mechanisms include direct competition between repressors and activators, interactions of the repressors with the basal transcriptional machinery, and recruitment of different chromatin modification activities (7, 9, 22, 25). When and how these distinct mechanisms of repression are employed remains poorly understood.
A variety of transcriptional repressors have been demonstrated to possess multiple repression activities. In a few cases, the distinct repression activities appear to block specific types of transcriptional activators. For example, the mammalian Zeb protein represses the muscle-specific activator MEF2C via a CtBP (C-terminal binding protein)-dependent domain and c-Ets, c-Myb, and other activators via a CtBP-independent activity (28). In other cases, such as that with the NRSF repressor, different repression activities have been found to be utilized at distinct promoters (19). Multiple repression activities might also be invoked to increase the overall level of repression activity at a given promoter, much as activators have been suggested to contact multiple promoter and cofactor targets to synergistically potentiate transcription.
A broad functional distinction between Drosophila melanogaster transcriptional repressors based on their range of repression activity has been made (9). Short-range repressors (e.g., Knirps, Giant, Snail, and Krüppel) are active when bound close (≤100 bp) to activators within enhancers, and in experimental settings, these proteins can directly repress basal promoter elements over the same distances (2, 11, 15). The local activity of the repressor proteins permits functional autonomy of different enhancers in complex promoters, with short-range repressors acting on the element only to which they are directly bound (34). Long-range repressors (e.g., Hairy, Engrailed) can function over distances of greater than 1 kbp and are able to repress several enhancer elements simultaneously (4). The different activities of each class of repressors probably reflect distinct mechanisms employed; however, the molecular events that differentiate one type of repression from the other are not well understood. Similar differences in the ranges of repression are likely to exist for repressors of other metazoans, although this issue has not been thoroughly explored outside of Drosophila.
A common property of short-range transcriptional repressors is their interaction with the evolutionarily conserved corepressor CtBP. This transcription factor was originally identified in human cells through its interaction with the adenovirus E1A oncoprotein (5, 32, 43). CtBP proteins are homologous to NAD-dependent d-hydroxyacid dehydrogenases and possess very similar overall structures to these enzymes, as revealed by crystallographic studies (16, 21). CtBP proteins, like the homologous dehydrogenases, are homodimers, with each CtBP subunit consisting of a nucleotide-binding domain (which binds NAD/NADH) and a substrate-binding domain. This latter region interacts with peptides containing a PXDLS sequence motif found in many CtBP interacting transcription factors (21, 42). Key residues required for catalytic activity in NAD-dependent d-hydroxyacid dehydrogenases are absolutely conserved in CtBP proteins. These include the residues involved in NAD/NADH binding (GXGXXGX17D) and the catalytic triad of the active site (H, E, and R). Recent studies have shown that CtBP has a weak dehydrogenase activity in vitro, although the physiological substrates of CtBP as well as the significance of this enzymatic activity in transcriptional repression remain unknown (3, 16).
CtBP is recruited to promoter elements through interactions with repressors containing a PXDLS motif, where it mediates repression through mechanisms not currently understood. CtBP itself interacts with chromatin-modifying factors, including histone deacetylases and histone methyltransferases (33, 39, 40). Thus, the major activity of CtBP might be to serve as a bridging molecule, recruiting chromatin- or factor-modifying enzymes to the promoter. Alternatively, or in addition, its dehydrogenase activity might provide an additional mode of transcriptional repression. Dehydrogenase activities have not yet been linked to transcriptional repression, but a coactivator complex of the Oct-1 transcription factor has been found to utilize a dehydrogenase enzyme for the activation of the histone H2B promoter (46). CtBP might also possess an alternative enzymatic activity similar to that of the NAD-dependent Sir2 repressor protein, which requires NAD as a substrate to mediate the deacetylation of histones (20).
CtBP protein is found not only in the nucleus but also in the cytoplasm, where it is thought to participate in other cellular processes. CtBP was identified in the Golgi structure as a protein ribosylated after treatment with the fungal toxin brefeldin A. CtBP is reported to possess an acylation activity involved in Golgi fission, converting lysophosphatidic acid to phosphatidic acid, increasing membrane curvature during Golgi fission (36, 44). Subcellular localization of mammalian CtBP can be influenced by the expression of CtBP-binding partners as well as by sumoylation, a process regulated by the Polycomb protein PC2 (14, 18, 29).
In addition to CtBP-dependent activity, Knirps and other short-range repressors possess additional repressor activities. Short-range repressors are capable of directly competing with transcriptional activators having overlapping binding sites, and they also possess CtBP-independent repression activities that do not require direct competition. Recent work has established that certain enhancers are not effectively repressed if CtBP-dependent pathways are blocked, while the CtBP-independent activity is by itself sufficient to repress other cis regulatory elements (15, 17, 22, 38). It is not known whether the difference in CtBP sensitivity of certain enhancers reflects mechanistic differences in transcriptional activation pathways or, rather, quantitative differences in the levels of transcriptional repression activity required. Consistent with a quantitative model, the CtBP-dependent and CtBP-independent repression activities of Knirps have been found to exhibit striking functional similarities in cell culture assays, indicating that they might utilize similar mechanisms of repression (31).
Here, we examine the functional properties of CtBP itself as a short-range repressor as well as Knirps subdomains that are CtBP dependent or CtBP independent, employing transcription assays with transgenic embryos, where target genes embedded in the native chromosomal milieu are acted upon by endogenous activators. We find that the range of CtBP activity is similar to that seen for short-range repressors, but CtBP is unable to recapitulate the full spectrum of activities exhibited by Knirps. A comparison of the CtBP-dependent and -independent activities of Knirps reveals that CtBP makes a quantitative contribution to Knirps activity levels. An analysis of CtBP mutants suggests that NAD binding plays a key role in CtBP-mediated repression, possibly independent of a dehydrogenase activity.
MATERIALS AND METHODS
The following oligonucleotides were used in the preparation of the reporter genes and Gal4 chimeric repressor genes: DA44 (5′ACGTGGATACGATTAAGTATGCATG3′), DA45 (5′GTACTGCACCTATGCTAATTCATAC3′), DA65 (5′TCCATGATAAACGCGTGCTAGACTATTGCAGGTACTGATCGAATGCCTCTGCATG3′), DA66 (5′GTACAGGTACTATTTGCGCACGATCTGATAACGTCCATGACTAGCTTACGGAGAC3′), DA217 (5′GCTGGCAAGCCACGTTTGGTGG3′), DA228 (5′CGGGGTACCGCTGCCGCTGCAGCGGCTTCTGCTGCCGATGCCGCT3′), DA243 (5′GGGTCGGTACCCTGCCCCCACACCTCCTCTTCCCAGGCTAC3′), DA414 (5′GGGGAATCTAGACTAACTAATTACTACTTGTCATCGTCATCCTTGTAATCAGCGGCCGCCGGCGCCTCCGTTGACTCGGCC3′), DA415 (5′GGGGAATCTAGACTAACTAATTACTACTTGTCATCGTCATCCTTGTAATCAGCGGCCGCCTTTTCTTGATTTGATATCATT3′), DA418 (5′GGCCGCTGATTACAAGGATGACGATGACAAGGC3′), DA419 (5′GGCCGCCTTGTCATCGTCATCCTTGTAATCAGC3′), DA420 (5′ACGGAGTACTGTCCTCCGGAATTCCGGAGGACTGTCCTCCGCATG 3′), DA421 (5′CGGAGGACAGTCCTCCGGAATTCCGGAGGACAGTACTCCGTCATG3′), DA428 (5′GGGTCGGTACCATGGACAAAAATCTGATGATGCCGAAGCGTTCGCGCATCGATGTC3′), DA448 (5′CGCGTCGCTGAAGAAGGCGGCTTGCGGTGTGCAAATCAGATTTGGGGC3′), DA452 (5′GATTTGCACACCGCAAGCCGCCTTCTTCAGCGACGCGTCCGCAACG3′), DA515 (5′ACTCGCATATGTTGAGCATATGTTTTGGGGGATTTTCCCAAATCGAGGGAAAACCCAAGCATG3′), DA516 (5′CTTGGGTTTTCCCTCGATTTGGGAAAATCCCCCAAAACATATGCTCAACATATGCGAGTCATG3′), DA575 (5′ GGGTCGGTACCGGATCCCGCTACGGACGTCGC3′), DA576 (5′GGGTCGGTACCCACCACCATCATCAGCAGCAGCAGCAGCAGCAGGTGCCGCGTC3′), DA578 (5′GGGGAATCTAGACTAACTAATTACTACTTGTCATCGTCATCCTTGTAATCAGCGGCCGCGGGCGACAAGCTCTGCATCTTG3′), DA579 (5′GGGGAATCTAGACTAACTAATTACTACTTGTCATCGTCATCCTTGTAATCAGCGGCCGCTCCTTCTTGAGCGGAAACGG3′), DA580 (5′GGGGAATCTAGACTAACTAATTACTACTTGTCATCGTCATCCTTGTAATCAGCGGCCGCCACCTCCACTTCTTGATCCTC3′), DA581 (5′GGGGAATCTAGACTAACTAATTACTACTTGTCATCGTCATCCTTGTAATCAGCGGCCGCGACACACACGAATATTCCCCTCA3′), DA623 (5′CTACCAATGCGGGCCAGTGCCACCAGACCCAAGGTGTCGCCGCGG3′), DA624 (5′CCTTGGGTCTGGTGGCACTGGCCCGCATTGGTAGCGCCGTGGCCC3′), DA633 (5′GGGTCGGTACCCATAGGCCGACTAGTGGATC3′), DA634 (5′GGGTCGGTACCAAGCTTGCATGACGGAGTAC3′), and T3 (5′ATTAACCCTCACTAAAG 3′).
Construction of lacZ reporter genes. (i) CRTU(−55).
The CRTU vector is based on the gt-55 lacZ reporter gene, a C4PLZ-based transposition vector that contains rhomboid and twist enhancer elements driving expression of a lacZ reporter from the transposase basal promoter, as described previously (11). To create CRTU(−55), gt-55 was digested with SphI to remove the Giant binding sites located at −55 bp from the transcription start site, and the annealed DA420 and DA421 oligonucleotides containing two upstream activation sequence (UAS) binding sites (underlined areas in the DA420 sequence) were introduced. To create CRT4U(−55) and CRT6U(−55), CRTU(−55) containing the SphI site 5′ of the UAS sites was digested with SphI, and one or two additional copies of the DA420-DA421 oligonucleotide were inserted.
(ii) CtdU(−55).
Oligonucleotides DA515 and DA516, containing two Twist (boldface type) and two Dorsal (underlined type) binding sites, were introduced into the SphI site of C4PLZ, retaining the 3′ SphI site. The vector created was digested with SphI, and the DA420-DA421 fragment containing two UAS sites was introduced to generate CtdU(−55).
(iii) CRTtdU(−55).
CRTU(−55) was digested with SphI, cleaving 5′ of the two UAS sites, and the DA515-DA516 oligonucleotide containing Twist and Dorsal sites was introduced, generating CRTtdU(−55).
(iv) Ctd(spacer)U(−55) derivatives and CtdU(−130) and Ctd(spacer)U(−130) derivatives.
A 55-bp spacer (DA65-DA66 fragment) was introduced at the SphI site into CtdU(−55) between the Dorsal/Twist activator sites and the 2xUAS site. Clones with single, double, and triple insertions generated Ctd(55)U(−55), Ctd(110)U(−55), and Ctd(165)U(−55), respectively. Oligonucleotides DA633 and DA634 were used to amplify fragments containing the Dorsal/Twist(spacer) 2xUAS sequences by PCR using CtdU(−55), Ctd(55)U(−55), Ctd(110)U(−55), and Ctd(165)U(−55) as templates. The fragments obtained were digested with KpnI and introduced into C4PLZ into its KpnI site, generating CtdU(−130), Ctd(55)U(−130), Ctd(110)U(−130), and Ctd(165)U(−130).
Construction of the Gal4-dCtBP fusion proteins.
The cDNA of the longer 479-amino-acid isoform of Drosophila CtBP (dCtBP) (dCtBPL) was obtained from Y. Nibu and sequenced to verify the sequence of the 3′ region, which has not been previously reported. This cDNA is derived from an mRNA that differs from that of the shorter 383-amino-acid isoform of dCtBP (dCtBPS) by an alternative splicing event, which involves a splice donor site within codon 373 (glycine) being joined to a coding sequence represented by two exons 3′ of the annotated CtBP gene (first exon, beginning with leucine codon, CG 8850538-8850770; second exon, CG 8851129-8851210). CtBPL coding sequence was PCR amplified from pGEX-5X-3-dCtBPL (a gift from Y. Nibu) by using the oligonucleotides DA217 and DA414 as primers. The reverse primer DA414 introduces a NotI site after the last codon of dCtBPL followed by a FLAG epitope tag with the sequence DYKDDDDK, four stop codons in three different reading frames, and an XbaI restriction site. The fragment generated by PCR was cloned between the KpnI and XbaI sites of pBS(SK+)-NotI−, a pBluescript (SK+) derivative where the NotI site had been previously removed by mung bean nuclease treatment. A second FLAG epitope tag coding a sequence comprised of DA418 and DA419 was inserted in the NotI site. The double FLAG-tagged dCtBPL (dCtBPL-FF) was then excised from pBS(SK+)NotI− with KpnI and XbaI and inserted into pKreg by using the same restriction sites (described in reference 23), which contain the cDNA coding for residues 1 to 93 of Gal4. DA428 and DA415 primers were used to obtain the dCtBPS cDNA from pGEX-5X-3-dCtBPS (from Y. Nibu) by PCR, and the double FLAG-tagged gene was constructed in a fashion similar to that for dCtBPL.
dCtBPL mutants.
Single- or double-point mutations were introduced into dCtBPL-FF to generate the dCtBPL-FF-H315Q and dCtBPL-FF-G181A, G183A mutants. The mutations were generated by QuikChange site-directed mutagenesis (Stratagene) with the primers DA448 and DA452 for the H315Q mutant and the primers DA623 and DA624 for the G181A, G183A mutant, with pBS(SK+)NotI−-dCtBPL-FF used as a template. The DNA fragments encoding the CtBP mutants were excised by KpnI/XbaI digestion and introduced into pKreg KpnI/XbaI sites.
Construction of Gal4-Knirps fusion proteins.
cDNAs encoding Knirps residues 75 to 429, 75 to 330, 139 to 330, and 202 to 358 were obtained by PCR using pTwiggy-Kni75-429 (15) as a template. The pair of oligonucleotides used in each case is as follows: DA575-DA581 (Kni75-429), DA575-DA579 (Kni75-330), DA576-DA579 (Kni139-330), and DA228-DA580 (Kni202-358). All reverse primers introduce a NotI site after the Knirps coding sequence, followed by a FLAG tag sequence, four stop codons, and a XbaI site, similarly to the primers used to amplify dCtBPL. The different cDNAs were introduced into pBS(SK+)NotI− in order to introduce a second FLAG sequence, as described above with regard to dCtBPL, and were then cloned into pKreg as KpnI/XbaI fragments. A Knirps cDNA with mutations in the dCtBP-binding motif (PMDL to AAAA) was obtained from pTwiggy-Kni75-429mut (15) by PCR using DA575 and DA581. The fragment was inserted into pBS(SK+)NotI− and digested with NotI, which released a fragment encompassing residues 330 to 429. After ligation of a FLAG sequence with NotI cohesive ends, the released fragment was reinserted 5′ of the FLAG epitope sequence to regenerate Kni75-429ΔPMDL-FF. A KpnI/XbaI fragment was released from pBS(SK+)NotI−-Kni75-429ΔPMDL-FF and ligated into pKreg. The cDNA encoding Knirps 189-330 was obtained by PCR from pBS-(SK+)NotI−-Kni139-330-FF with primers DA243 and T3. The fragment of DNA was digested with KpnI/XbaI and cloned into pKreg in the same restriction sites.
P-element transformation, RNA in situ hybridization of embryos, and antibody staining.
All the P-element transformation vectors containing the reporter genes and the chimeric proteins were introduced into the Drosophila germ line by injection of yw67 embryos as described previously (35). For each gene construct, at least three separate lines were tested, and similar results were obtained. RNA in situ hybridization experiments were performed as described previously (35) by using digoxigenin-UTP-labeled antisense RNA probes to lacZ. Embryos were fixed for antibody staining as described previously (35), and FLAG-tagged proteins were detected with antibody staining with M2 anti-FLAG-tag antibody (Sigma) by using an Elite PK-62000 Universal Vectastain ABC kit (Vector Laboratories, Inc.).
RESULTS
CtBPL and CtBPS demonstrate similar repression activities.
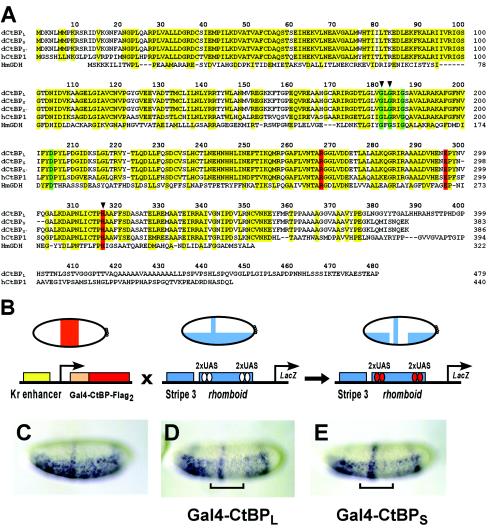
In Drosophila, there are at least three alternatively spliced transcripts of CtBP, predicted to produce proteins of 383, 386, and 479 amino acids in length, the longest form being a product of alternative splicing to two 3′ exons (23, 24, 27). Consistent with the presence of these transcripts, proteins of approximately 42 and 50 kDa are detected in embryo extracts by using α-CtBP polyclonal antibodies (P. Struffi, unpublished data). All forms of the protein share the highly conserved “dehydrogenase domain” also found in vertebrate CtBP proteins. The longer version of CtBP contains a nonconserved C-terminal extension of unknown function, which might be of regulatory importance. We have designated the 479-amino-acid isoform CtBPL and the 383-amino-acid isoform CtBPS (Fig. 1A).
FIG. 1.
CtBP protein sequences and functional assays in the Drosophila embryo. (A) Sequence alignment of CtBP proteins from D. melanogaster (the three dCtBP splice forms are designated L, S, and S′), hCtBP1, and the apoenzyme form of d-glycerate dehydrogenase from Hyphomicrobium methylovorum (HmGDH). Conserved residues in CtBP proteins, as well as those residues conserved in d-glycerate dehydrogenase, are indicated in yellow. Residues involved in NAD binding and the catalytic triad are displayed in green and red, respectively. Arrowheads indicate residues that were altered in the mutant forms of Gal4-CtBPL tested and shown in Fig. 7. (B) In vivo repression assay. Flies expressing Gal4-CtBPL or Gal4-CtBPS under the control of the Krüppel enhancer were crossed with flies containing the integrated eve stripe 3/rhomboid lacZ reporter transgene. Reporter gene expression was visualized in embryos by in situ hybridization with a lacZ antisense RNA probe. (C) Expression pattern of the unrepressed eve stripe 3/rhomboid lacZ reporter gene. (D and E) CtBPL (D) and CtBPS (E) exhibit similar levels of repression activity of the rhomboid enhancer (indicated by brackets). eve stripe 3 expression is unaffected, consistent with short-range repression. In this and subsequent figures, lateral or ventrolateral views of embryos are shown, anterior to left.
The CtBP corepressor is itself capable of mediating repression in cell culture assays and in the Drosophila embryo when tethered to the DNA via heterologous DNA-binding domains (6, 15, 16, 23, 24, 26, 42). To study the context dependence of CtBP repression, we created a series of P-element-based reporter gene targets that allowed us to test the repressor properties of CtBP in the native chromatin environment. Previous reports suggested that CtBPS might possess weaker repression activity than CtBPL, but these assays were conducted with different reporters, making a direct comparison difficult (23, 24). To directly compare the relative effectiveness of the CtBPL and CtBPS proteins, the Gal4 DNA-binding domain was fused to double FLAG epitope-tagged CtBPL and CtBPS, and proteins were expressed in a central domain of blastoderm embryos by using a Krüppel promoter element (Fig. 1B). The Gal4-CtBP proteins were assayed on lacZ reporter transgenes containing Gal4-binding UAS sites, allowing direct recruitment of Gal4-CtBP to the promoter. CtBPL and CtBPS were found to be equally effective at repressing a rhomboid enhancer containing internal Gal4 binding sites (Fig. 1C to E). The proteins did not inhibit an adjacent even-skipped stripe 3 element, indicating that they both exhibited short-range repression activity. As shown in Fig. 7, these proteins were expressed at similar levels and exhibited similar activity on other reporter transgenes, indicating that the presence or absence of the C-terminal extension does not affect repression activity in this assay.
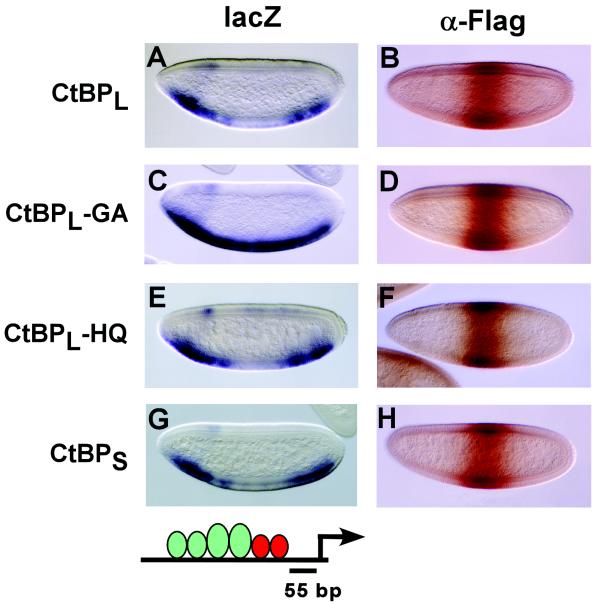
FIG. 7.
NAD binding is critical for repression by Gal4-CtBP, while the conserved histidine of the dehydrogenase active site is dispensable. The repression activities of Gal4-CtBPL, Gal4-CtBPS, Gal4-CtBPL-GA (G181A and G183A), and Gal4-CtBPL-HQ (H315Q), were assayed on the CtdU(−55) lacZ reporter. (A) The wild-type CtBP protein mediates robust repression activity in central regions of the embryo. (C) The mutation affecting NAD binding (G181 and G183) abolishes repression activity. (E) The mutation of the conserved histidine of CtBP does not affect repression. (G) Gal4-CtBPS effectively represses transcription of this reporter gene. (B, D, F, and H) Protein expression levels in central regions of the embryo were assayed by immunostaining with the M2 anti-FLAG antibody. At least four different lines of each CtBP mutant were analyzed for repression activity, and similar results were obtained in all cases.
CtBP is unable to mediate short-range repression from a promoter-proximal position.
Knirps and other short-range repressors are found to potently repress transcription when bound proximally to basal promoter elements, interfering with multiple enhancers that are located far beyond the previously identified range of short-range repression (∼100 bp). Under these circumstances, when the repressor sites are moved approximately 100 bp farther away from the transcription start site, repression is strongly attenuated or lost, suggesting that short-range repressors might be targeting the basal transcriptional machinery (2, 11, 15).
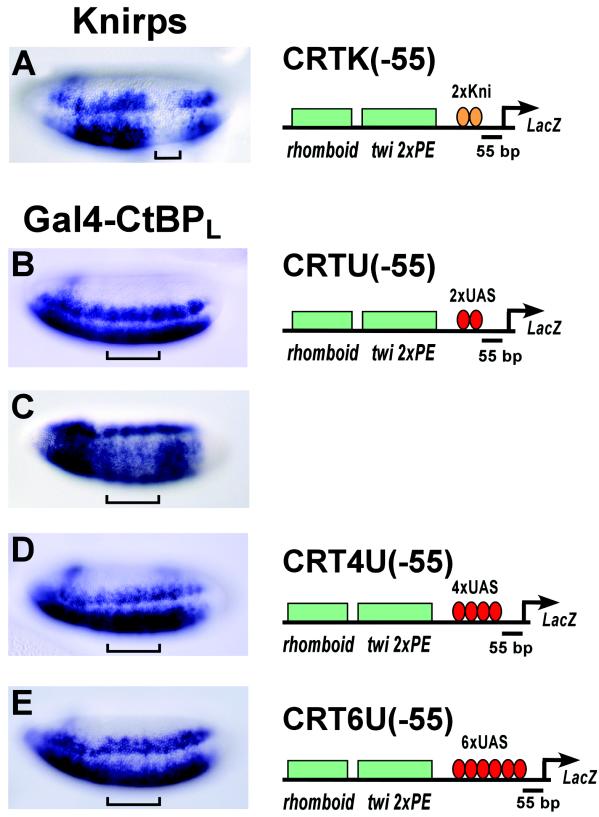
Endogenous Knirps protein bound to its cognate sites located at −55 bp mediated robust repression of a lacZ reporter gene, CRTK(−55), that was activated by the rhomboid and twist enhancer elements (Fig. 2A) (15). However, the Gal4-CtBPL protein (Fig. 2B) and the Gal4-CtBPS protein (data not shown) were virtually inactive when similarly positioned adjacent to the basal promoter in the CRTU(−55) reporter gene. Weak repression was detected in only a very small number of embryos (∼3%) (Fig. 2C). This result indicates that CtBP alone is incapable of recapitulating the full spectrum of Knirps activity. Many transcriptional activators show strong synergistic effects as the number of binding sites is increased. Repressors also can exhibit such effects. Repression by the Giant protein activity is enhanced by increasing the number of binding sites adjacent to a promoter (11). Therefore, we attempted to increase the quantity of Gal4-CtBPL protein bound to the promoter by increasing the number of Gal4 binding sites from 2 to 4 [CRT4U(−55)] or 6 [CRT6U(−55)]. However, Gal4-CtBPL repression activity was not markedly improved with these elements (Fig. 2D and E).
FIG. 2.
CtBP, unlike Knirps, fails to effectively repress from a promoter-proximal context. Knirps effectively represses transcription from a basal promoter when located −55 bp from the transcription start site (A). In the same context, Gal4-CtBPL shows no activity (B). Rarely, some embryos exhibited weak repression (C). Increasing the number of Gal4 binding sites in the promoter to 4xUAS or 6xUAS does not significantly enhance repression (D and E). Brackets indicate the positions where endogenous Knirps (A) and Gal4-CtBPL (B to D) are expressed in the embryos.
CtBP represses when situated proximally to transcriptional activators.
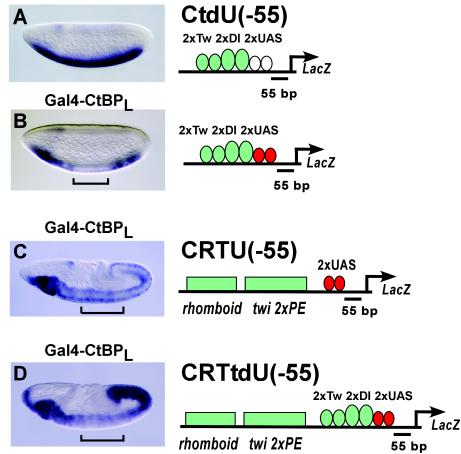
From the experiments shown above, we knew that Gal4-CtBP proteins were expressed and active in inhibiting the rhomboid regulatory element (Fig. 1D and E). However, the reporter genes tested (shown in Fig. 2) were not sensitive to repression, although they too are activated by Dorsal and Twist proteins. One potential difference between the two types of reporter transgenes is the distance of the Gal4 binding sites to the activators. In the case where we observe repression (Fig. 1), the most proximal Dorsal sites in the rhomboid enhancer are within 50 bp of the Gal4 binding sites, while the closest Dorsal site is >100 bp from the repressor sites shown in Fig. 2. We therefore created CtdU(−55), a lacZ reporter containing dual Twist/Dorsal binding sites immediately 5′ of the Gal4 binding sites. This transgene is driven in a robust ventral pattern (Fig. 3A), and in this situation, strong inhibition is seen by Gal4-CtBPL (Fig. 3B). Gal4-CtBPS was also active, as shown in Fig. 7. We conclude, therefore, that the position of the Gal4 binding sites near the basal promoter is permissive for Gal4-CtBP-mediated repression, as long as activators are located proximally to them. As demonstrated below, this repression is not due merely to steric hindrance or direct competition, for repression is maintained when repressor sites are moved 55 bp away, and inactive proteins fused to Gal4 do not repress.
FIG. 3.
CtBP repression is dependent on the presence of adjacent activators. (A) Expression pattern of the CtdU(−55) reporter containing two Twist and two Dorsal binding sites immediately 5′ of the UAS sites situated at −55 bp. (B) Gal4-CtBPL effectively represses transcription from the CtdU(−55) reporter, creating an unstained central gap (bracket). (C) Gal4-CtBPL does not repress transcription of the CRTU(−55) transgene in embryos at stages 6 and later in development. In the embryo shown (stage 8), staining posterior to the bracket is similar to that within the central domain, where Gal4-CtBP is expressed. (D) In embryos of similar age containing a reporter with proximal Twist and Dorsal sites in addition to the distal activator sites [CRTtdU(−55)], Gal4-CtBPL strongly represses Twist and Dorsal activities (bracket), leaving residual expression in the central region of the embryo, similar to that shown in panel C.
Because CtBP repression is enhanced by its immediate proximity to activators, we tested whether we could reverse the lack of CtBP activity observed in the CRTU(−55) reporter (Fig. 2B) by introducing additional proximal activator binding sites in this reporter. In the new reporter created, termed CRTtdU(−55), we introduced the Twist/Dorsal activator binding sites in the same position as in CtdU(−55) and compared the effect of CtBP-mediated repression on both of these reporter genes. Older germband-extended embryos containing the CRTU(−55) reporter, with only distal activator sites, exhibited weak lacZ expression that was not repressed by Gal4-CtBPL (Fig. 3C). The composite reporter CRTtdU (−55), which has activator binding sites arranged in both proximal and distal positions, showed no repression by both Gal4-CtBPL and Gal4-CtBPS in early blastoderm embryos (data not shown), but in older germband-extended embryos, attenuation of lacZ expression in central regions was evident in the presence of Gal4-CtBPL (Fig. 3D) and Gal4-CtBPS (data not shown). This result suggests that the proximal activator sites may be selectively repressed, leaving a level of signal typical of the more distal enhancer elements (Fig. 3D). This pattern of expression indicates that the two isoforms of the CtBP protein are capable of acting on local clusters of activators without directly repressing the basal promoter, as further experiments substantiate (see Materials and Methods).
Repression by Gal4-CtBP is dependent on proximity to activators, not to the basal promoter.
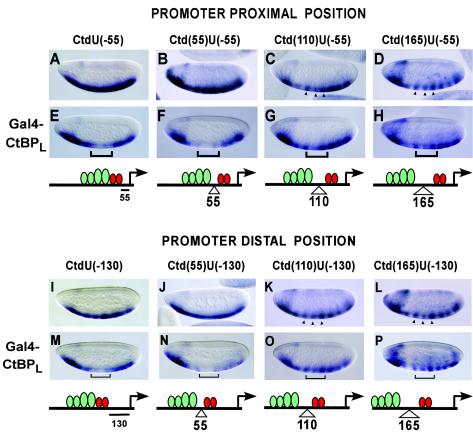
The activity of Gal4-CtBP shown on target genes containing adjacent activator sites suggests that this protein might be “quenching” the activators, that is, inhibiting their activity or their binding to DNA. To precisely determine the range of action of CtBP on neighboring activators, and to test whether the repression seen in the experiments shown in Fig. 3 depends on the proximity to activators or the basal promoter in a similar manner to short-range repressors, we used a series of reporter genes with variable spacing between activators and CtBP and between CtBP and the basal promoter. In the first series, the distance between CtBP and the promoter is maintained at −55 bp, while Twist/Dorsal binding sites are moved 55, 110, or 165 bp from the repressor by introducing 1, 2, or 3 copies of a 55-mer oligonucleotide between the activator binding sites and the UAS (Fig. 4A to H). This spacer DNA was used previously to study distance requirements for Giant and Knirps in a different promoter configuration (11, 15). In the second group, we tested activator distance dependence in a setting where CtBP is located at a more distal position, i.e., −130 bp from the transcription start site (Fig. 4I to P). This represents a distance where Knirps and Giant short-range repressors were found to be unable to effectively repress a basal promoter element (2, 11, 15). In these genes, Twist/Dorsal binding sites are located adjacent to or 55, 110, and 165 bp upstream of CtBP using the same 55-mer oligonucleotide.
FIG. 4.
Proximity to activators, but not to the basal promoter, is critical for CtBP repression activity. CtBP dependence on activators was tested on reporters containing two Twist and two Dorsal binding sites, where spacer DNAs (55, 110, and 165 bp) were introduced to move activator binding sites further 5′ of the two UAS sites. The activity of CtBP on this set of reporters was analyzed in both promoter-proximal (short-range) and promoter-distal (out of short-range) positions. (A to D and I to L) The embryos shown represent the expression pattern of the unrepressed reporter genes. (E, F, M, and N) CtBP-mediated repression is evident when activators sites are within 55 bp. (G, H, O, and P) Repression is absent when activators are 110 bp (G and O) or 165 bp (H and P) away from CtBP. Brackets indicate the region of the embryo where CtBP is expressed. Arrowheads show the central stripes of lacZ expression, which are not repressed by CtBP.
In a promoter-proximal position (−55 bp), Gal4-CtBPL mediates effective repression when activators are immediately adjacent or within 55 bp (Fig. 4E and F). The insertion of additional spacers, moving Twist/Dorsal activators 110 and 165 bp from the Gal4-CtBP binding sites, led to an unexpected striping in the expression pattern of the reporter gene (Fig. 4C and D), possibly due to the binding of pair-rule repressors to the inserted spacer sequences. However, individual portions of the expression pattern (Fig. 4) lie clearly in the central domain of the embryo where the Gal4-CtBP repressor is being expressed, allowing us to assay for repression in these regions. With these reporter genes, CtBP repression activity was strongly attenuated or absent, generally showing the same pattern as that of the reporter gene by itself (Fig. 4G and H).
With CtBP situated at a more distal position (−130 bp), where direct promoter-proximal effects are expected to be minimized, clear repression was again evident when activators were located within 55 bp (Fig. 4; compare panels I and J with M and N). No repression activity was detected when activators were moved 110 or 165 bp from the CtBP sites. Again, a strong striping in the reporter gene necessitated careful examination and comparison of the central stripes of expression (Fig. 4K, L, O, and P). Overall, similar patterns of repression are observed when Gal4-CtBPL is situated at both −55 and −130 bp from the transcription start site, suggesting that proximity to the activators, but not to the basal promoter, specifies CtBPL activity. These observations indicate that when CtBP is assayed out of its normal context (not as a component of a complex with DNA-bound transcriptional repressors that recruit it to the promoter), CtBP repression activity is dependent on the immediate proximity of activators.
Both CtBP-dependent and CtBP-independent repression activities contribute to Knirps promoter repression activity.
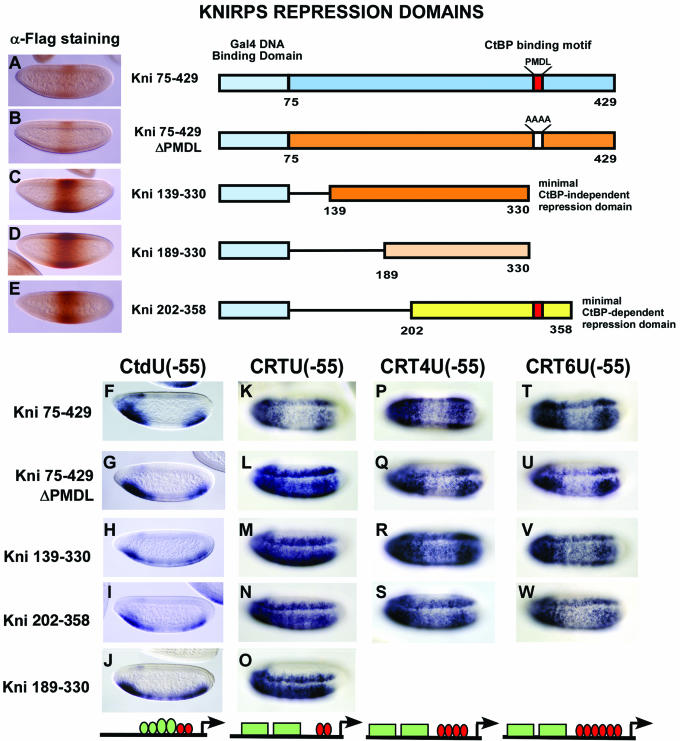
Knirps effectively represses transcription within enhancer elements and when situated adjacent to a basal promoter (Fig. 2A). Because the Knirps protein contains both CtBP-dependent and CtBP-independent repression activities, and CtBP itself fails to repress in the same context where Knirps is active (Fig. 2B), we postulated that Knirps might utilize its CtBP-independent activity to repress in a basal promoter context. To test this hypothesis, we assayed the CtBP-dependent and -independent domains of Knirps on the same reporters that were used to test Gal4-CtBP. Transgenic flies expressing Gal4-Knirps proteins representing either the CtBP-independent activity (Kni75-429ΔPMDL, Kni75-330, and Kni139-330), the CtBP-dependent activity (Kni202-358), or the full-length repression domain (Kni75-429) were analyzed. A negative control included a portion of Knirps previously shown to be inactive (Kni189-330).
The Gal4-Kni75-429 protein, containing the full-length repression domain, was able to repress the transcription of reporter genes containing adjacent activator binding sites as well as distal enhancer elements from a promoter-proximal position, [reporters CtdU(−55) and CRTU(−55), respectively], similar to the activity of the endogenous Knirps protein (Fig. 5F and K). In contrast, neither the CtBP-dependent nor the CtBP-independent domains by themselves were effective in blocking the activity of the rhomboid and twist enhancer elements in the CRTU(−55) reporter (Fig. 5L to N) although these repressors were active on the CtdU(−55) reporter with adjacent activator sites (Fig. 5G to I). Consistent with its encompassing the same CtBP-independent repression region of Knirps, Kni75-330 activity was similar to that of the minimal CtBP-independent domain Kni139-330 in all the reporters tested (data not shown).
FIG. 5.
Combined CtBP-dependent and CtBP-independent activities of Knirps mediate enhanced repression. (Top) Diagrams of the Gal4-Knirps repression domains analyzed are shown; the different regions that contain CtBP-independent (orange) and CtBP-dependent (yellow) repression activities or both (blue) are highlighted. Gal4-Kni189-330 (light orange) contains a portion of the CtBP-independent domain used as a negative control. The red box indicates the position of Knirps CtBP-binding motif (PMDL motif), which is mutated in Kni75-429ΔPMDL (PMDL→AAAA) and is shown as a white box. All constructs contain a double FLAG epitope tag at their C terminus. (A to E) Embryos expressing each of the Gal4-Knirps proteins indicated on their right were assayed for levels of protein by immunostaining with M2 anti-FLAG antibody. (F to W) Repression activities of Gal4-Knirps repressor genes containing proximal or distal activator sites. The full-length repression domain of Knirps as well as the individual repression domains were effective at mediating repression of the CtdU(−55) lacZ reporter containing proximal Twist and Dorsal activator sites (F to I). The full-length Knirps repression domain was effective, but the individual CtBP-dependent and CtBP-independent domains were ineffective, at repressing the CRTU(−55) reporter containing two UAS binding sites and distal Twist and Dorsal activator sites (K to N). Increasing the number of repressor binding sites from 2 to 4 or 6 within the CRTU lacZ reporter enhances repression by the weak CtBP-dependent and -independent domains (P to W). A portion of Knirps used as a negative control exhibited occasional weak activity on the CtdU(−55) reporter but not on CRTU(−55) (J and O). Activators are represented as green ovals and boxes, and repressors are represented as red ovals.
Repression of the rhomboid and twist enhancer elements by the Knirps subdomains in the context of the CRTU(−55) reporter was more effective when the number of Gal4 binding sites was increased from 2 to 4 or 6 (CRT4U and CRT6U reporter genes), suggesting a quantitative rather than qualitative deficiency (Fig. 5P to W). Repression by the subdomains, though, was still inferior to that of the full-length repression domain, judging by the frequency and extent of observed repression. Interestingly, the activity of the full-length repression domain which had been mutated to eliminate the CtBP binding motif, while weaker than the wild-type domain, was more effective than the minimal CtBP-independent repression domains, perhaps because this protein still retains some affinity for CtBP.
The portion of Knirps used as a negative control, Kni189-330, was previously shown to be inactive when assayed on a reporter containing even-skipped stripe enhancers (15) and, as expected, was inactive on CRTU. However, we observed occasional weak repression of the CtdU(−55) reporter (Fig. 5J), indicating that the protein may retain some residual activity, although at a very low level compared to those of the other Knirps constructs. This result also suggests that the CtdU(−55) reporter is particularly poised for repression. In other cases, proteins that were able to mediate some repression from multiple sites of the CRTU reporter were very active on the CtdU(−55) reporter; therefore, because of the poor activity on this reporter, we did not test the Kni189-330 construct on further CRTU reporter genes.
Differences in repression activity might reflect intrinsic activities of the proteins, or the differences might be a function of expression levels; therefore, we analyzed the expression levels of the Gal4-Knirps chimeras in the embryos (Fig. 5A to E). The staining intensities of the proteins indicated that the greater potency of the Knirps full-length repression domain (75-429) was not a function of higher levels of protein expression; in fact, these proteins were expressed at considerably lower levels than the individual subdomains (Fig. 5, compare A and B with C to E). A similar inverse relation between activity and expression levels was noted in a cell culture system expressing Tet-Knirps fusion proteins (31). mRNA levels from the different Gal4-Knirps genes were found to be similar by in situ hybridization analysis (data not shown), indicating lower translation levels or more rapid protein turnover of the Gal4-Kni75-429 and Gal4-Kni75-429ΔPMDL proteins.
In conclusion, neither individual repression domain showed preferential activity in a promoter-proximal position; rather, both domains demonstrated a restricted activity similar to that seen with Gal4-CtBP. These data contradict our original hypothesis that the CtBP-independent activity might explain the effectiveness of Knirps in a promoter-proximal position and suggest instead that the combination of the two activities provides a quantitatively superior level of repression required for effective regulation of some cis elements. This conclusion is supported by the ability of the CtBP-independent activity of Knirps to repress enhancers normally requiring CtBP when expressed at higher-than-normal levels (37).
Short-range repression mediated by the distinct Knirps subdomains.
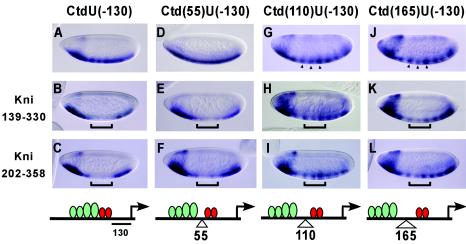
Previous studies have indicated that short-range repressors work over distances reaching approximately 100 bp to repress enhancers and basal promoters. The relevant distances have often been unclear, however, because assays have relied on elements with widely spaced activators and, in some cases, multiple repressor binding sites, making it difficult to determine which distances were relevant. To determine repression effectiveness on templates with more clearly defined activator-repressor positions, we tested the Knirps 139-330 and Knirps 202-358 subdomains on the lacZ reporters (shown in Fig. 4I to L) where repressor binding sites are situated at −130 bp and activators are located immediately 5′ or 55, 110, or 165 bp from the repressor binding sites (Fig. 6). Both CtBP-dependent and CtBP-independent portions of Knirps exhibited the same distance dependence seen with Gal4-CtBP. Repression was clearly observed with activators immediately adjacent to or within 55 bp of the repressors (Fig. 6A to F), and no repression was observed with more distally located activators (Fig. 6G to L). Recent studies have indicated that CtBP-dependent and -independent repression domains of Knirps exhibit similar properties on transiently transfected target genes in cell culture assays, possibly indicating a common mechanism of repression (31). The similar repressor activities of these domains on chromatinized reporter genes in transgenic embryos, both with respect to promoter context (Fig. 5) and activator proximity (Fig. 6), provide a further indication that the CtBP-dependent and -independent domains may act through similar repression pathways.
FIG. 6.
Knirps CtBP-dependent and CtBP-independent repression domains exhibit similar ranges of action. Gal4-Knirps proteins bound to UAS sites located at −130 bp were assayed for their ability to repress a lacZ reporter containing Dorsal and Twist activators at variable distances 5′ of the repressors. (A, D, G, and J) Reporter gene expression in unrepressed embryos. (B, E, H, and K) Repression activity of the Knirps minimal CtBP-independent domain 139-330. (C, F, I, and L) Repression activity of the Knirps minimal CtBP-dependent domain 202-358. Similar to the case with Gal4-CtBP, strong repression was noted only with activators located adjacent to or 55 bp from repressors but not at 110 or 165 bp, as expected for short-range activity. Brackets indicate the region of expression of the Gal4-Knirps proteins; arrowheads indicate the stripes of expression observed in the central regions for repression of 110- and 165-bp spacing constructs.
Mutation in the NAD binding domain of CtBP, but not in the catalytic site, abolishes transcriptional repression activity.
Previous work suggested that NAD binding to CtBP promotes multimerization of the protein in vitro (3) and permits interaction with the PXDLS-containing protein E1A, perhaps by facilitating a conformational change in the protein (16, 45). Mutations that interfere with NAD/NADH binding prevent CtBP from repressing a reporter gene activated by Gal4-E1A in cell culture, presumably because CtBP is not recruited to the promoter (16). A single glycine-to-glutamate point mutation of G183, the central glycine of the NAD binding motif (GxGxxG), interferes with NAD interaction, but it does not prevent CtBP interaction with the C-terminal region of E1A in glutathione S-transferase pull-down assays (21). In addition, when the same G183 is mutated to alanine, the mutant Gal4-CtBP protein can still repress transcription of a reporter activated by the simian virus 40 enhancer, as assayed in transient transfection assays performed in CtBP-knockout cells (10). Thus, at least in this context, NAD binding by CtBP appears to be dispensable for repression, but those previous studies did not address the question of whether NAD binding might be important for CtBP function on genes in their native chromatin environment. Therefore, we tested the repressor activity of a mutant form of Gal4-CtBPL containing alanine substitutions in two key glycine residues (G181A and G183A) (Fig. 1A), previously shown to be critical for NAD binding (21). This mutant form of CtBP was transcriptionally inactive compared to the wild-type protein (Fig. 7A and C). Its level of expression was comparable to that of the wild-type protein, indicating that a loss of activity in the NAD binding-defective mutant was not due to protein instability (Fig. 7 B and D).
A mutation that substitutes the catalytic histidine in d-lactate dehydrogenase destroys its enzymatic activity (41), and mutation of the conserved histidine in CtBP similarly abolishes its weak in vitro dehydrogenase activity (3). However, transient transfection assays in cell culture had not revealed any effect on CtBP repression when this residue was mutated, suggesting that CtBP may not require a dehydrogenase activity to mediate transcriptional repression (26, 42). We tested a mutant form of Gal4-CtBPL with a point mutation in the catalytic histidine (H315Q) (Fig. 1A). Consistent with the previous observations, this mutant protein was fully functional for repression (Fig. 7E) and was expressed at levels similar to those of the wild-type protein (Fig. 7 B and F). As in previous assays that tested mutant forms of CtBP in the presence of wild-type endogenous protein, we cannot rule out heterodimerization between mutant and wild-type forms of the protein, but such putative heterodimerization is clearly not able to rescue the activity of the NAD-binding mutant protein. As previously noted for the stripe3/rhomboid reporter, both CtBPL and CtBPS isoforms exhibited similar levels of repression activity in the CtdU(−55) reporter and were expressed at comparable levels (Fig. 7A, B, G, and H).
DISCUSSION
Quantitative role of CtBP in the multiple Knirps repression activities.
Knirps and other short-range repressors appear to be capable of mediating repression in at least three ways. One route of repression may consist of direct competition between repressors and activators with overlapping binding sites, as has been demonstrated for Knirps and Krüppel (13, 22). Although these experiments involved synthetic reporter genes, not complete endogenous regulatory regions, the close apposition of repressor and activator sites in native enhancers such as the even-skipped stripe 2 module indicates that competition might also be a factor in the regulation of endogenous elements. A clearly established second mode is the recruitment of the CtBP corepressor. In the absence of the CtBP protein or CtBP-binding capability of the repressor, Knirps, Krüppel, and other proteins are severely compromised in their ability to repress, and on some promoters or cis-regulatory elements, repression is abolished. This observation had led to the view that the only role of Knirps and Krüppel is to recruit CtBP. In this study, we show that Knirps subdomains, as does CtBP itself (Fig. 2, 3, and 4), can exhibit intermediate levels of activity that are sufficient for repression of some genes but not others; thus, the apparent complete dependence on CtBP for repression reflects repression activity dropping below a critical threshold rather than being totally absent. Finally, in addition to competition and CtBP recruitment, short-range repressors also possess CtBP-independent repression activities. Although this path has been suggested to involve only competition for DNA-binding sites, it is clear that the N-terminal region of Knirps contains an additional CtBP-independent repression activity that is not dependent on directly overlapping binding sites, as we have shown in this study and in previous work.
CtBP should be more realistically viewed as a major contributor to the complete spectrum of Knirps repression activities. Our experiments suggest that CtBP contributes quantitatively to the total repression output, allowing effective repression of particularly active cis-regulatory elements, while the CtBP-independent activity alone can suffice on less active elements. The combination of both repression domains makes a particularly potent repressor, demonstrating that both activities can be simultaneously mobilized at a single gene to effect repression, as shown in Fig. 5. On genes with a high ratio of activator sites to repressor sites, or in regions of the embryo where high activator concentrations saturate existing sites, both CtBP-dependent and CtBP-independent activities would be required (37). A similar situation exists for the Giant short-range repressor. CtBP-independent repression activity of Giant suffices for the repression of Krüppel in regions of limiting Bicoid activator concentration, while CtBP-dependent repression is required where Bicoid is more abundant (38).
Knirps repression activities: mechanisms and distances.
The similarities in ranges of action of both CtBP-dependent and -independent domains of Knirps as well as Gal4-CtBP itself (Fig. 4 and 6) suggest that these short-range repression activities might utilize similar molecular mechanisms of transcriptional repression, for example, targeting the same elements of chromatin. Such modifications might interfere with activator access to the DNA, resulting in a local inhibition. The weaker repression domains alone, including Gal4-CtBP itself, would still be effective against adjacent activator sites, which are apparently more sensitive to short-range activity. Endogenous Knirps protein or Gal4 fusion proteins containing both repression activities are also able to interfere with basal promoter function, suggesting that this context is a more demanding one for repressors, perhaps because basal factors are less sensitive to chromatin-mediated effects. Increasing the number of repressor binding sites allows even the weaker repressors to demonstrate some level of basal promoter repression, suggesting that the repression of activators and basal promoters might use similar mechanisms (Fig. 5). It is notable that the CtBP-dependent portion of Knirps is somewhat more potent than Gal4-CtBP itself (Fig. 2 and 5), possibly because this portion of Knirps is able to recruit CtBP in a more appropriate conformation or to interact with additional cofactors. We cannot distinguish at this level whether the same or different steps in the molecular process of repression are affected by each domain. The combination of the CtBP-dependent and CtBP-independent repressor domains, present in the full-length protein, produces higher levels of activity than the multimerization of the same type of domain. This synergy in repression suggests that the separate domains might also target separate processes during gene activation. However, because both domains act on similar distances (Fig. 6), both may be regulating the same process through common mechanisms. Additional mechanistic work will be required to determine if qualitative differences also distinguish the activity of the distinct repression domains and which molecular steps are regulated in each case.
The exact distance over which short-range repressors can interfere with activators has not been previously mapped because earlier distance dependence studies relied on the repression of groups of widely spaced activators, where relevant activator-repressor distances were not known. Other studies tested spacing from the start of transcription, potentially a less physiologically relevant situation, where it wasn't known from which element of the promoter the relevant distance should be measured. Our higher-resolution definition of distance requirements for Knirps repressor activity will serve a useful purpose for bioinformatic analysis of cis-regulatory sequences that are targeted by short-range repressors, allowing models to incorporate not only the presently utilized parameters of binding site density and affinity but also relevant activator-repressor spacing constraints.
Repression activity of CtBP isoforms.
The two isoforms of Drosophila CtBP that were analyzed (CtBPL and CtBPS) contain the conserved dehydrogenase domain and have similar repressor activities when tested as Gal4 fusion proteins, and both proteins are coexpressed during development. The possible significance of these different isoforms of the protein in Drosophila is not yet established. Vertebrate CtBPs encoded by distinct genes have been found to possess functional differences. Human CtBP1 (hCtBP1), but not hCtBP2, is sumoylated, a modification that results in nuclear localization of hCtBP1 (18). The mouse CtBP1 and CtBP2 genes are differentially regulated during embryonic development, and knockout mutants have different phenotypes, although some redundancy seems to exist between the two genes (12). Specific functions of CtBPL and CtBPS might be similarly regulated by distinct modifications or interactions involving the distinct C-terminal regions.
Role of NAD binding in CtBP repression activity: a catalytic role for CtBP?
Repression by CtBP might be mediated by associated corepressors, with CtBP functioning as a tethering molecule to deliver histone deacetylases, histone methyl transferases, and other transcription factors to the promoter (33). Alternatively, the protein might itself utilize a catalytic activity to modify chromatin or other factors and directly affect repression, possibly via the NAD-dependent dehydrogenase activity identified in vitro or by using NAD as a cofactor for histone deacetylation, similarly to the activity of Sir2 repressor proteins (20). The absolute conservation of catalytic residues in the presumptive active site in metazoan CtBPs speaks strongly for a catalytic function in some cellular process but not necessarily in transcription. For instance, CtBP is also found in the cytoplasm, where it has been suggested to function as an acyl transferase in the Golgi structure (44). In regard to its nuclear context, however, functional assays do not support a direct role for the active-site histidine of CtBP in transcriptional repression. Mutations affecting the catalytic histidine, analogous to those used to inactivate d-hydroxy acid dehydrogenases, do not inactivate chimeric CtBP proteins assayed for repression activity in cell culture assays (10, 26, 42). The histidine-to-glutamine mutant protein that we tested on chromosomally integrated reporter genes is similarly still functional for repression (Fig. 7). Our experiments cannot exclude the possibility that effects of the mutation were masked by heterodimerization with endogenous wild-type CtBP protein in the embryo. However, a CtBP protein mutated in its catalytic histidine retained the ability to repress endogenous target genes in CtBP knockout cells, showing that even in the absence of endogenous wild-type protein, the mutant protein can mediate its repressor function (10).
Extensive mutagenesis of CtBP affecting multiple residues in the catalytic site, dimerization interface, or NAD-binding surface results in a loss of interaction with the E1A C-terminal domain and, consequently, in the loss of repression of Gal4-E1A activated reporters (16). A limitation to these experiments was that in this study, there was no differentiation of effects of promoter targeting from repression activity. Relevant to this point, we observed a complete loss of transcriptional repression when two conserved glycines in the NAD-binding surface were mutated to alanine, within the context of Gal4-CtBP (Fig. 7). Mutations of the same glycine residues in homologous CtBP proteins have been shown to inhibit NAD binding (16, 21). Protein expression as assayed by antibody staining is not affected by this mutation, nor is it likely that the activity of the heterologous Gal4 DNA-binding domain would be sensitive to these point mutations in CtBP. In addition, a triple mutant containing substitutions of the conserved glycine residues is stable (16). Therefore, the lack of repression activity is very likely linked to a functional defect of some sort.
The inactivity of our Gal4-CtBP NAD binding mutant contrasts with results obtained from a recent study in which a Gal4-CtBP G183A mutant was found to retain function (10). The double mutant we tested might more effectively disrupt NAD binding, or the transient transfection might represent a more permissive environment for repression, than the chromosomally integrated genes tested in our study.
From our results, it is evident that NAD binding is also required for CtBP-mediated repression after its recruitment to the DNA. This requirement may reflect either an NAD-mediated enzymatic activity that is invoked during repression or NAD-induced allosteric changes in the protein that affect homodimerization or the recruiting of cofactors. Contrary to the previous results linking NAD binding to the ability of CtBP to contact the E1A protein, a recent report mapped the PXDLS binding motif to a region of CtBP located far from the NAD binding cleft. This study provided evidence that mutations that inhibit NAD binding do not necessarily prevent binding to the C-terminal region of the E1A protein (21). Therefore, the compromised activity of these mutant proteins that are unable to bind NAD might result from defective dimerization or cofactor recruiting, which would also be consistent with our results that NAD binding is critical at a step following CtBP recruitment to the DNA.
Goodman and colleagues provided evidence for a differential interaction of CtBP with NAD compared to that with NADH, suggesting that CtBP might function as a nuclear redox sensor, directly coupling oxidative states to gene regulation (8, 45). However, a marked difference in the binding of NAD to CtBP versus that of NADH to CtBP has not been observed in other in vitro studies (3, 16, 15). A similar redox-sensitive transcriptional switch mechanism has been proposed from studies of the Clock and c-Jun transcription factors as well as a recently described Oct-1 dehydrogenase coactivator protein (1, 30, 46). Therefore, whether NAD plays a role in allostery or in catalysis, it is possible that the CtBP contribution to short-range repression activity might be similarly regulated, resulting in a redox-responsive modulation of repression by Knirps and other factors.
Acknowledgments
We thank L. Kroos, Z. Burton, S. Triezenberg, and members of the Arnosti laboratory for comments on the manuscript; Y. Nibu and M. Levine for glutathione S-transferase-CtBP clones; and E. Fernandez-Villatoro for technical assistance, including image processing.
This study was supported by grant NIH GM56976 to D.N.A.
REFERENCES
- 1.Abate, C., L. Patel, F. J. Rauscher III, and T. Curran. 1990. Redox regulation of fos and jun DNA-binding activity in vitro. Science 249:1157-1161. [DOI] [PubMed] [Google Scholar]
- 2.Arnosti, D. N., S. Gray, S. Barolo, J. Zhou, and M. Levine. 1996. The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 15:3659-3666. [PMC free article] [PubMed] [Google Scholar]
- 3.Balasubramanian, P., L. J. Zhao, and G. Chinnadurai. 2003. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 537:157-160. [DOI] [PubMed] [Google Scholar]
- 4.Barolo, S., and M. Levine. 1997. hairy mediates dominant repression in the Drosophila embryo. EMBO J. 16:2883-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnadurai, G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 2:213-224. [DOI] [PubMed] [Google Scholar]
- 6.Criqui-Filipe, P., C. Ducret, S. M. Maira, and B. Wasylyk. 1999. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 18:3392-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deckert, J., and K. Struhl. 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol. Cell. Biol. 22:6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fjeld, C. C., W. T. Birdsong, and R. H. Goodman. 2003. Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc. Natl. Acad. Sci. USA 100:9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray, S., and M. Levine. 1996. Transcriptional repression in development. Curr. Opin. Cell Biol. 3:358-364. [DOI] [PubMed] [Google Scholar]
- 10.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. USA 100:4568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hewitt, G. F., B. S. Strunk, C. Margulies, T. Priputin, X. D. Wang, R. Amey, B. A. Pabst, D. Kosman, J. Reinitz, and D. N. Arnosti. 1999. Transcriptional repression by the Drosophila Giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development 126:1201-1210. [DOI] [PubMed] [Google Scholar]
- 12.Hildebrand, J. D., and P. Soriano. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22:5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoch, M., N. Gerwin, H. Taubert, and H. Jackle. 1992. Competition for overlapping sites in the regulatory region of the Drosophila gene Kruppel. Science 256:94-97. [DOI] [PubMed] [Google Scholar]
- 14.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 15.Keller, S. A., Y. Mao, P. Struffi, C. Margulies, C. E. Yurk, A. R. Anderson, R. L. Amey, S. Moore, J. M. Ebels, K. Foley, M. Corado, and D. N. Arnosti. 2000. dCtBP-dependent and -independent repression activities of the Drosophila Knirps protein. Mol. Cell. Biol. 20:7247-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar, V., J. E. Carlson, K. A. Ohgi, T. A. Edwards, D. W. Rose, C. R. Escalante, M. G. Rosenfeld, and A. K. Aggarwal. 2002. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol. Cell 10:857-869. [DOI] [PubMed] [Google Scholar]
- 17.La Rosee-Borgreve, A., T. Hader, D. Wainwright, F. Sauer, and H. Jackle. 1999. hairy stripe 7 element mediates activation and repression in response to different domains and levels of Kruppel in the Drosophila embryo. Mech. Dev. 89:133-140. [DOI] [PubMed] [Google Scholar]
- 18.Lin, X., B. Sun, M. Liang, Y. Y. Liang, A. Gast, J. Hildebrand, F. C. Brunicardi, F. Melchior, and X. H. Feng. 2003. Opposed regulation of corepressor CtBP by SUMOylation and PDZ binding. Mol. Cell 11:1389-1396. [DOI] [PubMed] [Google Scholar]
- 19.Lunyak, V. V., R. Burgess, G. G. Prefontaine, C. Nelson, S. H. Sze, J. Chenoweth, P. Schwartz, P. A. Pevzner, C. Glass, G. Mandel, and M. G. Rosenfeld. 2002. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298:1747-1752. [DOI] [PubMed] [Google Scholar]
- 20.Marmorstein, R. 2002. Dehydrogenases, NAD, and transcription—what's the connection? Structure (Cambridge) 10:1465-1466. [DOI] [PubMed] [Google Scholar]
- 21.Nardini, M., S. Spanò, C. Cericola, A. Pesce, A. Massaro, E. Millo, A. Luini, D. Corda, and M. Bolognesi. 2003. CtBP/BARS: a dual-function protein involved in transcription co-repression and Golgi membrane fission. EMBO J. 22:3122-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nibu, Y., K. Senger, and M. Levine. 2003. CtBP-independent repression in the Drosophila embryo. Mol. Cell. Biol. 23:3990-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nibu, Y., H. Zhang, E. Bajor, S. Barolo, S. Small, and M. Levine. 1998. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 23:7009-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nibu, Y., H. Zhang, and M. Levine. 1998. Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280:101-104. [DOI] [PubMed] [Google Scholar]
- 25.Nissen, R. M., and K. R. Yamamoto. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phippen, T. M., A. L. Sweigart, M. Moniwa, A. Krumm, J. R. Davie, and S. M. Parkhurst. 2000. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J. Biol. Chem. 275:37628-37637. [DOI] [PubMed] [Google Scholar]
- 27.Poortinga, G., M. Watanabe, and S. M. Parkhurst. 1998. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 17:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postigo, A. A., and D. C. Dean. 1999. Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol. Cell. Biol. 19:7961-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riefler, G. M., and B. L. Firestein. 2001. Binding of neuronal nitric-oxide synthase (nNOS) to carboxyl-terminal-binding protein (CtBP) changes the localization of CtBP from the nucleus to the cytosol: a novel function for targeting by the PDZ domain of nNOS. J. Biol. Chem. 276:48262-48268. [DOI] [PubMed] [Google Scholar]
- 30.Rutter, J., M. Reick, L. C. Wu, and S. L. McKnight. 2001. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293:510-514. [DOI] [PubMed] [Google Scholar]
- 31.Ryu, J. R., and D. N. Arnosti. 2003. Functional similarity of Knirps CtBP-dependent and CtBP-independent transcriptional repressor activities. Nucleic Acids Res. 31:4654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaeper, U., J. M. Boyd, S. Verma, E. Uhlmann, T. Subramanian, and G. Chinnadurai. 1995. Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc. Natl. Acad. Sci. USA 92:10467-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, Y., J. Sawada, G. Sui, E. B. Affar, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 34.Small, S., D. N. Arnosti, and M. Levine. 1993. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development 119:762-772. [PubMed] [Google Scholar]
- 35.Small, S., A. Blair, and M. Levine. 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 11:4047-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spano, S., M. G. Silletta, A. Colanzi, S. Alberti, G. Fiucci, C. Valente, A. Fusella, M. Salmona, A. Mironov, A. Luini, D. Corda, and S. Spanfo. 1999. Molecular cloning and functional characterization of brefeldin A-ADP-ribosylated substrate. A novel protein involved in the maintenance of the Golgi structure. J. Biol. Chem. 274:17705-17710. [DOI] [PubMed] [Google Scholar]
- 37.Struffi, P., M. Corado, M. Kulkarni, and D. N. Arnosti. 2004. Quantitative contributions of CtBP-dependent and -independent repression activities of Knirps. Development 131:2419-2429. [DOI] [PubMed]
- 38.Strunk, B., P. Struffi, K. Wright, B. Pabst, J. Thomas, L. Qin, and D. N. Arnosti. 2001. Role of CtBP in transcriptional repression by the Drosophila giant protein. Dev. Biol. 239:229-240. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian, T., and G. Chinnadurai. 2003. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 540:255-258. [DOI] [PubMed] [Google Scholar]
- 40.Sundqvist, A., K. Sollerbrant, and C. Svensson. 1998. The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett. 429:183-188. [DOI] [PubMed] [Google Scholar]
- 41.Taguchi, H., and T. Ohta. 1993. Histidine 296 is essential for the catalysis in Lactobacillus plantarum d-lactate dehydrogenase. J. Biol. Chem. 268:18030-18034. [PubMed] [Google Scholar]
- 42.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 17:5129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, J., and M. Crossley. 2001. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. BioEssays 23:683-690. [DOI] [PubMed] [Google Scholar]
- 44.Weigert, R., M. G. Silletta, S. Spano, G. Turacchio, C. Cericola, A. Colanzi, S. Senatore, R. Mancini, E. V. Polishchuk, M. Salmona, F. Facchiano, K. N. Burger, A. Mironov, A. Luini, and D. Corda. 1999. CtBP/BARS induces fission of Golgi membranes by acylating lysophosphatidic acid. Nature 402:429-433. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 295:1895-1897. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, L., R. G. Roeder, and Y. Luo. 2003. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114:255-266. [DOI] [PubMed] [Google Scholar]