Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome with at least 11 complementation groups (A, B, C, D1, D2, E, F, G, I, J, and L), and eight FA genes have been cloned. The FANCD1 gene is identical to the breast cancer susceptibility gene, BRCA2. The FA proteins cooperate in a common pathway, but the function of BRCA2/FANCD1 in this pathway remains unknown. Here we show that monoubiquitination of FANCD2, which is activated by DNA damage, is required for targeting of FANCD2 to chromatin, where it interacts with BRCA2. FANCD2-Ub then promotes BRCA2 loading into a chromatin complex. FANCD2−/− cells are deficient in the assembly of DNA damage-inducible BRCA2 foci and in chromatin loading of BRCA2. Functional complementation with the FANCD2 cDNA restores BRCA2 foci and its chromatin loading following DNA damage. BRCA2−/− cells expressing a carboxy-terminal truncated BRCA2 protein form IR-inducible BRCA2 and FANCD2 foci, but these foci fail to colocalize. Functional complementation of these cells with wild-type BRCA2 restores the interaction of BRCA2 and FANCD2. The C terminus of BRCA2 is therefore required for the functional interaction of BRCA2 and FANCD2 in chromatin. Taken together, our results demonstrate that monoubiquitination of FANCD2, which is regulated by the FA pathway, promotes BRCA2 loading into chromatin complexes. These complexes appear to be required for normal homology-directed DNA repair.
Fanconi anemia (FA) is an autosomal recessive disease characterized by cancer susceptibility and cellular hypersensitivity to DNA cross-linking agents, such as mitomycin C (MMC) and cisplatin. Eight FA genes have been cloned, corresponding to FA subtypes A, C, BRCA2/D1, D2, E, F, G, and L. The encoded FA proteins cooperate in a common pathway—the FA/BRCA pathway (6, 15). Six of the FA proteins (A, C, E, F, G, and L) assemble in a multisubunit nuclear complex (8, 18, 19), required for the activation (monoubiquitination) of the downstream FANCD2 protein (9). Whether the E3 ubiquitin ligase, BRCA1, also participates in the monoubiquitination of FANCD2 remains unclear (28). The activated FANCD2 protein is subsequently targeted to nuclear foci (9).
Whether BRCA2 participates with other FA proteins in this pathway has remained uncertain. On the one hand, BRCA2−/− patients share most of the clinical and cellular phenotypic features of other FA subtypes, suggesting a common pathway (13). On the other hand, BRCA2-deficient cells have a more severe defect in homologous recombination repair (21, 30), suggesting that BRCA2 may have functions independent of the FA pathway. Also, BRCA2−/− patients generally have a more severe clinical phenotype, with earlier onset of cancer (11). Although two-hybrid studies suggest that BRCA2 may interact with other FA proteins, such as FANCG (14), direct biochemical evidence linking BRCA2 to other FA proteins is lacking.
Unlike other FA proteins, BRCA2 has a well-defined role in homology-directed DNA repair (HDR). BRCA2 binds to single- and double-stranded DNA (31), interacts directly with RAD51 (4, 17), modulates RAD51 activity in vitro (7), and regulates the level of HDR (21, 27). BRCA2 is also required for the stabilization of stalled DNA replication forks (16), a process that could be related to the role of BRCA2 in HDR. Evidence of a biochemical interaction between BRCA2 and another FA protein may link the pathway to the BRCA2-mediated process of homologous recombination.
In the present study, we show that monoubiquitinated FANCD2 assembles with BRCA2 and another FA protein, FANCE, in a functional chromatin complex. DNA damage-inducible monoubiquitination of FANCD2 is required for its targeting to chromatin, and monoubiquitinated FANCD2 then promotes the assembly of ionizing radiation (IR)-inducible BRCA2 foci and chromatin loading of BRCA2. Moreover, the carboxy terminus of BRCA2 is required for the interaction of BRCA2 and FANCD2. An FA patient-derived mutant form of BRCA2 lacking amino acids 3226 to 3418 fails to colocalize or coimmunoprecipitate with FANCD2. The presence of BRCA2 and FANCD2 chromatin complexes suggests a link between the FA pathway and the process of homologous recombination DNA repair in mammalian cells.
MATERIALS AND METHODS
Cell culture.
The simian virus 40-transformed fibroblasts, PD20 (FA-D2), EUFA423 (FA-D1), and GM6914 (FA-A), as well as HeLa and U2OS cells, were grown in Dulbecco's modified Eagle's medium supplemented with 15% heat-inactivated fetal calf serum (FCS) in a humidified 5% CO2 incubator at 37°C. PD20 fibroblasts expressing either empty vector, full-length FANCD2 cDNA, or the FANCD2(K561R) mutant were described previously (9). AT22IJE-T fibroblasts (AT) expressing either empty vector or full-length ataxia telangiectasia mutated (ATM) cDNA (AT + ATM) were described previously (33). EUFA423 fibroblasts stably expressing empty vector or the hemagglutinin (HA) tag plasmid pcDNA3 HA-BRCA2 (EUFA423 + HA-BRCA2), and chromosome 13-transfected FA-D1 cells (EUFA423 + BRCA2) were described previously (13). BRCA2−/− CAPAN-1 cells were grown in Iscove's modified Dulbecco's medium with 15% heat-inactivated FCS. Epstein-Barr virus-transformed lymphoblasts PD7 (wild-type), HSC230 (FA-B), and EUFA130 (FA-E) were maintained in RPMI 1640 with 15% FCS. EUFA130 lymphoblasts expressing full-length HA-FANCE were described previously (23).
Generation of DNA damage.
Gamma irradiation was delivered with a Gammacell 40 apparatus (MDS Nordion, Ottawa, Canada). For MMC (Sigma Chemical) treatment, cells were continuously exposed to the drug for the indicated times. MMC sensitivity assays of human fibroblasts and lymphoblasts have been described previously (9).
Transient transfection.
EUFA423 (FA-D1) cells were transfected with the plasmids pcDNA3 and pcDNA3 HA-BRCA2 (13), using FuGENE 6 (Roche), according to the manufacturer's protocol.
Preparation of cellular fractions.
Cells were grown on 15-cm-diameter culture dishes and were exposed to IR (15 Gy, 15 h) or MMC (170 ng/ml, 15 h). Cells were trypsinized and washed with cold phosphate-buffered saline (PBS), aliquoted equally into four Eppendorf tubes, and collected by centrifugation at 1,200 rpm for 3 min at 4°C in a Sorvall RT 6000D centrifuge. One pellet, representing the whole-cell extracts (P1), was frozen in liquid N2. The remaining three pellets were resuspended in cold buffer A (10 mM PIPES, pH 7, 100 mM NaCl, 1 mM EGTA, 300 mM sucrose, 0.5 mM sodium orthovanadate, 50 mM sodium fluoride, 10-μg/ml aprotinin, 10-μg/ml leupeptin, 10-μg/ml pepstatin A, 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 0.5% Triton X-100 and were incubated at room temperature (RT) for 2 min to permeabilize the cells. The suspension was then centrifuged as described above, and the supernatant (S2; detergent-soluble nuclear proteins) was collected and frozen. Pellets were washed with cold buffer A. One pellet (P2; detergent-insoluble nuclear proteins) was frozen. Nuclei were then digested with RNase-free DNase I (200 U/ml) (Roche) in buffer A for 30 min at RT. The residual pellet was collected by centrifugation as described above. The supernatant (S3; DNase I-sensitive proteins) from one tube was frozen, and all pellets were washed with cold buffer A. One residual pellet (P3; DNase I-resistant protein) was frozen. The remaining residual pellet was extracted with cold buffer A containing 250 mM ammonium sulfate for 5 min at RT to extract the remaining chromatin. The supernatant (S4; chromatin) was collected following centrifugation at 3,000 rpm for 3 min and was then frozen. Cell-equivalent volumes from each cell fraction were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotted with antibodies.
Immunoblotting.
Cells were fractionated, and whole-cell extracts were subjected to 6% SDS-PAGE or 3 to 8% NuPAGE Tris-acetate (Invitrogen) gel electrophoresis, transferred to nitrocellulose membranes, and subjected to Western blot analysis (9). The following antibodies were used: anti-BRCA2 (monoclonal Ab-1 and polyclonal Ab-2) (Oncogene Research Products), anti-FANCD2 (affinity-purified E35 polyclonal antibody and FI-17; Santa Cruz Biotech.), anti-HA (HA.11; Babcock), anti-histone H4 (Santa Cruz Biotech.), anti-FANCE (a generous gift from Grover Bagby, Oregon Health and Science University, Portland), and anti-ATM (Novus Biologicals).
Coimmunoprecipitation.
For immunoprecipitation from whole-cell extracts, cells were lysed with lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with protease inhibitors (1-μg/ml leupeptin and pepstatin, 2-μg/ml aprotinin, 1 mM PMSF) and phosphatase inhibitors (1 mM sodium orthovanadate, 10 mM sodium fluoride). Immunoprecipitation of protein (6 mg) from whole-cell extracts or of chromatin proteins extracted as described above was performed by using the monoclonal antibody to BRCA2 (Ab-1), polyclonal antibody to BRCA2 (H-300) (Santa Cruz Biotech.), or polyclonal antibodies to FANCD2 (E35). As a negative control, preimmune rabbit serum or normal mouse immunoglobulin G (IgG) was used. The lysates were precleared with protein A-Sepharose for 30 min at 4°C. Cleared extracts were then incubated with antibodies overnight at 4°C before protein A-Sepharose was added for 2 to 4 h. The immunoprecipitated materials were washed extensively with lysis buffer and analyzed by 6% SDS-PAGE or 3 to 8% NuPAGE Tris-acetate gel electrophoresis, followed by immunoblot analysis with the indicated antibodies and enhanced chemiluminescence detection with the ECL system.
Detection of monoubiquitinated FANCD2 in chromatin.
HeLa cells were transfected with a HA-tagged ubiquitin expression vector (pMT 123) using FuGENE 6 (Roche) as previously described (9). After transfection, cells were treated with the indicated dose of IR and MMC and then were fractionated into S2 (soluble nuclear proteins) and S4 (chromatin) fractions. These two fractions from HeLa cells were subjected to immunoprecipitation with a polyclonal antibody (E35) to FANCD2. Immune complexes were run on 6% SDS-PAGE transferred to nitrocellulose, and immunoblotted with anti-FANCD2 (FI-17) or anti-HA (HA.11) monoclonal antibodies.
Immunoprecipitation and phosphatase assay.
To determine whether shifts of BRCA2 mobility on immunoblots corresponded to protein phosphorylation, BRCA2 was immunoprecipitated with monoclonal antibody (Ab-1) and treated with alkaline phosphatase as previously described (9). Samples were separated by 3 to 8% NuPAGE Tris-Acetate (Invitrogen) gel electrophoresis and immunoblotted with polyclonal antibody to BRCA2 (Ab-2).
Immunofluorescence microscopy.
Preparation of cells for immunofluorescence microscopy was performed essentially as previously described (9). Cells were prepermeabilized with 0.25% Triton X-100 in PBS for 1 min on ice and then fixed with 4% paraformaldehyde in PBS for 15 min, followed by permeabilization with 0.5% Triton X-100 in PBS for 1 min. For immunofluorescence microscopy, fixed cells were incubated with specific primary antibodies at the appropriate dilution in 3% bovine serum albumin-PBS for 1 h at RT. FANCD2 was detected with the affinity-purified E35 polyclonal antibody or monoclonal antibody FI-17. BRCA2 was detected with monoclonal antibody (Ab-1) or polyclonal antibody (Ab-2). Anti-HA monoclonal antibody (HA.11) was used to detect overexpressed HA-tagged BRCA2; RAD51 was detected with anti-RAD51 monoclonal antibody (Oncogene Research Products). Cells were subsequently washed three times in PBS and incubated for 1 h at RT with species-specific fluorescein (Jackson Immunoresearch) or Texas red-conjugated (Amersham) secondary antibodies diluted in 3% bovine serum albumin in PBS. Cells were counterstained with 4′6-diamidine-2-phenylindole dihydrochloride (Roche) to visualize nuclei. Slides were mounted in Vectashield (Vector Laboratories). Images were acquired using an Axioplan 2 imaging microscope (Carl Zeiss) equipped with a digital camera and processed by using Openlab software.
RDS assay.
The radioresistant DNA synthesis (RDS) assay was performed as described previously (25). Briefly, cells were plated in 60-mm-diameter plates, incubated with 10 nCi of [14C]thymidine for 24 h, and cultured in the absence of labeling medium for 24 h. The cells were then treated in replicate with 0, 4, or 8 Gy of IR. At 30 min following DNA damage, cells were incubated with 2.5-μCi/ml [3H]thymidine for 15 min. Cells were fixed, loaded onto glass fiber filters, and counted as previously described (20).
RESULTS
DNA damage activates the colocalization, cofractionation, and coimmunoprecipitation of BRCA2 and FANCD2.
In the absence of DNA damage, BRCA2 and FANCD2 were expressed diffusely in the nuclei of HeLa cells (Fig. 1A). IR at 2 or 15 Gy, or MMC exposure, activated the assembly of BRCA2 and FANCD2 foci, and these foci showed a >80% colocalization (Fig. 1A, merge). This strong colocalization of FANCD2 and BRCA2 following IR or MMC treatment suggested that the proteins may be loaded together into chromatin complexes, perhaps at sites of DNA damage. Consistent with this notion, phosphorylated histone H2AX also colocalized with FANCD2 and BRCA2 following exposure to IR or MMC (data not shown).
FIG. 1.
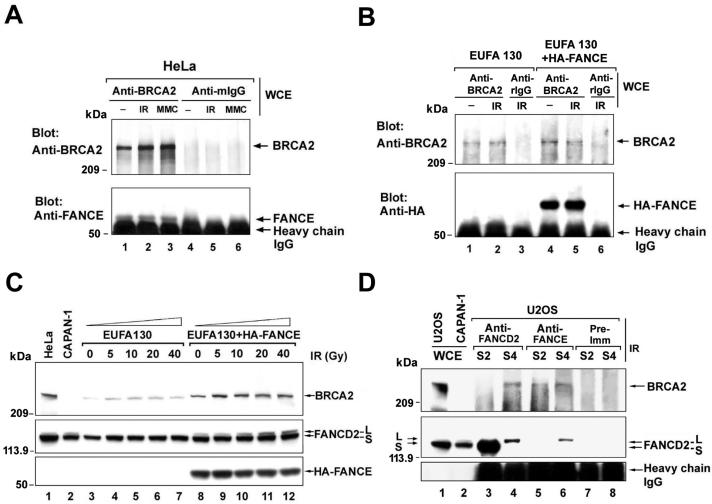
Colocalization, cofractionation, and coimmunoprecipitation of monoubiquitinated FANCD2 and BRCA2 in chromatin. (A) Colocalization of FANCD2 and BRCA2 in DNA damage-inducible foci. HeLa cells were either untreated, exposed to IR (2 or 15 Gy), or treated with MMC (80 ng/ml), as indicated. HeLa cells were double stained with polyclonal anti-FANCD2 (E35) (red) and monoclonal anti-BRCA2 (Ab-1) (green) antibodies after the indicated time of treatment. Magnification, ×630. (B) Protocol utilized for nuclear fractionation of cells. Cytoplasm and nucleoplasm were extracted by permeabilization with detergent, the resulting nuclei were DNase I digested, and chromatin was extracted with ammonium sulfate (NH2SO4). (C) U2OS cells, either untreated or exposed to IR (15 Gy) or MMC (170 ng/ml), were fractionated 15 h after the initiation of DNA damage. Supernatants (S) and pellets (P) were subjected to Western blot analysis with the indicated antibodies: E35 for FANCD2 and Ab-1 for BRCA2. The presence of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody. (D) S2 (soluble nuclear proteins) and S4 (chromatin) fractions of U2OS cells were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 antibody (Ab-1) or a control mouse antibody (mIgG) and then immunoblotted with either polyclonal anti-BRCA2 (Ab-2) or anti-FANCD2 (E35) antibodies. Heavy chain IgG was used as a loading control. (E) HeLa cells were transfected with a cDNA encoding HA-ubiquitin, as indicated. After transfection, cells were treated with the indicated dose of IR or MMC. S2 (soluble nuclear proteins) and S4 (chromatin) fractions of HeLa cells were immunoprecipitated (IP) with a polyclonal antibody (E35) to FANCD2, as indicated. Immune complexes were run on SDS-PAGE and immunoblotted with anti-FANCD2 (FI-17) or anti-HA (HA.11) monoclonal antibodies. WCE, whole-cell extract.
Previous studies have indicated that FANCD2 exists as two isoforms: an unubiquitinated isoform (FANCD2-S) and a monoubiquitinated isoform (FANCD2-L) (9). Cellular exposure to IR, MMC, or other genotoxic agents results in the FA complex-dependent conversion of FANCD2-S to FANCD2-L (9). We next determined the cellular localization of FANCD2-S and FANCD2-L by cellular fractionation. Nuclei from either untreated or treated U2OS cells were fractionated into detergent-soluble (S2) and insoluble (P2) nuclear fractions and chromatin (S4) components, as outlined in Fig. 1B. Interestingly, FANCD2-S was nearly exclusively extracted with the soluble nuclear fraction (Fig. 1C, lane 4), while the IR-inducible or MMC-inducible FANCD2-L isoform was selectively retained in the chromatin fraction (lane 8). In contrast, little FANCD2-L protein was present in the chromatin fraction derived from untreated U2OS cells. These results suggest that the monoubiquitin tag of FANCD2-L may function as a chromatin targeting signal.
We also determined the cellular localization of BRCA2 following DNA damage (Fig. 1C, anti-BRCA2 immunoblots). An increased amount of BRCA2 was detected in the chromatin fraction (S4) following treatment with IR or MMC (lane 8), as compared to that in untreated cells. A fraction of BRCA2 cofractionated with chromatin-associated monoubiquitinated FANCD2 (lane 8), suggesting that the proteins may interact in chromatin.
To test for an interaction between BRCA2 and FANCD2, we next immunoprecipitated BRCA2 from the soluble nuclear (S2) and chromatin (S4) fractions derived from U2OS cells (Fig. 1D). Although more BRCA2 was detected in the S2 fraction (lanes 3 and 4, anti-BRCA2 immunoblot), FANCD2-S did not coimmunoprecipitate with it. In contrast, FANCD2-L in the S4 (chromatin) fraction selectively coimmunoprecipitated with BRCA2 (lanes 7 and 8), and this association was enhanced by IR (compare lanes 7 and 8 of FANCD2-L). The BRCA2 protein had a slightly slower electrophoretic mobility in chromatin (i.e., increased mass) after IR treatment, suggesting that BRCA2 in chromatin is posttranslationally modified following IR (Fig. 1D, compare lanes 7 and 8). A reciprocal coimmunoprecipitation with an antibody to FANCD2, followed by an immunoblot with anti-BRCA2 antibodies, confirmed the interaction of FANCD2-L and BRCA2 in the chromatin (S4) fraction (see Fig. 7D, lanes 4).
FIG. 7.
Interaction of FANCE and BRCA2. (A) Whole-cell extracts (WCE) from HeLa cells, either untreated or exposed to IR (15 Gy, harvested 15 h later) or MMC (40 ng/ml, harvested 24 h later), were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 antibody (Ab-1) or a control mouse antibody (mIgG). Heavy chain IgG was used as a loading control. (B) Whole-cell extracts from unirradiated or irradiated EUFA130 (FA-E), and EUFA130 cells stably expressing HA-FANCE (EUFA130 + HA-FANCE), were subjected to immunoprecipitation with polyclonal anti-BRCA2 antibody (H-300) or a control rabbit antibody (rIgG) and then immunoblotted with anti-BRCA2 (Ab-1) or anti-HA antibodies. Heavy chain IgG was used as a loading control. (C) EUFA130 (FA-E) lymphoblasts and EUFA130 cells stably expressing HA-FANCE (EUFA130 + HA-FANCE) were either untreated or exposed to IR at different doses, as indicated, and harvested after 4 h. Western blotting was performed with anti-BRCA2 (Ab-1), anti-FANCD2 (E35), or anti-HA antibodies. (D) Reciprocal coimmunoprecipitation of FANCD2 and BRCA2 with other antibodies. The indicated fractions (S2 and S4), prepared from irradiated U2OS cells, were immunoprecipitated with antibody to FANCD2 (E35), FANCE, or a control nonimmunized rabbit serum (Pre-imm), and the immune complexes were immunoblotted with anti-BRCA2 (Ab-1) or anti-FANCD2 (FI-17) antibodies. Heavy chain IgG was used as a loading control. (E) Schematic model of the ATM-BRCA2-FANCD2-mediated DNA damage response. IR activates ATM, resulting in the phosphorylation of BRCA2, FANCD2, and several other protein substrates. Activated BRCA2 is then recruited to chromatin by FANCD2, which is activated by monoubiquitination. The interaction of BRCA2 and FANCD2 requires both the C terminus of BRCA2 and FANCD2 monoubiquitination. Following its loading onto chromatin, BRCA2 then functions downstream of monubiquitinated FANCD2.
To confirm that the FANCD2 protein in the S4 (chromatin) fraction is indeed monoubiquitinated, we transfected HeLa cells with a cDNA encoding HA-ubiquitin, and endogenous FANCD2 was then immunoprecipitated (Fig. 1E). As expected, IR and MMC treatment activated the monoubiquitination (HA labeling) of FANCD2 (lanes 9 and 10, respectively). FANCD2 in the S2 (soluble nuclear proteins) fraction was nonubiquitinated, as demonstrated by the absence of HA-ubiquitin in the FANCD2 immunoprecipitate (lanes 3 to 5).
Monoubiquitination of FANCD2 promotes the assembly of IR-inducible BRCA2 foci.
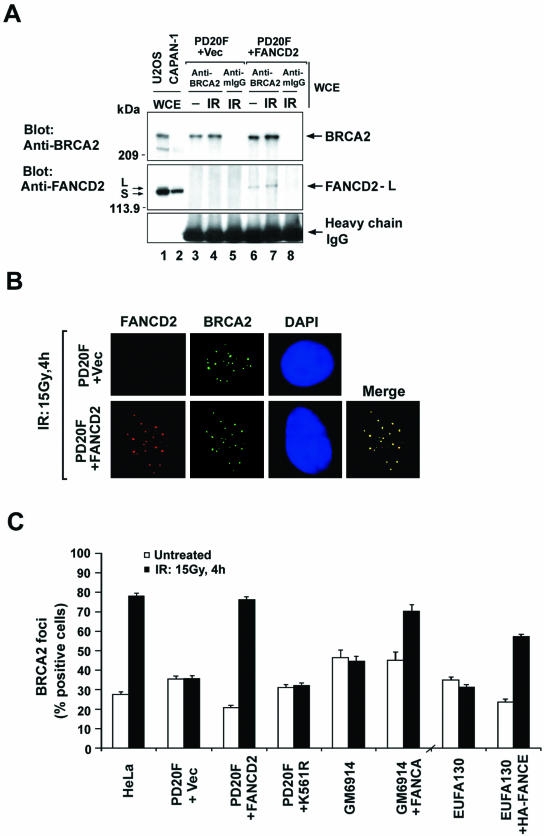
To determine the functional importance of the interaction between BRCA2 and FANCD2, we examined an MMC-sensitive patient-derived FANCD2−/− cell line (FA-D2), PD20F (Fig. 2). As expected, FANCD2 was not detected in anti-BRCA2 immune complexes immunoprecipitated from whole-cell extracts of FA-D2 cells (Fig. 2A, lanes 3 and 4). The interaction between BRCA2 and FANCD2 was observed after stable transfection with the FANCD2 cDNA (Fig. 2A, lanes 6 and 7).
FIG. 2.
Monoubiquitinated FANCD2 promotes IR-induced assembly of BRCA2 foci. (A) Whole-cell extracts of PD20 (FA-D2) fibroblasts stably expressing empty vector (PD20F + Vec) and PD20 fibroblasts corrected with FANCD2 (PD20F + FANCD2) were subjected to immunoprecipitation with mouse monoclonal anti-BRCA2 (Ab-1) or a control mouse antibody (mIgG) and then immunoblotted with either anti-FANCD2 (E35) or anti-BRCA2 (Ab-2) antibodies. Heavy chain IgG was used as a loading control. (B) Formation of subnuclear FANCD2 and BRCA2 foci in response to IR treatment. PD20 (FA-D2) fibroblasts, stably expressing empty vector alone (upper panel) or full-length FANCD2 cDNA (lower panel), were either untreated or treated with IR (15 Gy) and fixed 4 h later. Cells were double stained with polyclonal anti-FANCD2 (E35) (red) and monoclonal anti-BRCA2 (Ab-1) (green) antibodies and analyzed by immunofluorescence microscopy. Magnification, ×630. (C) Quantification of BRCA2 foci. PD20 (FA-D2) fibroblasts stably expressing empty vector alone (PD20F + Vec), FANCD2 (PD20F + FANCD2), or FANCD2-K561R mutant (PD20F + K561R), GM6914 (FA-A) fibroblasts and corrected GM6914 fibroblasts stably expressing FANCA (GM6914 + FANCA), EUFA130 (FA-E) lymphoblasts and corrected EUFA130 lymphoblasts (EUFA130 + HA-FANCE), and HeLa cells were either untreated or treated with IR (15 Gy) and fixed 4 h later. Cells with more than four distinct foci were counted as positive. A total of 200 cells/sample were analyzed. The values shown are the mean ± standard deviation from three separate experiments.
PD20F (FA-D2) fibroblasts had no FANCD2 foci, as previously described (9) (Fig. 2B). Although these cells did exhibit BRCA2 foci (Fig. 2B), the percentage of cells with BRCA2 foci did not increase upon cellular exposure to IR (Fig. 2C). Correction of these cells with the FANCD2 cDNA restored the formation of IR-inducible BRCA2 foci, as measured by the percentage of cells with BRCA2 foci following IR treatment (Fig. 2C). A nonubiquitinated mutant of FANCD2 (FANCD2-K561R) (9) failed to complement the cells and failed to increase the percentage of cells with IR-activated BRCA2 foci (Fig. 2C and Table 1). FA-A (complementation group A) and FA-E fibroblasts also exhibited no increase in the percentage of cells with BRCA2 foci following IR. Functional complementation of these cells, with either the FANCA or FANCE cDNA, respectively, restored FANCD2 monoubiquitination and MMC resistance (Table 1) and the ability to assembly IR-inducible BRCA2 foci (Fig. 2C). Taken together, these results indicate that the generation of monoubiquitinated FANCD2 by the functional FA/BRCA pathway promotes the assembly of DNA damage-inducible BRCA2 foci.
TABLE 1.
Characterization of FA and corrected FA cell lines
| Cell line/plasmid | FA group | MMC sensitivitya | IR induction of focib:
|
FANCD2/BRCA2 complex in chromatin | |
|---|---|---|---|---|---|
| BRCA2 | FANCD2 | ||||
| Fibroblasts | |||||
| HeLa | Wild type | R | ↑ | ↑ | + |
| GM6914 | A | S | − | − | − |
| GM6914 + FANCA | A | R | ↑ | ↑ | + |
| PD20 + Vec | D2 | S | − | − | − |
| PD20 + FANCD2 | D2 | R | ↑ | ↑ | + |
| PD20 + K561R | D2 | S | − | − | − |
| EUFA423 | D1 | S | ↑c | ↑↑↑ | − |
| EUFA423 + BRCA2 | D1 | R | ↑ | ↑ | + |
| Lymphoblasts | |||||
| EUFA130 | E | S | − | − | − |
| EUFA130 + HA-FANCE | E | R | ↑ | ↑ | + |
R, resistant; S, sensitive.
↑, increase; −, no change.
The C terminus-truncated BRCA2 in EUFA423 (FA-D1) cells was present in both cytoplasm and nucleus.
The carboxy terminus of BRCA2 is required for the functional interaction of BRCA2 and FANCD2.
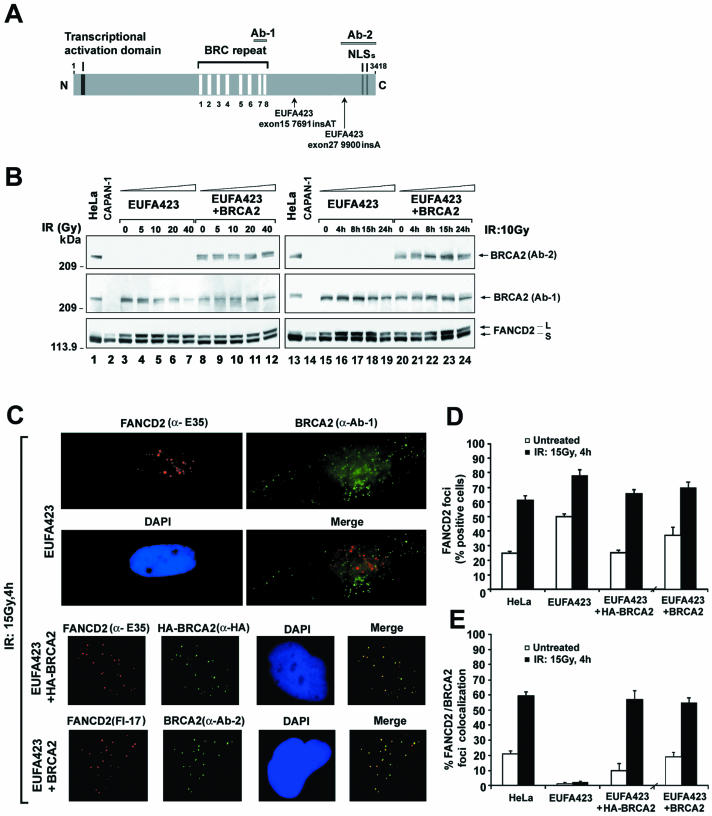
To further appreciate the functional importance of BRCA2 and FANCD2 complexes, we examined a FA-D1 (BRCA2−/−) fibroblast line, EUFA423 (Fig. 3). These cells have two mutant alleles of BRCA2 (13). The cells express a truncated BRCA2 protein, encoded by one of the mutant alleles (exon 27, 9900insA) (13), and this mutant protein lacks the carboxy-terminal 192 amino acids of BRCA2 (Fig. 3A). The truncated BRCA2 protein can be detected with anti-BRCA2 antibody (Ab-1) but not with Ab-2. Uncorrected EUFA423 cells or EUFA423 cells stably transfected with the full-length BRCA2 cDNA were examined for FANCD2 monoubiquitination (Fig. 3B). Uncorrected EUFA423 cells were hypersensitive to DNA damage (Table 1) and upregulated the monoubiquitination of FANCD2 at lower doses (5 Gy) (Fig. 3B, compare lanes 4 and 9) and shorter time points (4 h) after IR than corrected EUFA423 cells (Fig. 3B, compare lanes 16 and 21). The increase in FANCD2 monoubiquitination in EUFA423 cells probably results from the greater accumulation of DNA damage in these DNA repair-deficient cells.
FIG. 3.
FANCD2 and BRCA2 foci fail to associate in FA-D1 (BRCA2−/−) cells. (A) Schematic diagram of human BRCA2 protein, indicating mutations in EUFA 423 (FA-D1) cells. The positions of epitopes for the anti-BRCA2 antibodies Ab-1 and Ab-2 are shown. (B) EUFA423 (FA-D1) fibroblasts and corrected cells stably transfected with human chromosome 13 (EUFA423 + BRCA2) were exposed to IR, either at different doses (left) or for different periods of time (right). Western blotting was performed with anti-BRCA2 (Ab-1 or Ab-2) or anti-FANCD2 (E35) antibodies. (C) Immunofluorescent localization of BRCA2 and FANCD2 following treatment with IR (15 Gy) was examined in EUFA423 (FA-D1) fibroblasts, EUFA423 fibroblasts transiently transfected with pcDNA3 HA-BRCA2 (EUFA423 + HA-BRCA2), and EUFA 423 fibroblasts stably transfected with human chromosome 13 (EUFA423 + BRCA2). Cells were double stained with the indicated antibodies and analyzed by immunofluorescence microscopy. Magnification, ×630. (D and E) Quantification of the percentage of cells with FANCD2 foci (D) and the percentage of cells with colocalization of FANCD2 and BRCA2 foci (E) in HeLa cells, EUFA423 fibroblasts, EUFA423 fibroblasts transiently expressing HA-BRCA2 (EUFA423 + HA-BRCA2), and EUFA423 fibroblasts stably expressing human chromosome 13 (EUFA423 + BRCA2). Cells were either untreated or treated with IR (15 Gy) and fixed 4 h later for immunofluorescence microscopy. Cells with more than four distinct foci were counted as positive. Two hundred and 100 cells/sample were analyzed in panels D and E, respectively. The values shown are the mean ± standard deviation from three separate experiments. (F) S2 (soluble nuclear proteins) and S4 (chromatin) fractions of irradiated EUFA423 cells and EUFA423 cells stably expressing human chromosome 13 (EUFA423 + BRCA2) were subjected to immunoprecipitation with rabbit polyclonal anti-FANCD2 antibody (E35) or a control nonimmunized rabbit serum (Pre-imm) and then immunoblotted with either monoclonal anti-BRCA2 (Ab-1) or anti-FANCD2 (FI-17) antibodies. Heavy chain IgG was used as a loading control. WCE, whole-cell extract.
We next examined BRCA2 foci and FANCD2 foci in these FA-D1 cells. Uncorrected EUFA423 cells exhibited IR-inducible BRCA2 foci (Fig. 3C) and an abnormally high percentage of cells with FANCD2 foci, both with and without IR treatment (Fig. 3D). The increase in the percentage of FA-D1 cells with FANCD2 foci is consistent with the increased monoubiquitination of FANCD2 observed on immunoblots in Fig. 3B. In contrast, BRCA2-corrected EUFA423 cells exhibited similar percentages of cells with FANCD2 foci as HeLa cells (Fig. 3D). Interestingly, based on counts, the FANCD2 foci in FA-D1 cells failed to colocalize with BRCA2 foci (Fig. 3E). Colocalization of BRCA2 and FANCD2 was restored in corrected EUFA423 cells (Fig. 3E). Importantly, using an antibody to FANCD2, FANCD2-L and BRCA2 from the damaged chromatin of BRCA2-corrected EUFA423 cells coimmunoprecipitated (Fig. 3F, lane 8) but C-terminally truncated BRCA2 from damaged chromatin of EUFA423 cells did not coimmunoprecipitate with FANCD2-L (Fig. 3F, lane 7). Taken together, these results indicate that FA-D1 cells assemble elevated FANCD2 foci, but that the carboxy-terminal 192 amino acids of BRCA2 are required for the functional interaction of BRCA2 and FANCD2.
FA-D1 and FA-D2 cells are defective in the assembly of IR-inducible RAD51 foci.
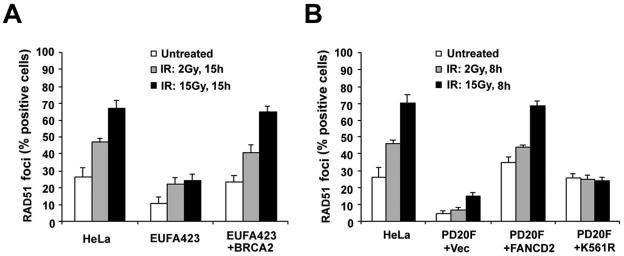
Previous studies have indicated that BRCA2 binds RAD51 and that BRCA2-deficient cell lines are defective in RAD51 focus formation (4, 17, 32). We therefore examined IR-induced RAD51 foci in FA-D1 and FA-D2 cells (Fig. 4). EUFA423 (BRCA2-deficient) cells were defective in IR-inducible RAD51 focus assembly, as demonstrated by a decreased percentage of cells with RAD51 foci, either with or without IR treatment, relative to HeLa cells or corrected EUFA423 cells (Fig. 4A). Similarly, a much lower percentage of PD20 (FANCD2-deficient) cells had IR-inducible RAD51 foci, either with or without exposure to IR, than PD20 cells corrected by the expression of wild-type FANCD2. PD20 cells expressing the FANCD2-K561R mutant were also defective in the assembly of IR-inducible RAD51 foci (Fig. 4B). Taken together, these results suggest that at least one function of the FA/BRCA pathway is to assemble DNA damage-inducible RAD51 foci.
FIG. 4.
FA-D1 and FA-D2 cells are defective in the assembly of IR-inducible RAD51 foci. (A) Quantification of RAD51 foci in EUFA423 (FA-D1) fibroblasts, EUFA423 fibroblasts stably transfected with human chromosome 13 (EUFA423 + BRCA2), and HeLa cells. Cells were either untreated or treated with IR (2 or 15 Gy) and fixed 15 h later. Cells were stained with monoclonal anti-RAD51 antibody and analyzed by immunofluorescence microscopy. Cells with more than four distinct foci were counted as positive. Two hundred cells/sample were analyzed. The values shown are the mean ± standard deviation from three separate experiments. (B) Quantification of RAD51 foci in PD20 (FA-D2) fibroblasts stably expressing empty vector alone (PD20F + Vec), full-length FANCD2 cDNA (PD20F + FANCD2), or the FANCD2 K561R mutant (PD20F + K561R), and HeLa cells. Cells were either untreated or treated with IR (2 or 15 Gy) and fixed 8 h later. Immunofluorescence microscopy and counts were performed as described above for panel A.
Monoubiquitination of FANCD2 promotes IR-inducible accumulation of BRCA2 in chromatin.
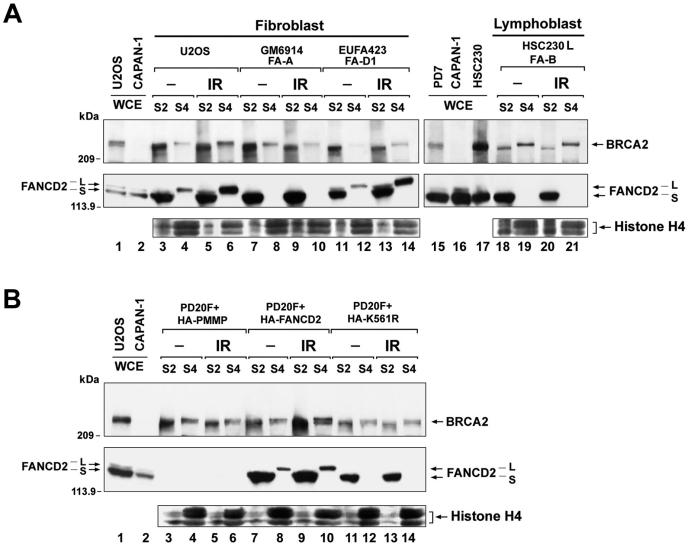
We next fractionated cells from multiple FA subtypes (Fig. 5) and analyzed the recruitment of BRCA2 and FANCD2-L to chromatin. In U2OS cells, which have a functional FA pathway, IR activated the recruitment of monoubiquitinated FANCD2 and BRCA2 to chromatin (Fig. 5A, compare lanes 4 and 6). In FA-A cells and FA-B cells, no monoubiquitinated FANCD2 was observed (lanes 10 and 21), and there was no increase of BRCA2 in chromatin (S4) fractions following IR (compare lanes 8 and 10 and 19 and 21 for FA-A and FA-B cells, respectively).
FIG. 5.
Monoubiquitination of FANCD2 promotes IR-inducible accumulation of BRCA2 in chromatin. (A) S2 (soluble nuclear proteins) and S4 (chromatin) fractions were prepared from U2OS and FA cells, which were either untreated or treated with IR (15 Gy, fractionated after 15 h). Fractions were subjected to Western blot analysis with anti-FANCD2 (E35) or anti-BRCA2 (Ab-1) antibodies. The extraction of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody, as indicated. The FA cells are represented by GM6914 (FA-A) fibroblasts, EUFA423 (FA-D1) fibroblasts, and HSC230 (FA-B) lymphoblasts. (B) S2 (soluble nuclear proteins) and S4 (chromatin) fractions were prepared from PD20 (FA-D2) fibroblasts stably expressing empty vector alone (PD20F + HA-PMMP), HA-FANCD2 (PD20F + HA-FANCD2), or FANCD2-HA-K561R mutant (PD20F + HA-K561R), which were either untreated or treated with IR (15 Gy, fractionated after 6 h). Fractions were subjected to Western blot analysis with anti-FANCD2 (E35) or anti-BRCA2 (Ab-1) antibodies. The extraction of chromatin in the S4 fraction was confirmed by blotting with anti-histone H4 antibody.
FA-D2 cells were also defective in IR-inducible loading of BRCA2 onto chromatin (Fig. 5B, compare lanes 4 and 6). Wild-type FANCD2 (PD20F + HA-FANCD2), but not the nonubiquitinated mutant of FANCD2 (PD20F + HA-K561R), corrected IR-inducible BRCA2 chromatin loading (compare lanes 8 and 10 and 12 and 14 for PD20 cells expressing wild-type HA-FANCD2 and the FANCD2-K561R mutant HA-K561R, respectively). These results confirm that monoubiquitinated FANCD2 promotes the loading of BRCA2 onto damaged chromatin. Failure to load BRCA2 onto damaged chromatin is a common feature of all FA subtypes examined in this study (FA subtypes A, B, D1, D2, and E) and was consistently seen in different experiments in this study.
Even though FANCD2 is strongly monoubiquitinated in FA-D1 cells and efficiently transported to chromatin (Fig. 5A, compare lanes 12 and 14), these cells exhibited decreased colocalization of BRCA2 and FANCD2 foci (Fig. 3E) and were deficient in the interaction of monoubiquitinated FANCD2 and BRCA2 in chromatin (Fig. 3F). Therefore, the monoubiquitination and targeting of FANCD2 onto chromatin occur independently of the carboxy terminus of BRCA2. The carboxy terminus of BRCA2 is required, however, for the efficient interaction of BRCA2 and FANCD2 in chromatin.
IR activates the ATM-dependent phosphorylation of BRCA2, resulting in activation of an intra-S-phase checkpoint.
BRCA2 has multiple SQ phosphorylation consensus sites and may therefore be a direct substrate of ATM or ATR (1). Given that BRCA2 in chromatin migrates more slowly following IR (Fig. 1D, compare lanes 7 and 8), we tested whether this electrophoretic change represents an IR-dependent phosphorylation of chromatin-derived BRCA2 (Fig. 6A). Irradiation activated an upward shift of BRCA2 in chromatin (S4) fractions (lane 4), and in vitro phosphatase treatment reversed this shift (lane 5). Taken together, these results indicate that IR activates the phosphorylation of BRCA2 and that phosphorylated BRCA2 accumulates in the chromatin of IR-damaged cells. To test whether ATM is involved in this process, we examined ATM−/− fibroblasts or ATM cDNA-corrected cells (Fig. 6B). IR failed to activate BRCA2 phosphorylation in chromatin in AT (ATM−/−) cells (compare lanes 3 to 5), suggesting that ATM may be the kinase for this reaction. Functional complementation of these cells with the ATM cDNA restored the IR-dependent phosphorylation of BRCA2 (Fig. 6B, compare lanes 6 to 8). Also, IR activated the association of ATM protein with the S4 (chromatin) fraction, consistent with previous studies (2).
FIG. 6.
ATM is required for IR-activated phosphorylation of BRCA2 in chromatin. (A) BRCA2 immunoprecipitated (Ab-1) from chromatin fractions was treated either with or without λ-phosphatase and phosphatase inhibitors, as indicated. Chromatin fractions were derived either from untreated U2OS cells or 15 h following IR treatment (15 Gy). The mobility shift of BRCA2 is shown in lanes 4 and 6. WCE, whole-cell extract. (B) BRCA2 derived from the chromatin fraction of corrected AT (AT + ATM) fibroblasts at time points after treatment with IR (15 Gy) undergoes a mobility shift, while BRCA2 from the chromatin fraction of AT (ATM−/−) fibroblasts does not. Chromatin fractions were immunoblotted with either anti-BRCA2 (Ab-1), anti-ATM, or anti-FANCD2 (E35) antibodies. (C) EUFA423 (FA-D1) and PD20 (FA-D2) cells are defective in an IR-inducible S-phase checkpoint. RDS was assessed 30 min after delivery of IR to the indicated cell lines.
The IR-activated, ATM-dependent phosphorylation of several proteins, including NBS1 (10), BRCA1 (5), and FANCD2 (25), triggers an intra-S-phase checkpoint. Cells deficient in any of these ATM substrates lack this checkpoint and exhibit RDS. Interestingly, EUFA423 (FA-D1) cells have a defect in the intra-S checkpoint response (Fig. 6C), similar to the defect observed in FA-D2 (PD20F) cells (25). Correction of EUFA423 with wild-type BRCA2 restored the intra-S checkpoint, further indicating that BRCA2 is an ATM substrate that participates in the intra-S checkpoint. We have previously reported that while the K561 mutant of FANCD2 is still functional for the intra-S checkpoint in PD20 cells, it is associated with sensitivity to MMC (25). Thus, the interaction between monoubiquitinated FANCD2 and BRCA2 in chromatin appears to be required for establishing the MMC resistance of cells; however, this interaction is not required for establishment of the S-phase checkpoint.
Interaction of BRCA2 with other FA proteins.
Recent studies have indicated that the FANCD2 protein binds directly to FANCE, a subunit of the FA complex (A/C/E/F/G/L complex) (22). We next determined whether FANCE is present in BRCA2 and FANCD2 immune complexes (Fig. 7). An antibody to BRCA2 coimmunoprecipitated FANCE from either untreated or DNA-damaged HeLa whole-cell extracts (Fig. 7A). As expected, BRCA2 failed to coimmunoprecipitate with FANCE from whole-cell extracts of FA-E (FANCE deficient) lymphoblasts (Fig. 7B, lanes 1 and 2). Coimmunoprecipitation of BRCA2 and HA-FANCE was observed after functional complementation with the HA-FANCE cDNA (Fig. 7B, lanes 4 and 5). Moreover, HA-FANCE complementation of FA-E cells promoted the monoubiquitination of FANCD2 and the stability of BRCA2 (Fig. 7C, lanes 8 to 12). BRCA2, FANCD2-Ub, and FANCE coimmunoprecipitated from S4 (chromatin) fractions (Fig. 7D). These results suggest that BRCA2 interacts, either directly or indirectly, with FANCE and perhaps other components of the FA pathway.
DISCUSSION
Interaction of BRCA2 and FANCD2 in a common DNA repair pathway.
Our results demonstrate that BRCA2 is indeed a protein component of the FA pathway and that it functions downstream in this pathway. BRCA2 colocalizes, cofractionates, and coimmunoprecipitates with monoubiquitinated FANCD2 from chromatin. According to one possible model (Fig. 7E), DNA damage by IR activates the FA complex-dependent monoubiquitination of FANCD2, resulting in its targeting to chromatin. Monoubiquitinated FANCD2 in chromatin then promotes the loading of BRCA2 onto chromatin. The carboxy-terminal 192 amino acids of BRCA2 are required for the functional interaction of BRCA2 and FANCD2 in nuclear foci. Whether FANCD2-L binds to BRCA2 directly, or indirectly via other protein intermediates, remains unknown. Recent studies using the two-hybrid system suggest a direct interaction between BRCA2 and FANCD2 (C. Mathew, personal communication). Importantly, loss of the interaction between monoubiquitinated FANCD2 and BRCA2 in chromatin is a common feature of other FA subtypes. This defect correlates with the chromosome instability and abnormal homology-directed DNA repair of FA cells (30).
Our results are consistent with those of Bruun et al. (3), which suggest that BRCA2 is epistatic with other FA genes and functions downstream in the pathway. According to that study, disruption of BRCA2 expression by small inhibitory RNA did not further increase MMC-induced chromosome breakage in FA-A cells.
While it has been recently reported that BRCA2 interacts with the FA protein, FANCG, by two-hybrid analysis (14), here we present the first direct evidence of an intracellular interaction of endogenous levels of BRCA2 with an upstream FA protein, FANCE, and with downstream monoubiquitinated FANCD2. The interaction of FANCE with BRCA2 appears to be independent of FANCD2, since it occurs in PD20 (FA-D2) cells lacking FANCD2 and in EUFA423 (FA-D1) fibroblasts (data not shown), in which FANCD2 does not bind to BRCA2 (Fig. 3F). This may be of functional significance for BRCA2 loading onto chromatin. FANCE binds to both FANCD2 and FANCC (22) and thus may link FANCD2 to the machinery for monoubiquitination. As such, the constitutive interaction of FANCE with BRCA2 may facilitate the targeting of BRCA2 to chromatin. The finding that BRCA2, like FANCD2, is phosphorylated in chromatin in an ATM-dependent manner may also be of significance to the regulation of BRCA2 loading onto chromatin (Fig. 6). Whether ATM-phosphorylated FANCD2 and BRCA2 cooperate in the S-phase checkpoint response remains unknown.
Possible functions of FANCD2-L in homology-directed DNA repair.
The function of the chromatin complex of monoubiquitinated FANCD2 (FANCD2-L) and BRCA2 remains unknown, but increasing evidence suggests that the complex may play a role in HDR. While the requirement for BRCA2 in HDR is well known (7, 17, 21, 31), the function of FANCD2-L is less clear. FANCD2-L may be required for the proper pairing of homologous chromosomes (or homologous regions of sister chromatids), thus allowing BRCA2 to unload RAD51 at appropriate sites (31). Consistent with this view, meiotic cells isolated from Fancd2−/− mice exhibit increased chromosome mispairing (12). In support of such a role for FANCD2-L, FANCD2 colocalizes with RAD51 during S phase (24). Alternatively, FANCD2-L may contribute to the efficiency or timing of repair, but not to the actual strand invasion or processing of Holliday junctions. Accordingly, FA-G cells, which lack monoubiquitinated FANCD2, have only a mild defect in DNA repair (30) compared to the more profound eightfold deficit observed in BRCA2−/− cells (21).
FA cells are specifically impaired in the repair of interstrand cross-links. Cross-link repair requires the generation of a double-strand break (DSB) intermediate and subsequent HDR activity. The activated (monoubiquitinated) FANCD2 protein is therefore likely to contribute to the sensing of the DNA cross-link or the processing of the cross-link to a DSB. The interaction of FANCD2-L with BRCA2/RAD51 complexes suggests that FANCD2-L may either (i) redirect the BRCA2/RAD51 complexes to specific sites of cross-link lesions or (ii) enable the BRCA2/RAD51 complexes to process and repair such lesions. How the chromatin-associated FANCD2-L enables BRCA2 to repair DNA cross-links remains a central unanswered question in FA research.
Germ line or somatic disruption of the FA/BRCA pathway in cancer.
The interaction of the breast/ovarian cancer susceptibility gene BRCA2/FANCD1 in a common pathway with other FANC genes suggests that inherited or acquired defects in this pathway may result in cancer in the general (non-FA) population. Several lines of evidence support this notion. First, epigenetic inactivation of FANCF (26) accounts for the chromosome instability and cisplatin hypersensitivity of a subset of ovarian tumors. Second, germ line mutations in BRCA2, FANCC, or FANCG are found in individuals with inherited pancreatic cancer (29). Third, germ line disruption of the murine Fancd2 gene results in mice with ovarian and breast epithelial cancers (12). We predict that inherited (germ line) mutations or polymorphisms in other FANC genes may account for an increased cancer risk of individuals in the general (non-FA) population. The actual cancer risk and tumor spectrum may depend on the presence of specific mutant FA alleles.
Acknowledgments
We thank M. Buchwald for the primary FA-B lymphoblast line, HSC230. We thank G. Bagby for the anti-FANCE antibody. We thank L. Moreau for chromosome breakage analysis.
This work was supported by National Institutes of Health grants RO1HL52725, RO1DK43889, and PO1HL54785 (A.D.D.). P.R.A. is a Special Fellow of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Andegeko, Y., L. Moyal, L. Mittelman, I. Tsarfaty, Y. Shiloh, and G. Rotman. 2001. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 276:38224-38230. [DOI] [PubMed] [Google Scholar]
- 3.Bruun, D., A. Folias, Y. M. Akkari, Y. Cox, S. Olson, and R. Moses. 2003. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair 2:1007-1013. [DOI] [PubMed] [Google Scholar]
- 4.Chen, P. L., C. F. Chen, Y. Chen, J. Xiao, Z. D. Sharp, and W. H. Lee. 1998. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. USA 95:5287-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 6.D'Andrea, A. D., and M. Grompe. 2003. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 3:23-34. [DOI] [PubMed] [Google Scholar]
- 7.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273-282. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Higuera, I., Y. Kuang, D. Näf, J. Wasik, and A. D. D'Andrea. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19:4866-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 10.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, K. Spring, S. Kozlov, M. F. Lavin, R. A. Gatti, P. Concannon, and K. Khanna. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch, B., A. Shimamura, L. Moreau, S. Baldinger, M. Hag-Alshiekh, B. Bostrom, S. Sencer, A. D. D'Andrea. 2004. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood 103:2554-2559. [DOI] [PubMed] [Google Scholar]
- 12.Houghtaling, S., C. Timmers, M. Noll, M. J. Finegold, S. N. Jones, M. S. Meyn, and M. Grompe. 2003. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17:2021-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz, C. De Die-Smulders, N. Persky, M. Grompe, H. Joenje, G. Pals, H. Ikeda, E. A. Fox, and A. D. D'Andrea. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297:606-609. [DOI] [PubMed] [Google Scholar]
- 14.Hussain, S., E. Witt, P. A. Huber, A. L. Medhurst, A. Ashworth, and C. G. Mathew. 2003. Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1. Hum. Mol. Genet. 12:2503-2510. [DOI] [PubMed] [Google Scholar]
- 15.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-457. [DOI] [PubMed] [Google Scholar]
- 16.Lomonosov, M., S. Anand, M. Sangrithi, R. Davies, A. R. Venkitaraman. 2004. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 17:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marmorstein, L. Y., T. Ouchi, and S. A. Aaronson. 1998. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc. Natl. Acad. Sci. USA 95:13869-13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medhurst, A. L., P. A. Huber, Q. Waisfisz, J. P. de Winter, and C. G. Mathew. 2001. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol. Genet. 10:423-429. [DOI] [PubMed] [Google Scholar]
- 19.Meetei, A. R., S. Sechi, M. Wallisch, D. Yang, M. K. Young, H. Joenje, M. E. Hoatlin, and W. Wang. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23:3417-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan, S. E., C. Lovly, T. K. Pandita, Y. Shiloh, and M. B. Kastan. 1997. Fragments of ATM which have dominant-negative or complementing activity. Mol. Cell. Biol. 17:2020-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moynahan, M. E., A. J. Pierce, and M. Jasin. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7:263-272. [DOI] [PubMed] [Google Scholar]
- 22.Pace, P., M. Johnson, W. M. Tan, G. Mosedale, C. Sng, M. Hoatlin, J. de Winter, H. Joenje, F. Gergely, and K. J. Patel. 2002. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21:3414-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taniguchi, T., and A. D. D'Andrea. 2002. The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of FANCC. Blood 100:2457-2462. [DOI] [PubMed] [Google Scholar]
- 24.Taniguchi, T., I. Garcia-Higuera, P. R. Andreassen, R. C. Gregory, M. Grompe, and A. D. D'Andrea. 2002. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414-2420. [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi, T., I. Garcia-Higuera, B. Xu, P. R. Andreassen, R. C. Gregory, S. T. Kim, W. S. Lane, M. B. Kastan, and A. D. D'Andrea. 2002. Convergence of the Fanconi anemia and ataxia telangiectasia signaling pathways. Cell 109:459-472. [DOI] [PubMed] [Google Scholar]
- 26.Taniguchi, T., M. Tischkowitz, N. Ameziane, S. V. Hodgson, C. G. Mathew, H. Joenje, S. C. Mok, and A. D. D'Andrea. 2003. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat. Med. 9:568-574. [DOI] [PubMed] [Google Scholar]
- 27.Tutt, A., D. Bertwistle, J. Valentine, A. Gabriel, S. Swift, G. Ross, C. Griffin, J. Thacker, and A. Ashworth. 2001. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 20:4704-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandenberg, C. J., F. Gergely, C. Y. Ong, P. Pace, D. L. Mallery, K. Hiom, and K. J. Patel. 2003. BRCA1-independent ubiquitination of FANCD2. Mol. Cell 12:247-254. [DOI] [PubMed] [Google Scholar]
- 29.Van Der Heijden, M. S., C. J. Yeo, R. H. Hruban, and S. E. Kern. 2003. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 63:2585-2588. [PubMed] [Google Scholar]
- 30.Yamamoto, K., M. Ishiai, N. Matsushita, H. Arakawa, J. E. Lamerdin, J.-M. Buerstedde, M. Tanimoto, M. Harada, L. H. Thompson, and M. Takata. 2003. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol. Cell. Biol. 23:5421-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]
- 32.Yuan, S. S., S. Y. Lee, G. Chen, M. Song, G. E. Tomlinson, and E. Y. Lee. 1999. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59:3547-3551. [PubMed] [Google Scholar]
- 33.Ziv, Y., A. Bar-Shira, I. Pecker, P. Russell, T. J. Jorgensen, I. Tsarfati, and Y. Shiloh. 1997. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene 15:159-167. [DOI] [PubMed] [Google Scholar]