Abstract
Simple and predictable trans-acting regulatory tools are needed in the fields of synthetic biology and metabolic engineering to build complex genetic circuits and optimize the levels of native and heterologous gene products. Transcription activator-like effectors (TALEs) are bacterial virulence factors that have recently gained traction in biotechnology applications due to their customizable DNA binding specificity. In this work we expand the versatility of these transcription factors to create an inducible TALE system by inserting tobacco-etch virus (TEV) protease recognition sites into the TALE backbone. The resulting engineered TALEs maintain transcriptional repression of their target genes in Escherichia coli, but are degraded following the induction of the TEV protease, thereby promoting expression of the previously repressed target gene of interest. This TALE-TEV technology enables both repression and induction of plasmid or chromosomal target genes in a manner analogous to traditional repressor proteins but with the added flexibility of being operator agnostic.
Keywords: TALEs, trans-regulators, synthetic biology, induction systems, TEV protease, LacI
Introduction
The specific repression and subsequent induction of one or more gene(s) is a common methodology used for a wide variety of purposes, including heterologous protein production, gene overexpression studies, and metabolic engineering. Traditionally this is accomplished by using one or more well-characterized repressor proteins, such as LacI, TetR, or λ repressor (e.g. cI857) to repress target gene expression; target genes are then expressed following the addition of an appropriate inducer, such as isopropyl β-D-1-thiogalactopyranoside (IPTG), anhydrotetracycline (aTc), or an increase in temperature, respectively1. These and other transcriptional repressors have been and continue to be the stalwarts of numerous molecular and synthetic biology gene expression schemes2. However, for all the reliability of these repressor proteins, they are each limited by a common feature; each repressor recognizes a single specific DNA target sequence. The ability to target any specific DNA sequence with the same repression-induction characteristics as these canonical repressors would be useful in a variety of applications, such as constructing intricate and combinatorial gene circuits, regulating heterologous genes and operons, and the targeting of native promoters on the chromosome.
The last several years have seen an explosion in the understanding and application of trans-acting regulators, such as transcriptional activator-like effectors (TALEs) and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system as tools for genome engineering3, 4. These trans-regulators are popular due to the ease with which they can be tailored to target and interact with a specific contiguous DNA sequence of interest5, 6. With further modification, TALEs and CRISPR/Cas have successfully been used as tools for bactericidal applications7, 8, DNA labeling9, 10, and transcriptional repression and activation11-15. We sought to develop a system that would blend the repression/induction properties of canonical repressors with the customizable DNA target specificity of trans-regulators. TALEs have proven amenable to manipulation, particularly by fusion of nucleases and eukaryotic transcriptional regulators to the C-terminus, making them an ideal platform for constructing our engineered regulators16, 17. Efforts in this area have generated ligand- and light-responsive TALE induction systems for activating gene expression in mammalian cells18, 19.
Here we present the development of an operator agnostic regulatory system for use in Escherichia coli that relies upon proteolytic degradation of an engineered TALE to activate gene expression (Fig. 1). Briefly, a TALE designed to repress bacterial transcription initiation at a specific locus12 is modified to contain tobacco-etch virus (TEV) protease recognition sites. Expression of this modified TALE inhibits transcription initiation; following chemical induction of TEV protease, the TALE is post-translationally degraded, thereby promoting expression of the originally repressed TALE target gene. We demonstrate that this TALE-TEV induction platform works for both plasmid and chromosomal based target genes and is independent of the utilized TALE repeat domain. We envision this system finding widespread use in probing genes of unknown function in their native context, enabling the construction of temporal gene knockouts and knockdowns, and permitting the design of intricate regulatory networks for heterologous gene expression in the fields of metabolic engineering and synthetic biology.
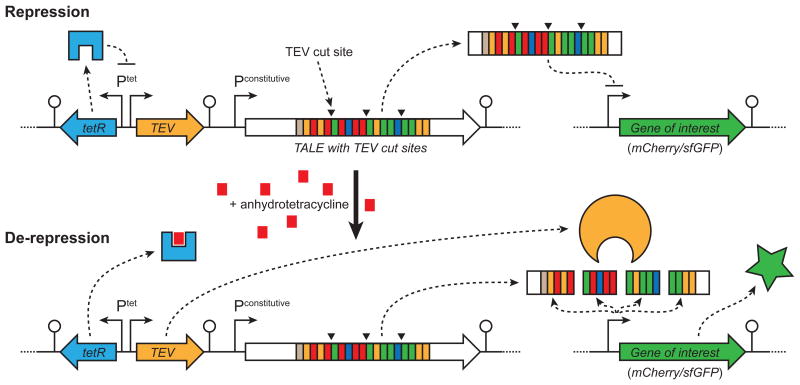
Figure 1.
Cartoon of the TALE-TEV repression-induction system. A constitutively expressed TALE, containing tobacco etch virus (TEV) protease cut sites, binds to a promoter proximal region of the target gene and inhibits its expression. Induction of the TetR-regulated TEV protease following addition of anhydrotetracycline (aTc) results in proteolytic cleavage of the modified TALE, alleviating target gene repression.
Results
Construction of a TALE-TEV repressor-induction system
Engineered TALEs are capable of repressing transcription initiation and elongation in Escherichia coli12. To mimic the function of common allosteric regulators (e.g. LacI), we sought to make a TALE that could both repress transcription but also be functionally inactivated by proteolytic degradation to de-repress target gene expression (Fig. 1). To operate properly this system would need to: (i) express a specific protease with tunable and tight regulation and (ii) utilize a modified TALE susceptible to proteolytic degradation, but still capable of high fidelity DNA recognition. The catalytic domain of the nuclear inclusion protease from tobacco etch virus (TEV) was selected for our proof-of-concept system as it is a fairly small protein (27 kDa), recognizes a seven amino acid epitope of the general motif E-X-X-Y-X-Q-(G/S), and has been successfully expressed in E. coli20. We placed the gene encoding TEV protease in our system under the regulation of the Ptet promoter to minimize leaky expression and ensure induction following addition of aTc21.
Locations for incorporation of TEV protease recognition sites into the TALE backbone were motivated by previous structure-function data5, 22-24, our observation that TALEs with a reduced number of repeat domains repress poorly (Supplementary Results, Supplementary Fig. 1), and analysis of homology models (Supplementary Fig. 2, 3). We hypothesized that proteolytic cleavage of a TALE into multiple pieces would generate fragments with fewer repeat domains, some lacking full length N- or C-termini, thereby reducing the likelihood that these fragments would maintain target gene repression. As a proof-of-concept, we inserted the ENLYFQG TEV protease recognition sequence into three locations within the TlacO repeat domain, between the 4th-5th, 9th-10th, and 14th-15th 34-amino acid repeats (Supplementary Fig. 2, 3). Complete cleavage of this modified TlacO protein (referred to as A1) by TEV protease would generate four individual protein fragments.
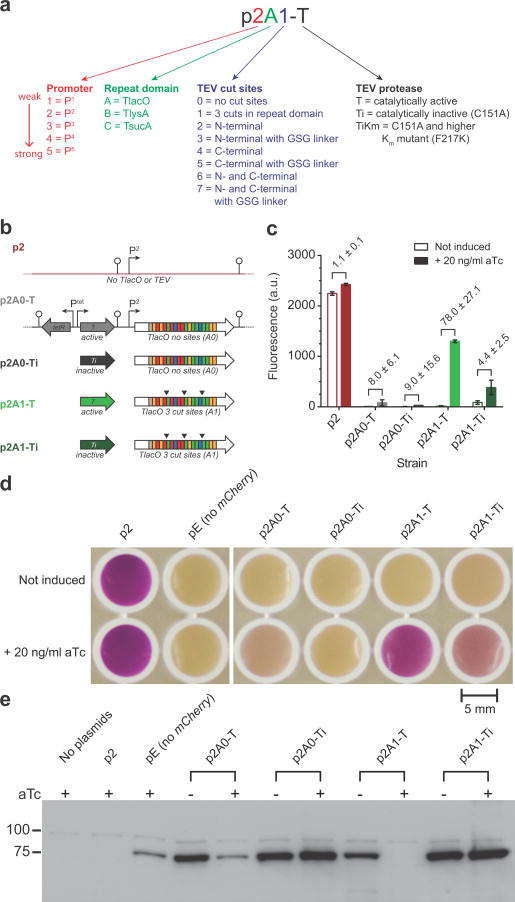
We constructed a two plasmid system consisting of an expression and reporter plasmid to assay the functionality of the proof-of-concept system in E. coli MG1655 ΔlacI (Fig. 2a, b). A low-copy number expression plasmid contained cassettes for constitutive expression of the repressor (either an unmodified TlacO, i.e. A0, or a variant containing three TEV site insertions, i.e. A1) and aTc inducible expression of tev protease. A medium copy number reporter plasmid contained a gene encoding either mCherry or superfolder GFP (sfGFP) expressed from a strong promoter containing the lac O1 operator 3′ of the -10 sequence. When cells harboring the engineered repressor (i.e. A1) were induced with aTc, red fluorescence increased 78-fold (Fig. 2c and Supplementary Fig. 4a, b, p2A1-T) relative to the uninduced state. Conversely, strains expressing a TALE without TEV protease cut sites, such as plasmids p2A0-T and p2A0-Ti, maintained repression following aTc addition. As expected, in the absence of tev protease induction (i.e. no aTc added) we do not see significant mCherry expression in strains containing any of the TALE-TEV expression plasmids. These results are also easily noticeable to the naked eye (Fig. 2d) and demonstrate that the proof-of-concept is working as intended; a TALE targeted to the lac operator and modified to contain TEV protease recognition sites represses gene expression and can be de-activated following expression of the protease, enabling expression of the previously repressed reporter gene. Interestingly, a qualitative immunoblot of whole cell lysates probed for full length TALE A0 and A1 demonstrated that, while only the A1 construct is completely degraded in the presence of active TEV protease, the A0 TALE appears to be partially degraded upon tev protease induction despite the fact this protein has no identifiable TEV protease cleavage sites (Fig. 2e); we confirmed this observation by performing a semi-quantitative Western blot for full length TALE (Supplementary Fig. 4c-f). Nevertheless, the complete lack of full length TALE A1 in both blots corroborates the proposed mechanism of de-repression, and, importantly, repression of the reporter gene is maintained despite the partial degradation of TALE A0 (Fig. 2c). Surprisingly, we also observed in these proof-of-concept studies that a protease rendered catalytically inactive (C151A25) was also able to facilitate de-repression of mCherry expression (Fig. 2c). Importantly, expression of the different system components (e.g. TALE, TEV) does not appear to impair cell growth relative to control strains, when cells are permitted a brief period of growth prior to induction (Supplementary Fig. 4b).
Figure 2.
Characterization of the proof-of-concept TALE-TEV system. (a) Legend for the vector nomenclature system used throughout this manuscript. (b) Schematic of the different TALE-TEV expression vectors. Both TALES A0 and A1 are constitutively expressed from the P2 promoter and are designed to bind the lac operator. The A1 construct contains three TEV protease recognition sites, denoted by (▼), in its repeat domain. Each expression vector contains either an active (i.e. T) or catalytically inactive (i.e. Ti) tev protease gene regulated by the Ptet promoter. The p2 vector is a null control vector devoid of both a TALE and TEV protease. (c) Mean mCherry fluorescence values of E. coli MG1655 ΔlacI cells containing one of the five expression vectors and the p5Cherry reporter vector after 24 hours of growth (n = 3, see Supplementary Fig. 4 for full time course). Induction of tev protease with aTc (20 ng/ml) 2.5 hours after inoculation results in de-repression of the mCherry reporter gene in strain p2A1-T expressing the A1 TALE and an active protease. Numbers adjacent to brackets indicate the fold induction. Error bars represent the standard deviation. (d) Photograph of samples from the 24 hour time point following centrifugation and resuspension in 300 ul of media. The pE strain contains a null reporter vector. (e) Western blot probed with an anti-FLAG primary antibody to observe full length A0 or A1 TALEs after 24 hours. All data shown is from cultures grown at 30°C.
Comparison of TALE-TEV system to alternative strategies
To probe the dynamic range of our A1 TALE-TEV construct, we varied the concentration of aTc, responsible for tev protease induction, from 0.2 to 200 ng/mL (Supplementary Fig. 5). The greatest difference in expression, as quantified by mCherry fluorescence, between the induced and uninduced states was seen with 20 ng/mL aTc resulting in fold inductions ranging from 144 ± 12.1 (8 hr time point) to 14.5 ± 1.0 (24 hr time point); this concentration of aTc was used for all further experiments. In addition, induction by TALE degradation is reversible – that is, repression can be restored by diluting fully induced cells into fresh medium without aTc (Supplementary Fig. 6).
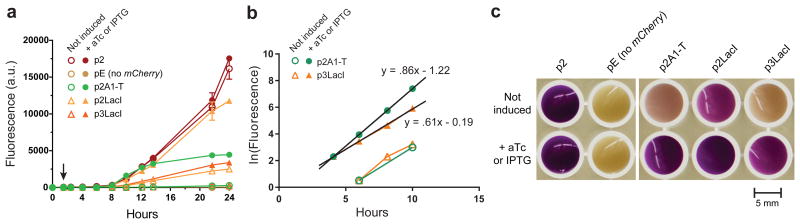
To benchmark the kinetics of our TALE-TEV system to that of the paradigmatic LacI-IPTG system, we expressed lacI in the place of the TALE with either the same P2 constitutive promoter used to express the A1 TALE, or the P3 promoter, which is approximately four times stronger (see Online Methods). We discovered that, following induction with aTc, our TALE-TEV system achieves a level of mCherry fluorescence in-between that observed for LacI driven by the two different promoters and induced with IPTG (Fig. 3a-c). The dynamics of induction for the p2A1-T construct were nearly equivalent to that of p2LacI despite the indirect mechanism of de-repression in our system; that is, aTc does not directly inhibit TALE binding, but induces the synthesis of a protease that then mediates TALE degradation and subsequent relief of repression. The TALE driven by the weaker P2 promoter not only maintains comparable repression in the absence of an inducing agent than P3 driven LacI, but also accumulates mCherry fluorescence slightly faster than p3LacI (Fig. 3a, b).
Figure 3.
Comparison of the TALE-TEV system to LacI-IPTG. (a) Mean mCherry fluorescence values of E. coli MG1655 ΔlacI cells containing either the p2A1-T vector (
 ,
,
 ) or the p2- or p3LacI expression vectors and the p5Cherry reporter vector (n=3). LacI is constitutively expressed, like the A1 TALE, from either the P2 promoter (
) or the p2- or p3LacI expression vectors and the p5Cherry reporter vector (n=3). LacI is constitutively expressed, like the A1 TALE, from either the P2 promoter (
 ,
,
 ) or from the approximately 4-fold stronger P3 promoter (
) or from the approximately 4-fold stronger P3 promoter (
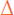 ,
,
 ). Induction of TEV protease with aTc (20 ng/ml) or LacI with IPTG (500 μM) occurred 1.5 hours after inoculation, denoted with (↓).Error bars represent the standard deviation. (b) The natural logarithm of a subset of the data from (a). Kinetics of induction with construct p2A1-T are comparable to that of p3LacI despite both constructs appearing equally leaky in the absence of inducer. (c) Photograph of samples from the 24 hour time point following centrifugation and resuspension in 300 ul of media.
). Induction of TEV protease with aTc (20 ng/ml) or LacI with IPTG (500 μM) occurred 1.5 hours after inoculation, denoted with (↓).Error bars represent the standard deviation. (b) The natural logarithm of a subset of the data from (a). Kinetics of induction with construct p2A1-T are comparable to that of p3LacI despite both constructs appearing equally leaky in the absence of inducer. (c) Photograph of samples from the 24 hour time point following centrifugation and resuspension in 300 ul of media.
In addition, we sought to compare our system to an alternative setup where induction occurs solely by dilution of the repressor (i.e. through a reduction in intracellular TALE concentration over time by growth without TALE production) in order to determine if proteolytic degradation of our TALE was necessary to achieve the observed kinetics. To create such a system, TALE A0 was cloned into a plasmid downstream of the aTc inducible PLtetO1 promoter such that addition of aTc to the culture medium would cause the repression, rather than the induction, of our reporter gene. De-repression with this construct was facilitated by washing and resuspending cells in medium lacking aTc. As expected, proteolytic degradation of the TALE resulted in superior induction characteristics, as the protein production rate was approximately three times higher than for the system operating by dilution (Supplementary Fig. 7, see methods for explanation of protein production rates). Further, a major advantage of our TALE-TEV system over the alternative operating through a dilution mechanism is that proteolytic degradation enables induction even during an early stationary phase where the growth medium will not permit a large number of population doublings and concomitant dilution of the TALE repressor (Supplementary Fig. 8).
Making a universal TALE-TEV scaffold
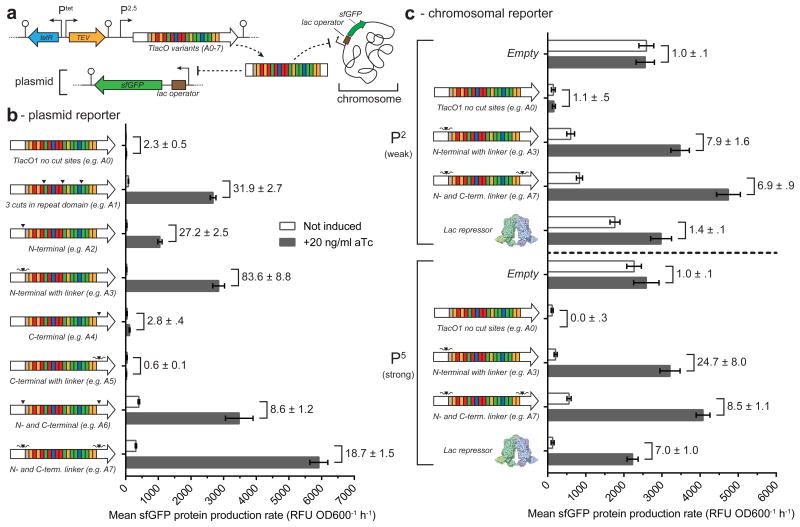
As engineering TEV sites into the repeat domain of each newly synthesized TALE would be laborious, we sought to improve our system with the goal of enabling the end user to substitute their repeat domain of interest into a new TALE scaffold already containing TEV protease recognition sites. Specifically, we engineered A0 (i.e. TlacO) to encode a recognition site in the conserved N- and/or C-terminal regions of the TALE backbone. We also made an additional version of each of these constructs that included Gly-Ser-Gly flanking residues on both sides of the ENLYFQ/S TEV site to potentially improve TEV accessibility and cleavage (Supplemental Fig. 3). The new TALE constructs, A2-A7, were subcloned into the TALE-TEV expression plasmid and grown and assayed in a plate reader for their ability to both repress and de-repress a fluorescent reporter protein (Fig. 4a). Our results, presented as mean sfGFP protein production rates (RFU OD600-1 h-1), demonstrate that constructs containing both N- and C-terminal TEV sites (e.g. A6, A7) led to the most absolute sfGFP expression following TEV induction, but had reduced fold induction levels compared to the original A1 and the new A2 and A3 constructs, due to leakier expression of the reporter gene in the absence of aTc (Fig. 4b). Interestingly, constructs with only C-terminal TEV protease cleavage sites (e.g. A4, A5) did not de-repress; this result was unexpected considering the addition of a C-terminal TEV site increased the level of de-repression in the A6 and A7 variants. Addition of flanking Gly-Ser-Gly linkers improved both total fluorescence and fold induction levels relative to companion constructs (i.e. A3 vs. A2 and A7 vs. A6), suggesting that increasing flexibility around the TEV protease recognition site is important for promoting de-repression, presumably by making it easier for TEV protease to recognize and cleave without impacting TALE repression.
Figure 4.
Characterization of TALE scaffolds with alternative TEV cut sites. (a) Schematic of the two different reporter systems used to assess the new TALE scaffolds. TALE variants are constitutively expressed with Ptet regulating expression of a catalytically active tev protease. New scaffolds have the TEV recognition site inserted in either the N- or C-terminus or at both loci. Recognition sites are either insertions of the native seven amino acid TEV recognition site (▼) or contain flanking glycine-serine-glycine residues (
 ) (see Supplementary Fig. 2-3). (b) Mean sfGFP protein production rates for each of the examined TALE constructs (A0-A7) targeting a plasmid encoding sfGFP (n=3). TALE constructs A0-A7 were all expressed from the P2 promoter. (c) Mean sfGFP protein production rates for constructs A0, A3, and A7 or LacI targeting a chromosomal sfGFP reporter (n=3). All constructs were expressed from the P2 or P5 promoters. Numbers adjacent to brackets indicate the fold induction. All error bars represent the standard error.
) (see Supplementary Fig. 2-3). (b) Mean sfGFP protein production rates for each of the examined TALE constructs (A0-A7) targeting a plasmid encoding sfGFP (n=3). TALE constructs A0-A7 were all expressed from the P2 promoter. (c) Mean sfGFP protein production rates for constructs A0, A3, and A7 or LacI targeting a chromosomal sfGFP reporter (n=3). All constructs were expressed from the P2 or P5 promoters. Numbers adjacent to brackets indicate the fold induction. All error bars represent the standard error.
To further explore the dynamics of the universal TALE-TEV scaffolds, we selected the A7 construct for semi-quantitative gene expression, Western blot, and promoter tuning experiments. We harvested cells from shake flask cultures prior to and following aTc induction in order to monitor gene expression levels of A7, tev protease, and sfGFP. Following induction, tev protease is highly expressed, which leads to a corresponding temporal increase in sfGFP reporter gene expression (Supplementary Fig. 9a-c). An increase in TALE mRNA levels can also be seen following addition of aTc. This is consistent with transcriptional read-through of the terminator between the TALE and tev protease genes from the tev protease promoter. Examination of the Western blot probing for full length TALE shows that the A7 variant is rapidly degraded one hour after TEV induction (Supplementary Fig. 9d), consistent with the proposed mechanism of de-repression. Interestingly, the A7 TALE appears to be less stable than the proof-of-concept A1 TALE, which correlates with the observed leaky sfGFP fluorescence in the p2A7-T strain in the absence of aTc induction. To address leaky target gene expression and explore the range of achievable fold inductions for the A7-T system we substituted the P2 promoter driving TALE expression for four other characterized constitutive promoters (e.g. P1, P3-P5). Increasing TALE expression leads to a nearly threefold increase in fold induction, demonstrating that the TALE-TEV system can be tailored to help meet the design needs of synthetic regulatory networks (Supplementary Fig. 10).
Targeting chromosomal loci and multiplexing
To probe the ability of our TALE-TEV system to regulate chromosomal targets, a fluorescent reporter gene (Ptrc-sfGFP) was integrated into the lacIZYA locus and targeted with plasmid borne A0, A3, or A7 TALE constructs driven by either a weak (P2) or strong (P5) promoter (Fig. 4c). As with the proof-of-concept studies, the native LacI repressor was examined in parallel. Trends between plasmid- and chromosomal-based sfGFP protein production rates are similar, but the magnitude of the fold inductions were larger when a plasmid-based reporter was targeted (e.g. 29.9 ± 3.0 (see Supplementary Fig. 10b) vs. 6.9 ± 0.9 with p2A7-T targeting plasmid-borne or chromosomal sfGFP, respectively). Both the A3 and A7 TALE variants outperform LacI when expressed from a weak promoter, and A7 and LacI demonstrate near identical fold inductions from the stronger P5 promoter (Fig. 4c).
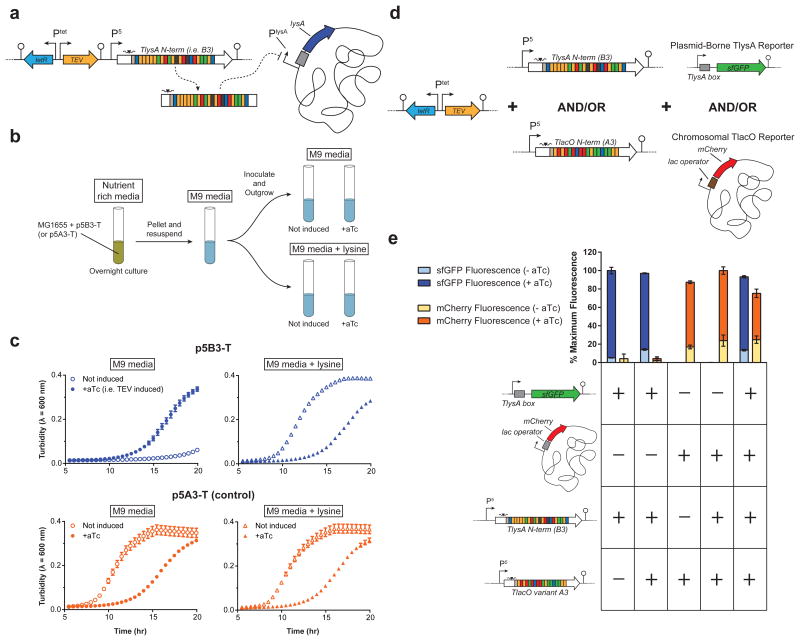
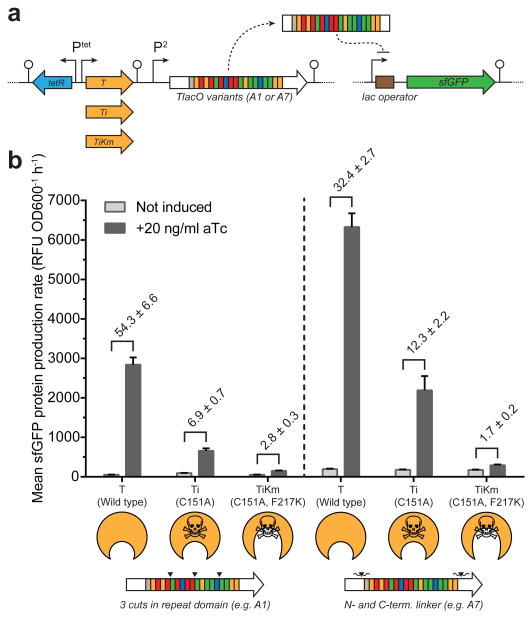
To demonstrate the modularity of our regulators, the DNA-binding domain from the A3 and A7 constructs was replaced with a repeat domain targeting 22 base pairs proximal to the lysA promoter (Supplementary Fig. 11a). LysA, encoding diaminopimelate decarboxylase, is necessary for growth on minimal media without lysine26. We verified that the new TALE-TEV plasmids containing the lysA targeting repeat domain (e.g. B3 and B7) functioned equivalently to the former TlacO constructs by assaying repression in an analogous plasmid-based reporter system (Supplementary Fig. 11b, c). In addition, the A3 repeat domain was also replaced by a repeat domain targeting the sucA σs promoter-proximal region, making TALE C3, to further demonstrate the universality of our TALE scaffold (Supplementary Fig. 11). The p5B3-T construct was then used to demonstrate that the TALE-TEV system can temporally silence native genes and, in this case, impose a reversible auxotrophy (Fig. 5a). Wildtype E. coli MG1655 containing p5B3-T were grown in nutrient rich media, washed of residual lysine, and resuspended in M9 + glucose or M9 + glucose + lysine (Fig. 5b). Cell outgrowth was monitored following induction of tev protease to determine if: i) the B3 TALE construct would approximate an auxotrophy in a wild type background; and ii) growth could be recovered by expressing tev protease. Cells in minimal media harboring p5B3-T did not grow for nearly 20 hours after inoculation, demonstrating that TlysA can effectively mimic a genetic lysA knockout for an extended period. Importantly, this auxotrophy could be reversed by adding aTc to induce tev protease and alleviate lysA repression (Fig. 5c). As expected, in media supplemented with lysine growth occurs despite constitutive expression of B3; however, we see an increase in the duration of lag phase when tev protease is induced in this supplemented media. In control strains expressing the A3 TALE (Fig. 5c, bottom panel) we also observe this increase in lag phase when tev protease is expressed. This phenomenon occurs whether or not the TEV protease being produced is catalytically active (Supplementary Fig. 12). These data demonstrate that induction of TEV before cells reach very early exponential phase (i.e. the time of aTc addition used throughout these studies in rich media) can negatively burden the cells and impact growth, as opposed to induction following entry into exponential (Supplementary Fig. 4b). Importantly, this study establishes that the TALE-TEV system can be used to make transient “knockouts” of essential genes, which may prove useful for probing genes of unknown function, developing counter-selection schemes, or building temporal regulation into industrial strains to improve production.
Figure 5.
Targeting chromosomal loci (a-c) and multiplexing (d-e) using the TALE-TEV system. (a) A TALE targeted to the native E. coli lysA promoter proximal region containing an N-terminal TEV cut site (TALE B3) is constitutively expressed from the P5 promoter. (b) Cartoon of the experimental setup employed to determine if (i) B3 can inhibit cell growth and (ii) if expression of an active TEV protease can alleviate repression to promote growth. Overnight nutrient rich cultures were supplemented with lysine then washed and outgrown in each of the medias shown. Cultures were induced with 20 ng/ml aTc 1.5 hours after inoculation as appropriate. (c) Growth curves of the p5B3-T and the p5A3-T control strains in minimal media with or without lysine (n=3). (d) A three-plasmid system was used to express two TALEs (A3 and B3) with TEV cleavage sites and their corresponding reporter genes. The TlysA sfGFP reporter was harbored on a plasmid while the TlacO mCherry reporter was present on the chromosome. (e) Strains containing plasmids with each aforementioned genetic element (+) or the appropriate empty-vector control (-) were grown in shake tubes for 24 hours with or without addition of aTc before taking fluorescence measurements, which were normalized to the maximum fluorescence intensity for each reporter (n=3). aTc-inducible tev protease was always present on the plasmid containing TlysA or the corresponding negative control plasmid but was omitted from the plasmid harboring TlacO and the corresponding empty vector control. Error bars represent the standard deviation.
In order to verify that multiple TALEs could be used simultaneously in the same strain, we constructed a system using TALEs A3 and B3 to repress mCherry and sfGFP, respectively (Fig. 5d). As expected, when both TALEs are present, aTc induction of tev protease results in expression of both reporter genes (Fig. 5e). These results demonstrate that our TALE-TEV system can be multiplexed. However, reporter expression was noticeably leakier when both TALEs were present as compared to controls containing a single TALE. This could be explained by a reduction in the amount of each individual TALE synthesized when both TALE genes are present.
De-repression can be mediated by TALE sequestration
In our initial proof-of-concept studies we noticed that mCherry was de-repressed following induction of a catalytically inactive TEV protease (Fig. 2). We hypothesized that the inactive protease was able to promote de-repression by binding and sequestering TALEs with TEV protease recognition sites without actually cleaving the peptide backbone. Western blots probing for full length TALE confirm that A1 is not degraded following induction of the catalytically inactive protease (Fig. 2e). To explore this sequestration hypothesis further, we cloned a tev protease variant that is not only catalytically inactive (i.e. C151A), but also has a nearly 8-fold reduced affinity for its target substrate (i.e. mutant F217K Km = 0.466 ± 0.057 vs Wild-type Km = 0.061 ± 0.01027) into our TALE-TEV expression vector (Fig. 6a).
Figure 6.
De-repression by sequestration of the TALE. (a) Schematic of the TALE-TEV system using the A1 and A7 constructs constitutively expressed from the P2 promoter and one of three different tev protease genes controlled by the Ptet promoter. TEV variants studied include the catalytically active (i.e. T), catalytically inactive (C151A, i.e. Ti) and a mutant with an increased Km that is also catalytically inactive (C151A, F217K, i.e. TiKm). (b) Mean sfGFP production rates for both TALE constructs with each of the TEV protease variants (n=3). Skull and crossbones denotes a catalytically inactive protease and an enlarged binding pocket on the cartoon proteases depict an increased Km value relative to wildtype. Fold induction values are displayed above the brackets. Error bars represent the standard error.
Consistent with prior experiments, the catalytically inactive TEV protease mutant, Ti, facilitated de-repression of the sfGFP gene repressed by either A1 or A7 TALEs (Fig. 6b). However, when inducing the TiKm variant, sfGFP is barely expressed over the uninduced samples, suggesting that the ability of the F217K protease to sequester the TALE is significantly impaired relative to the C151A mutant. These data reveal that simple binding of the TALE in our system can mediate de-repression, albeit not as efficiently as both binding and proteolytic cleavage by an active protease. De-repression by binding, rather than destroying, may be a simple approach to adapt other trans-regulators, such as zinc fingers and CRISPR/Cas, into induction platforms.
Discussion
TALEs have rapidly become established tools of molecular biology28, 29. Here, we expand upon the use of TALEs as regulators of gene expression by making an induction platform akin in function to more conventional systems like LacI-IPTG or TetR-aTc, but one where the TALE can be re-programmed to target any DNA sequence of interest. A modified TALE containing TEV recognition sites represses its target gene, and, upon protease induction, the TALE is degraded, de-repressing the target gene (Fig. 1). Interestingly, a TALE without TEV cleavage sites is degraded upon expression of active tev protease, albeit to a lesser extent (Fig. 2e and Supplementary Fig. 4c,d), resulting in a negligible effect on target gene repression (Fig. 2c). These observations may be explained by the open structure TALEs adopt during the DNA search process rendering the protein more susceptible to cleavage despite the lack of specific TEV protease recognition sequences.30 Though TEV protease activity does not seem to have a major impact on growth or reporter gene expression, the apparent non-specific degradation of TALEs without cleavage sites highlights the potential risk of secondary effects caused by tev protease expression, which might be exacerbated in organisms with larger genomes due to the increased number of potential cleavage targets. These concerns may be alleviated in future iterations of this system by screening for alternative proteases that do impact stability of non-engineered TALEs.
We found that a universal induction scaffold could be built by engineering a TEV cut site into the N-terminus of the TALE. This is explained by a body of results demonstrating that the non-canonical repeats at the N-terminus make substantial contributions to DNA binding, whereas those non-canonical repeats in the C-terminus appear less important.29 Specifically, TALEs without cryptic N-terminal repeats were observed to have poor or non-existent DNA binding activity, while the N-terminus itself is capable of binding DNA nonspecifically.30-32 Other studies demonstrate that interactions between the more N-terminal repeats and their cognate DNA base contribute more to DNA binding than those near the C-terminus.33-36 Collectively, these data corroborate our results showing that cleavage of the TALE in the N-terminus is the more effective method to post-translationally disrupt TALE binding and promote de-repression.
Trans-regulators are also attractive tools for building multiplexed and orthogonal regulatory systems (Fig. 5d, e) for use in complex genetic circuits due to their reprogrammable DNA binding and demonstrated fidelity13, 34, 37-39. The TALE-TEV system adds to the suite of available tools; by expressing several different TALEs with TEV recognition sites and inducing the corresponding protease, one could simultaneously repress and then de-repress multiple genes at a time. These multiplexed TALEs could be made orthogonal by introducing other sets of specific proteases and their cognate tag on each corresponding TALE, using proteases such as the recently described mf-lon system and its variants40. Alternatively, orthogonal induction systems could be built by expanding on the concept of repressor sequestration by fusing epitope tags to trans-regulators coupled to regulated expression of their cognate antibodies41. While the small size of many bacterial genomes (including that of E. coli) should abet the design of orthogonal TALEs with minimal off-target binding, it should be noted that this is more challenging as the size of the host genome increases34.
Induction systems are frequently used in molecular biology, most commonly for the overexpression of recombinant proteins and tight regulation of toxic products. The TALE-TEV system is not intended to replace, but rather complement these classic systems and expand their versatility by harnessing the re-programmable nature of trans-regulators. While the work presented here was demonstrated in the genetically amenable host E. coli, we envision exciting applications of this technology to probe gene function and dynamics in other less tractable bacterial hosts or even eukaryotes.
Online Methods
Strains and reagents
The E. coli K-12 strain MG1655 ΔlacI12 was used in all experiments , except for chromosomal reporter studies as described below. Unless otherwise noted, E. coli were grown in lysogeny broth (LB) at 37 °C with shaking at 250 rpm, and tev protease was induced with 20 ng/ml anhydrotetracycline (aTc). The antibiotics kanamycin (50 μg/ml) and ampicillin (100 μg/ml) were added to the media as appropriate to maintain selection of the expression and reporter plasmids, respectively. All chemicals and culture media used were from Fisher Scientific or Sigma Aldrich. Enzymes for plasmid and strain construction were purchased from Promega and New England Biolab. Oligonucleotides were acquired from Integrated DNA Technologies.
Plasmid construction
All plasmids created for this study were made using Gibson assembly42 and sequence confirmed via Sanger sequencing (Functional Biosciences, Madison, WI). Representative sequences of an expression and reporter plasmid and all interchangeable ORF and regulatory components are included in the Supplementary Note. The expression plasmids were all built using plasmid pBT-2 as a backbone43. The TlacO gene was subcloned into the expression vector platform from plasmid pA0 (formerly pMK-RQ_2-TALElacO112). The TlysA gene was made by gene synthesis by GeneArt (Life Technologies, Grand Island, NY). The gene was codon-optimized for expression in E. coli MG1655 using Optimizer44 and was sequence verified by GeneArt. The lacI gene was isolated by PCR from chromosomal DNA from E. coli strain MG1655. Constitutive promoters used to express TALE variants and lacI were from the iGEM distrubtion (http://parts.igem.org/Promoters/Catalog/Anderson). The tev protease gene was subcloned from vector pRK79327. The catalytically inactive (i.e. C151A) and increased Km mutants (i.e. F217K) were made by Quikchange mutagenesis (Agilent Technologies). The divergent Ptet promoters and TetR gene were isolated from pBbB2K-GFP45. The reporter plasmids were constructed using plasmid pMSB-6 as a backbone12. The mCherry gene was amplified from ptrcCherry (formerly pMSBmCherry12). sfGFP was isolated from PRM-GFP46. The TlysA binding site was substituted for the lac operator via Gibson assembly.
Chromosomal sfGFP reporter strain construction
A recombineering strategy utilizing the thymidylate synthase (thyA) gene of E. coli as a selectable and counter-selectable marker was used to integrate sfGFP or mCherry into the chromosome of E. coli K-12 MG165547. First, the native copy of thyA was eliminated from a strain of E. coli K-12 MG1655 harboring the recombineering plasmid pKD4648 by electroporation with a linear dsDNA PCR product containing regions of homology upstream and downstream of thyA generated by overlap extension PCR. The desired recombinants lacking thyA were isolated by plating on minimal medium containing the counter-selective agent trimethoprim (20 μg/mL), thymine (100 μg/mL), and ampicillin (100 μg/mL). A cassette was then assembled containing thyA and Ptrc-sfGFP (or Ptrc-mCherry) between two 500 bp regions of homology flanking the lacIZYA locus in a plasmid with the pKD13 backbone48, giving plasmids pMP004-sfGFP-thyA and pMP004-mCherry-thyA. These cassettes were then amplified using primers rMP145 and rMP150 and electroporated into E. coli K12 MG1655 ΔthyA pKD46 to give MG1655 ΔthyA lacIZYA∷sfGFP-thyA pKD46 and MG1655 ΔthyA lacIZYA∷mCherry-thyA pKD46. Correct recombinants were selected by plating on minimal medium lacking thymine. MG1655 ΔthyA lacIZYA∷Ptrc-sfGFP pKD46 and the analogous mCherry integrant were then generated by electroporating the aforementioned strains with primer rMP08449 and plating on minimal medium with 20 μg/mL trimethoprim and 100 μg/mL thymine. thyA was then reintegrated at its native locus by transforming with a linear dsDNA PCR product containing the thyA open reading frame (generated using primers rMP019 and rMP020) and selecting on minimal medium without thymine. This yielded the strains MG1655 lacIZYA∷Ptrc-sfGFP and MG1655 lacIZYA∷Ptrc-mCherry after curing the plasmid pKD46 by growing at 42°C in the absence of ampicillin. All loci of interest were verified by sequencing. All linear recombination cassettes were digested with DpnI and purified prior to transformation. Transformations were performed by outgrowing the strain of interest in 50 mL SOB medium (0.5% [w/v] yeast extract, 2% [w/v] tryptone, 8.5 mM NaCl, 2.5 mM KCl, 20 mM MgSO4, pH 7.0) with appropriate antibiotics/supplements, washing three times in 10% v/v ice-cold glycerol, electroporating, outgrowing in SOC medium (SOB media + 20 mM glucose), and plating on appropriate selective medium.
Fluorescence and OD600 time-course measurements
E. coli strain MG1655 ΔlacI was transformed with the appropriate expression and reporter plasmids and incubated overnight at 37 °C on LB agar plates containing kanamycin and ampicillin. Colonies were picked and used to inoculate LB with antibiotics. Inoculated cultures were incubated overnight at 30 °C with shaking at 250 rpm. Saturated overnight cultures were normalized and used to inoculate 6 ml of fresh LB media containing antibiotics to an ∼OD600 = 0.03 in shake tubes. Cultures were outgrown in a shaking (250 rpm) waterbath at 37 °C for 1.5 hours and then induced with aTc to a final concentration of 20 ng/ml except where noted otherwise. Uninduced cultures were given an equivalent volume of a 50/50 EtOH/water solution. Strains harboring lacI containing expression plasmids were induced with either 500 μM or 1 mM IPTG as noted. Cultures were vortexed and dispensed (100 μl) into black, flat, clear bottom 96-well plates (Corning, Inc., Corning, NY). Samples were pipet mixed before fluorescence and optical density measurements were taken in a Tecan M1000 plate reader. Fluorescence measurements for cultures containing the superfolder GFP or mCherry reporter proteins were at excitation/emission wavelengths of 485/510 nm or 587/610nm, respectively. Optical density measurements were taken at 600 nm. For continuous cultivation experiments used to determine protein production rates, lids were taped onto the 96-well plates and the plates were then placed into a Tecan M200 plate reader. Cells were grown and fluorescence emission and OD600 were measured every 15 to 30 minutes, depending on the experiment, at the same wavelengths as above for 22 hours at 37 °C with shaking (orbital, 3 mm amplitude) in the instrument between readings. Protein production rates (PPR) were calculated as described previously50. PPR calculations were derived from data in the linear range of a plot of the fluorescence values as a function of OD600. An sfGFP maturation rate constant of 7.39 h-1 51 was used for calculations. Error was propagated into the calculated PPR values from parameter standard errors in both the slope of the fluorescence vs. OD600 line and the specific growth rate. Error was also propagated from the relevant quantities into the fold induction values throughout this manuscript. All data were analyzed using Microsoft Excel and plotted using GraphPad Prism.
Photographs of time-course cultures
Images were captured in RAW format using a Nikon D7000 digital-SLR camera (16.2 megapixels) equipped with a 35 mm (f/1.8) lens and processed in Photoshop (CS6). Digital images were cropped, leveled, and contrast/brightness adjusted in Adobe Photoshop.
RNA isolation and quantitative PCR
Cultures (50 ml in LB media) for quantitative PCR were grown in 250 ml flasks at 37 °C in a shaking waterbath (250 rpm). At appropriate time points prior to and following induction with aTc (final [25 ng/ml]) 1 volume of cells were harvested and immediately mixed with 2 volumes of Qiagen RNAprotect bacteria reagent (Qiagen, Valencia, CA). Following centrifugation (10 min at 5000 × g), cell pellets were snap frozen with liquid nitrogen and stored at -80 °C until processed. Total RNA was isolated from thawed cell pellets using an RNeasy Mini kit following the appropriate manufacturer's instructions. Residual DNA was digested using a TURBO DNA-free kit (Life Technologies, Carlsbad, CA). Total RNA was quantified with a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE) and converted to cDNA using an iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA). Quantitative PCR was performed using an iQ SYBR Green Supermix kit (Bio-Rad) with primers for the genes of interest (A7, tev, sfGFP) and reference (cysG, rrsA) as shown in Supplementary Table 3. Primers were designed using Primer3Plus52. PCR was performed using a Bio-Rad CFX Connect Real-Time system with the following protocol (95 °C for 3 min, 95 °C for 15 sec, 55.5 °C for 20 sec, 72 °C for 30 sec, repeat 2-4, 39 more times, followed by a melt curve). Cq values were determined by regression analysis using CFX Manager 2.1 software. A standard curve for each primer pair of interest was generated over six orders of magnitude using the same qPCR primers and conditions as described above in order to calculate amplification efficiencies. Linear double-stranded DNA templates of approximately 600 bp for each gene of interest were generated for use as standards using the primers shown Supplementary Table 3. Presented average fold changes were determined using the normalized expression methodology (i.e. ΔΔCq) using the geometric mean of the reference genes cysG and rrsA as the normalization factor.
Western blotting
Cell samples for Western blotting were collected at appropriate time points, as indicated in Fig. 1 and Supplementary Fig. 5, and centrifuged for 10 min at 5000 × g. Pellets were snap frozen in liquid nitrogen and stored at -20 °C. Each sample pellet was resuspended in phosphate buffered saline and normalized to the same OD600. In preparation for SDS-PAGE, samples were diluted 1:1 with Laemmli sample buffer containing 2-mercaptoethanol, heated at 95 °C for 10 minutes and subjected to electrophoresis on 12% polyacrylamide gels. An equivalent number of OD600 milliliters was loaded in each lane for each sample. Proteins were transferred to Immobilon PVDF for 70 min at 100 V. Following transfer, the PVDF was rinsed and blocked overnight at 4 °C in TBST (20 mM Tris, 137 mM NaCl, 1% [vol/vol] Tween 20, pH 7.6) containing 5% nonfat dry milk. Blots were washed 3 times for 5 min each in TBST and then incubated with a 1:5000 dilution of mouse anti-FLAG primary antibody (Sigma-Aldrich, F3165-.2 mg) in TBST containing 5% bovine serum albumin for 1 hr at room temperature. Blots were again washed as above and incubated for 1 hr at room temperature with goat anti-mouse IgG secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology, Inc., sc-2005) diluted 1:5000 in 2.5% nonfat dry milk in TBST. Blots were then washed 3 times with TSBT, for 5 min each. The detection reagent SuperSignal West Dura Extended duration substrate was applied to the surface of each blot following the manufacturer's instructions (Thermo Scientific). Blots were imaged with an ImageQuant LAS 4000 instrument (GE Healthcare). Digital blot images were cropped, leveled, and brightness/contrast adjusted with Adobe Photoshop.
Lysine auxotrophy assay
E. coli K-12 strain MG1655 was transformed with the expression plasmids p5B3-T and p5A3-T (as a control) and plated on LB agar containing kanamycin (50 μg/ml). Colonies were picked and incubated overnight with shaking at 30 °C in LB broth containing kanamycin and 0.4 mM lysine. Saturated cultures were pelleted (5 min at 4000 × g) and resuspended in an equivalent volume of minimal M9 media + glucose (0.4% [w/v]) + kanamycin. Cultures were then diluted to an OD600 = 0.03 in either M9 media + glucose + kanamycin or M9 media + glucose + kanamycin + lysine (0.4 mM) in shake tubes. Inoculated cultures were incubated with shaking (250 rpm) in a waterbath at 37 °C for 1.5 hours and then induced with aTc to a final concentration of 20 ng/ml or given an equivalent volume of a 50/50 EtOH/water solution. Induced cultures were then dispensed into a clear, flat bottom 96-well plate to measure their growth for 22 hours in a Tecan M200 plate reader as described above in the time-course measurements methods section.
Multiplexing Experiment
Cultures of E. coli K-12 strain MG1655 ΔlacI and E. coli K-12 strain MG1655 lacIZYA∷Ptrc-mCherry transformed with plasmids p5B3-T, p5A3-15A, and p5lysAsfGFP or a combination of the appropriate negative control plasmids (pTEV, pBAD33, and pBAD24, respectively) were each used to inoculate six aliquots of 6 mL of LB medium with kanamycin (50 μg/ml), ampicillin (100 μg/ml), and chloramphenicol (34 μg/ml) to an initial OD600 of .03. These cultures were then outgrown in a water bath with shaking (250 rpm) at 37°C for 1.5 hours prior to addition of 20 ng/mL anhydrotetracycline (final concentration) to three of the replicates for each strain. The remaining three cultures for each strain received an equivalent amount of 50% v/v ethanol. All cultures were then returned to the incubator, and samples were taken 24 hours after inoculation to measure OD600 and mCherry/sfGFP fluorescence values.
Supplementary Material
Acknowledgments
This work was funded by the National Science Foundation (CBET-114678). M.F.C. was supported by NHGRI Genomic Sciences Training Program (T32 HG002760). A.L.M. was supported by NSF SEES fellowship (GEO-1215871). M.C.P. was supported by an NIH Biotechnology Training Program (T32 GM08349). We are grateful to David Waugh for providing guidance on TEV protease variants. We would also like to thank Jeffrey Cameron for assistance with taking the photographs that appear in this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
M.F.C. and B.F.P. conceived the study. M.F.C. and M.C.P.designed and performed the experiments with the following exceptions. A.L.M. proposed and helped design and perform the TEV Km mutant experiment. C.B.J. assisted with preliminary experimental work. M.F.C. and M.C.P. analyzed the data. M.F.C., M.C.P., and B.F.P. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annual review of biomedical engineering. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- 2.Brophy JA, Voigt CA. Principles of genetic circuit design. Nature methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnology and bioengineering. 2013;110:1811–1821. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 5.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 6.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature biotechnology. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bikard D, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature biotechnology. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nature structural & molecular biology. 2013;20:1321–1324. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- 10.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Politz MC, Copeland MF, Pfleger BF. Artificial repressors for controlling gene expression in bacteria. Chem Commun (Camb) 2013;49:4325–4327. doi: 10.1039/c2cc37107c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen AA, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Molecular systems biology. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Pinera P, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Meth. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boissel S, et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic acids research. 2013 doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF., Iii Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS synthetic biology. 2013 doi: 10.1021/sb400114p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucast LJ, Batey RT, Doudna JA. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques. 2001;30:544–546. doi: 10.2144/01303st06. 548, 550 passim. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand KP, Postle K, Wray LV, Jr, Reznikoff WS. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene. 1983;23:149–156. doi: 10.1016/0378-1119(83)90046-x. [DOI] [PubMed] [Google Scholar]
- 22.Lienert F, et al. Two- and three-input TALE-based AND logic computation in embryonic stem cells. Nucleic acids research. 2013;41:9967–9975. doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Streubel J, Blücher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nature biotechnology. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 24.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phan J, et al. Structural basis for the substrate specificity of tobacco etch virus protease. The Journal of biological chemistry. 2002;277:50564–50572. doi: 10.1074/jbc.M207224200. [DOI] [PubMed] [Google Scholar]
- 26.Dewey DL, Work E. Diaminopimelic Acid and Lysine: Diaminopimelic Acid Decarboxylase. Nature. 1952;169:533–534. doi: 10.1038/169533a0. [DOI] [PubMed] [Google Scholar]
- 27.Kapust RB, et al. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 28.Copeland MF, Politz MC, Pfleger BF. Application of TALEs, CRISPR/Cas and sRNAs as trans-acting regulators in prokaryotes. Current opinion in biotechnology. 2014;29:46–54. doi: 10.1016/j.copbio.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Lange O, Binder A, Lahaye T. From dead leaf, to new life: TAL effectors as tools for synthetic biology. Plant J. 2014;78:753–771. doi: 10.1111/tpj.12431. [DOI] [PubMed] [Google Scholar]
- 30.Cuculis L, Abil Z, Zhao H, Schroeder CM. Direct observation of TALE protein dynamics reveals a two-state search mechanism. Nature communications. 2015;6:7277. doi: 10.1038/ncomms8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 32.Gao H, Wu X, Chai J, Han Z. Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell research. 2012;22:1716–1720. doi: 10.1038/cr.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meckler JF, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic acids research. 2013;41:4118–4128. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic acids research. 2012 doi: 10.1093/nar/gks404. 10.1093/nar/gks1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Quintero AL, et al. An improved method for TAL effectors DNA-binding sites prediction reveals functional convergence in TAL repertoires of Xanthomonas oryzae strains. PloS one. 2013;8:e68464. doi: 10.1371/journal.pone.0068464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology. 2013 doi: 10.1038/nbt.2675. 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Na D, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nature biotechnology. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 38.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nature methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zalatan JG, et al. Engineering Complex Synthetic Transcriptional Programs with CRISPR RNA Scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cameron DE, Collins JJ. Tunable protein degradation in bacteria. Nature biotechnology. 2014;32:1276–1281. doi: 10.1038/nbt.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guglielmi L, Martineau P. Expression of single-chain Fv fragments in E. coli cytoplasm. Methods Mol Biol. 2009;562:215–224. doi: 10.1007/978-1-60327-302-2_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature methods. 2009;6:343–U341. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Lynch MD, Gill RT. Broad host range vectors for stable genomic library construction. Biotechnology and bioengineering. 2006;94:151–158. doi: 10.1002/bit.20836. [DOI] [PubMed] [Google Scholar]
- 44.Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic acids research. 2007;35:W126–131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee TS, et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. Journal of biological engineering. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang D, Holtz WJ, Maharbiz MM. A genetic bistable switch utilizing nonlinear protein degradation. Journal of biological engineering. 2012;6:9. doi: 10.1186/1754-1611-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stringer AM, et al. FRUIT, a scar-free system for targeted chromosomal mutagenesis, epitope tagging, and promoter replacement in Escherichia coli and Salmonella enterica. PloS one. 2012;7:e44841. doi: 10.1371/journal.pone.0044841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellis HM, Yu DG, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leveau JH, Lindow SE. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J Bacteriol. 2001;183:6752–6762. doi: 10.1128/JB.183.23.6752-6762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nature biotechnology. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 52.Untergasser A, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic acids research. 2007;35:W71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.