Abstract
The Pim family of proto-oncogenes encodes a distinct class of serine/threonine kinases consisting of PIM1, PIM2, and PIM3. Although the Pim genes are evolutionarily highly conserved, the contribution of PIM proteins to mammalian development is unclear. PIM1-deficient mice were previously described but showed only minor phenotypic aberrations. To assess the role of PIM proteins in mammalian physiology, compound Pim knockout mice were generated. Mice lacking expression of Pim1, Pim2, and Pim3 are viable and fertile. However, PIM-deficient mice show a profound reduction in body size at birth and throughout postnatal life. In addition, the in vitro response of distinct hematopoietic cell populations to growth factors is severely impaired. In particular, PIM proteins are required for the efficient proliferation of peripheral T lymphocytes mediated by synergistic T-cell receptor and interleukin-2 signaling. These results indicate that members of the PIM family of proteins are important but dispensable factors for growth factor signaling.
The Pim family of proto-oncogenes consists of three members: Pim1, Pim2, and Pim3. They constitute a distinct class of protein kinases with specificity towards phosphorylation on serine/threonine residues (34), although the researchers conducting two studies have reported on additional tyrosine kinase activity displayed by human and Xenopus PIM, respectively (32, 39). Pim1 was originally identified as a target for proviral activation in Moloney murine leukemia virus (M-MuLV)-induced T-cell lymphomas (7). In the mouse, Pim1 encodes two proteins with short half-lives: a 33-kDa protein and a 44-kDa protein initiated from an upstream alternative (CUG) start codon (34). Pim1 mRNA has a short half-life due to the presence of five copies of the AUUU(A) destabilization motif in the 3′ untranslated region (UTR) (34). Pim2 and Pim3 show strong homology to Pim1 (2, 4, 12, 44). At the amino acid level PIM2 and PIM3 show 61 and 71% identity to PIM1, respectively. PIM2 shares many of the properties of PIM1: the mRNA and protein are labile, Pim2 transcription can be initiated from upstream CUG codons (44), and EμMyc EμPim2 double transgenic mice develop B-lymphoid tumors comparable to those arising in EμMyc EμPim1 compound transgenic mice (3, 45). In addition, Pim2 can effectively substitute for the Pim1 oncogene in lymphomagenesis, as illustrated by the high number of activating proviral insertions near Pim2 in MuLV-induced tumors in Pim1-deficient EμMyc transgenic mice (44). Since the 3′ UTR of Pim3 also contains mRNA-destabilizing motifs and since Pim3 can substitute for Pim1 and Pim2 in MuLV-induced lymphomagenesis, it is likely that PIM3 has activities that overlap with those of PIM1 and PIM2 (2, 25).
PIM proteins are widely expressed; together, they cover most if not all tissues. The highest Pim1 mRNA levels are found in thymus and testis (35), whereas Pim2 mRNA expression is the highest in brain and thymus (3) and Pim3 mRNA is the most abundant in kidney (12). The three Pim members appear to be coexpressed during mouse development, although in some tissues a tendency towards expression of Pim1 and Pim3 or of Pim2 and Pim3 exists (11). The transcription of Pim1 and Pim2 is induced by a wide range of growth factors and mitogens: interleukin-2 (IL-2) (3, 8), IL-3 (3, 8), granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF (22), IL-4 (3), IL-5 (40), IL-6 (22), IL-7 (3, 9), IL-9 (3), IL-12, IL-15, alpha interferon (24), gamma interferon (3, 48), erythropoietin (26), thrombopoietin (TPO) (26), prolactin (6), concanavalin A (3), lipopolysaccharide (3), and phorbol myristate acetate (47). The majority of these factors transduce their primary signal through the JAK/STAT pathway, indicating that this cascade is probably instrumental in regulating the expression of the Pim genes. This notion is underscored by the presence of STAT-responsive elements in the Pim1 promoter and was directly proven for the STAT3-mediated transactivation of Pim1 induced by IL-6 (36).
The generation of Pim1 mutant mice illustrated a role for PIM1 in growth factor signaling. Although no in vivo abnormalities were shown (20), PIM1-deficient pre-B cells and bone marrow-derived mast cells exhibit impaired in vitro proliferation in response to the presence of IL-7 and IL-3, respectively (9, 10). An important role for PIM1 in cytokine signaling was further implied by the PIM1-mediated rescue of a thymic cellularity defect in mice that lack the IL-2 receptor common-γ-chain. This rescue appears to be dependent on a functional CD3 complex, since a similar phenotype observed in CD3γ-deficient mice could not be rescued by the Pim1 transgene (18). The absence of a significant phenotype in Pim1-deficient mice could be explained by a potential functional redundancy of PIM family members.
We generated compound Pim1−/− Pim2−/− Pim3−/− mice to address the role of the PIM family in mammalian development. While these mice are viable and fertile, their body size is reduced at birth and throughout postnatal life. Proliferation of hematopoietic cells in response to growth factors is impaired in vitro as well as in vivo in the absence of PIM. In addition, PIM proteins appear to be required for efficient cell cycle induction of peripheral T cells in response to synergistic T-cell receptor (TCR) and IL-2 signaling.
MATERIALS AND METHODS
Generation of compound-Pim1-, Pim2-, and Pim3-KO mice.
The construction of the Pim1 neo59 knockout (KO) line has been described previously (20, 41). Pim2- and Pim3-deficient animals were generated as follows: mouse genomic DNA fragments containing the complete Pim2 or Pim3 gene were isolated from a genomic λ-phage (λFIX) library derived from 129OLA inbred mice. For the Pim2 targeting construct, genomic BamHI fragments encompassing Pim2 exons 1, 2, and 3 were replaced with the hygromycin resistance gene (Pgp) controlled by the human PGK promoter. The Pim3 KO construct was generated by replacing a genomic BstEII fragment, containing a part of exon 3, exons 4 to 5, and part of exon 6, with an internal ribosome entry site-βGeo (IRES-βGeo) cassette. The KO constructs were introduced into 129Ola-derived embryonic stem cells by electroporation and drug-resistant clones were selected using hygromycin and G418, respectively. Homologous recombinants were identified by Southern blot analysis using DNA probes located internally or externally with respect to the deleted region. Selected clones with normal karyotypes were injected into C57BL/6 blastocysts. Chimeric mice born from these embryos were crossed to FVB/N females to produce heterozygous mutant F1 offspring. Pim2+/− F1 mice were back-crossed to inbred Pim1neo59 FVB/N mice for 10 generations, and F11 animals were intercrossed, yielding compound Pim1−/− Pim2−/− mice. Pim3+/− mice were bred to FVB/N mice for two generations and subsequently crossed to 11th-generation FVB/N Pim1−/− Pim2−/− mice. Progeny intercrosses yielded wild-type and all single KOs, as well as triple mutant animals. All mice were maintained under specific-pathogen-free conditions. All animal experiments were approved by the Dutch Animal Research Committee.
DNA and RNA analysis.
Pim2 and Pim3 KO alleles were identified initially by Southern analysis of tail DNA restricted with EcoRI (Pim2) or SspI (Pim3); full-length mouse Pim2 cDNA or a genomic DNA fragment 3′ to Pim3 was used as a probe. Subsequently, Pim mutants were identified by PCR: the Pim1 alleles were detected using Pim1 and Neo primers (Pim1For, 5′-AAGCACGTGGAGAAGGACCG-3′; Pim1Rev, 5′-GACTGTGTCCTTGAGCAGCG-3′; NeoA, 5′-CGTCCTGCAGTTCATTCAGG-3′), the Pim2 alleles with Pim2 and Pgp primers (Pim2For, 5′-CACCGCGTCACGGATAGACG-3′; Pim2Rev, 5′-CCCACCTTCCACAGCAGCG-3′; HygroFor, 5′-AGCACTCGTCCGAGGGCAAAGG-3′; Pim2Rev2, 5′-CACGGTGGACCAGCCTAGC-3′), and the Pim3 alleles with Pim3 and lacZ primers (Pim3Sp1, 5′-CTGGACCAAATTGCTGCCCAC-3′; Pim3Sp4, 5′-GGATCTCTGGTTCAAGTATCC-3′; LacZ1, 5′-CGTCACACTACGTCTG-AACG-3′; LacZ2, 5′-CGACCAGATGATCACACTCG-3′).
For the tissue-specific expression of the different Pim members, total RNA was isolated using TriZol (Gibco BRL). Poly(A)+ RNA was extracted from total RNA with PolyATract (Promega), separated by agarose gel electrophoresis, and blotted to Hybond-N membranes (Amersham). Northern blots were hybridized sequentially to full-length mouse Pim1, Pim2, and Pim3 and human GAPDH cDNA probes. The full-length Pim2 and Pim3 cDNAs were cloned from a size-fractionated concanavalin A blast-derived cDNA λ-phage library.
We analyzed the expression of the Pim1, Pim2, and Pim3 transcripts in splenocytes by reverse transcription-PCR (RT-PCR). For this, total spleen RNA was isolated using TriZol (Gibco BRL), treated with DNase, and reverse transcribed with random hexamer primers (Promega) and avian myeloblastosis virus-reverse transcriptase (Roche). The cDNA was amplified in a single PCR using Pim1For and Pim1Rev, Pim2For and Pim2Rev, or Pim3Sp1 and Pim3Sp4 primer combinations. As a control, Gapdh was amplified in the same PCR using Gapdh1 (5′-ACCGGATTTGGCCGTATT-3′) and Gapdh2 (5′-TCTGGGATGGAAATTGTGAG-3′).
Bone marrow colony assays.
Mice (8 to 12 weeks of age) were sacrificed, and both femurs were flushed with Hank's balanced medium containing 50 mM HEPES (pH 7.4), 50 μM β-mercaptoethanol, and 20% fetal calf serum (FCS) (BioWhittaker). Nucleated bone marrow cells were counted in cell lysis buffer (StemCell Technologies). Cells were first diluted in Iscove's modified eagle medium (Gibco BRL) containing 50 μM β-mercaptoethanol and 2% FCS and were thereafter plated in duplicate in methylcellulose (M3234) (StemCell Technologies) containing the appropriate cytokines and growth factors. The cells were cultured at 37°C and 5% CO2 under specific assay conditions as follows: for CFU-GM assays, 1.5 · 104 cells/ml, 10 ng of recombinant murine GM-CSF (Peprotech)/ml, 12 days; for CFU-Meg assays, 4 · 105 cells/ml, 40 ng of recombinant human TPO (R&D Systems)/ml, 8 days; for CFU-mix assays, 1.5 · 104 cells/ml, 10 ng of recombinant murine IL-3 (rmIL-3) (Peprotech)/ml, 12 days; for CFU-Eos assays, 2 · 105 cells/ml, 20 ng of rmIL-5 (R&D Systems)/ml, 15 days; for CFU-mix assays, 2 · 105 cells/ml, 50 ng of recombinant murine stem cell factor (SCF) (Peprotech)/ml, 12 days. After counting of the colonies, cultures were fixed in 4% formaldehyde and stained with a 2% Giemsa solution to determine the cell types.
T- and B-cell proliferation assays.
Splenocytes and bone marrow cells were isolated from 8- to 12-week-old mice and counted in a Casy 1 counter (Schärfe Systems). Splenocytes were cultured in round-bottom 96-well plates at 106 cells per ml in complete medium (RPMI-1640 containing penicillin and streptomycin, glutamine, 50 μM β-mercaptoethanol, and 10% FCS) in the presence of recombinant human IL-2 (rhIL2) (R&D Systems) (concentrations as indicated in figures) and/or coated αCD3 (2C11) (Pharmingen) (concentrations as indicated in figures) for 48 h followed by 16 h with 0.5 μCi of [3H]thymidine. Bone marrow cells were cultured in flat-bottom 96-well plates at 107 cells/ml in complete medium with 50 or 250 U of rmIL-7 (Peprotech)/ml for 3 days and for an additional 18 h in the presence of 0.5 μCi of [3H]thymidine.
Flow cytometry.
Thymocytes, splenocytes, and bone marrow cells from mice 6 to 8 weeks of age were isolated and analyzed by flow cytometry using fluorochrome or biotin-tagged antibodies (all Pharmingen) to CD3ɛ, Mac1, CD69, B220, BP1, CD62l, TCRβ, CD4, CD8, CD25 (IL2Rα), CD117, IL2Rβ, common-γ-chain, IL-7Rα, Sca1, heat-stable antigen (HSA), Gr-1, immunoglobulin M (IgM), IgD, and NK1.1. Biotinylated antibodies were detected using antigen-presenting-cell-conjugated streptavidin (Molecular Probes). Cells were stained in phosphate-buffered saline with 1% bovine serum albumin, 0.05% sodium azide, and 2% normal mouse serum.
Splenocytes were isolated from 8- to 12-week-old mice and cultured at a density of 106 cells/ml in the presence or absence of hIL-2 (200 U/ml) in 48-well plates coated with 0.3 μg of αCD3 (Pharmingen)/ml for the indicated time periods. Carboxyfluorescein diacetate succinimidyl ester (CFSE) (Molecular Probes) staining was performed according the manufacturer's protocol. Cells were labeled for 1 h after 24 h of culture with 10 μM bromodeoxyuridine (BrdU; La Roche). All cells were stained with αTCRβ or CD3 antibodies (Pharmingen) and, where appropriate, fixed in 0.25% paraformaldehyde and permeabilized in 0.1% Tween-phosphate-buffered saline. αBrdU-fluorescein isothiocyanate (La Roche) was used to detect BrdU incorporation, and cell-cycle profiles were analyzed using 7-aminoactinomycin D (7-AAD; Molecular Probes). Apoptotic cells were detected by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling staining according the manufacturer's protocol (La Roche) or with biotinylated annexin V (Molecular Probes), antigen-presenting-cell-conjugated streptavidin (Pharmingen), and propidium iodide (Molecular Probes). All cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson).
Western blot analysis.
Splenocytes isolated from 8- to 12-week-old mice were cultured at 106 cells per ml in starvation medium (RPMI 1640 containing penicillin and streptomycin, glutamine, 50 μM β-mercaptoethanol, and 0.5% FCS) for 16 h. Cells were then treated with rhIL2 (R&D Systems) for 20 min (concentrations as indicated in legends to the figures), and whole cell lysates were prepared using standard 0.5% NP-40 lysis buffer. Lysates were separated using 10% polyacrylamide gel electrophoresis and blotted to nitrocellulose. STAT5 protein levels and STAT5 tyrosine phosphorylation were detected using antibodies from Transduction Labs-Becton Dickinson and Cell Signaling Technology (Beverly, Mass.), respectively.
RESULTS
Generation and initial characterization of Pim mutant mice.
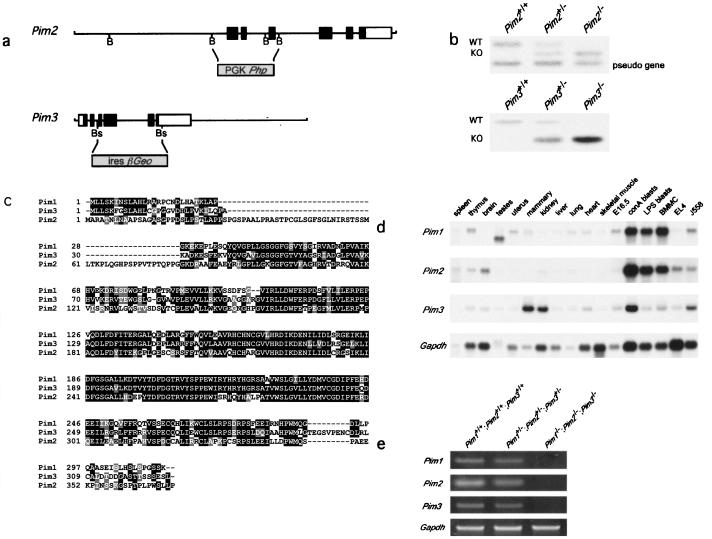
To inactivate Pim2 and Pim3 we designed targeting vectors that disrupted the kinase domains. The Pim2 targeting vector contained the hygromycin resistance gene under the control of the human PGK promoter. Homologous recombination deletes exons 1, 2, and 3 (Fig. 1a). Since Pim3 is expressed in embryonic stem cells, the Pim3 targeting vector was constructed with the promoterless IRES-βGeo cassette (29), which replaces part of exon 3, exons 4 to 5, and part of exon 6 (Fig. 1a). Correctly targeted, karyotypically normal embryonic stem clones were injected into blastocysts to generate chimeric mice and, subsequently, homozygous Pim2 or Pim3 mutant mice (Fig. 1b). Both PIM2- and PIM3-deficient mice are viable, healthy, and fertile and do not show any gross abnormalities (data not shown).
FIG. 1.
The generation of Pim1 Pim2 Pim3 compound KO mice. (a) Targeting constructs for Pim2 and Pim3. PGK-Php was introduced into the BamHI (B) sites of Pim2, thereby deleting exons 1, 2, and 3. For Pim3, a part of exon 3, exons 4 and 5, and part of exon 6 were deleted through introduction of the promoterless IRES-βGeo cassette into the BstEII (Bs) sites. (b) Genotype analysis of Pim2 and Pim3 KO mice by Southern blotting using full-length Pim2 cDNA or the 3′ UTR of Pim3 as a probe. WT, wild type. (c) Comparison of the amino acid sequences of PIM1, PIM2, and PIM3. Identical residues are shown in white characters on a black background; similar residues are shown in white characters on a grey background. (d) Expression of Pim1, Pim2, and Pim3 mRNA. The results are shown for bone marrow-derived mast cells grown on IL-3 (BMMC), EL4 (T-cell line), and J558 (plasmacytoma). LPS, lipopolysaccharide. (e) RT-PCR analysis of Pim1, Pim2, and Pim3 expression in wild-type, Pim heterozygous, and Pim homozygous mutant splenocytes.
The lack of an obvious phenotype in both the Pim2 and Pim3 mutants could be a consequence of redundancy with the remaining Pim genes, which is suggested by the amino acid homology and the overlap in expression of PIM1, PIM2, and PIM3 (Fig. 1c and 1d). Therefore, the Pim2 and Pim3 KO mice were crossbred with the previously described Pim1 mutant mice, generating compound Pim1−/− Pim2−/− Pim3−/− mutants. Homozygous triple mutant mice were observed at a frequency expected for normal Mendelian segregation (data not shown). mRNA of the three Pim genes was undetectable by RT-PCR in the spleens of these mice, in contrast to the results seen with spleens from wild-type and Pim1+/− Pim2+/− Pim3+/− mice, confirming the functional deletion of the genes (Fig. 1e).
Mice deficient for Pim1, Pim2, and Pim3 are smaller.
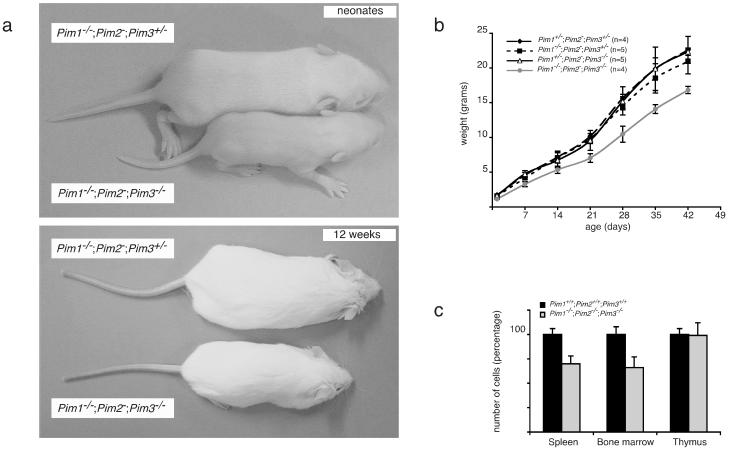
Pim genes can act as potent oncogenes (3, 45). Whereas oncogene overexpression promotes cellular growth either by an increase in cell size or by an increase in cell number, the opposite may be expected when these genes are disrupted. Several oncogene KOs, including Myc and CyclinD1, do indeed display a size reduction of certain tissues (37, 43). The size of Pim1 (20), Pim2, or Pim3 single-KO mice is unaltered in comparison to that of wild-type mice. However, both mature male and mature female Pim1−/− Pim2−/− Pim3−/− mice are approximately 30% smaller than their Pim1+/− Pim2−/− Pim3−/−, Pim1−/− Pim2−/− Pim3+/−, Pim1+/− Pim2−/− Pim3+/−, and wild-type littermates (Fig. 2a and b and data not shown). These size differences are already apparent at birth, as Pim1−/− Pim2−/− Pim3−/− neonates weigh 1.15 (± 0.10) g (n = 4) whereas their Pim1+/− Pim2−/− Pim3−/−, Pim1−/− Pim2−/− Pim3+/−, and Pim1+/− Pim2−/− Pim3+/− littermates weigh 1.63 (± 0.05) (n = 4), 1.66 (± 0.22) (n = 3), and 1.70 (± 0.10) (n = 6) g, respectively. These results demonstrate a functional redundancy between the three Pim genes with respect to their effect on body size. Analysis of Pim1−/− Pim2−/−, Pim2−/− Pim3−/−, and Pim1−/− Pim3−/− compound KO mice revealed that loss of PIM1 and PIM3 contributes predominantly to body size reduction. Still, Pim1−/− Pim2−/− Pim3−/− mice are reduced in size compared to Pim1−/− Pim3−/− mice, indicating an additional PIM2 effect (data not shown).
FIG. 2.
PIM-deficient mice display a growth defect. (a) Comparison of male neonatal and mature Pim1−/− Pim2− Pim3−/− mice with Pim1−/− Pim2− Pim3+/− littermates (upper and lower panel, respectively). (b) Growth curve of male Pim1+/− Pim2− Pim3+/−, Pim1−/− Pim2− Pim3+/−, Pim1+/− Pim2− Pim3−/−, and Pim1−/− Pim2− Pim3−/− littermates. The weight of Pim1+/− Pim2− Pim3+/− mice is similar to that of wild-type mice. (c) Cell numbers in spleens, bone marrows, and thymuses from 8-week-old wild-type (n = 9) and Pim1−/− Pim2−/− Pim3−/− (n = 9) mice.
Except for the thymus, all organs displayed a size reduction corresponding to the decrease in body weight. To address whether the decreased body weight can be attributed to a decrease in cell number or cell size or both, we analyzed the relative sizes and numbers of cells in spleen and bone marrow. Whereas the cell size was only slightly reduced in triple-KO (TKO) mice, the cell number in PIM-deficient bone marrow and spleen was decreased to the same extent as the body size, indicating that the reduction in body weight is mainly the result of a decrease in cell number (Fig. 2c).
Disruption of the Pim genes causes subtle changes in hematopoietic differentiation.
The high levels of Pim expression in hematopoietic tissues and the oncogenic activity of PIM in hematopoietic tumors prompted us to investigate the effect of PIM inactivation on the development of the hematopoietic lineages. Whole-blood analysis did not show significant differences between PIM-deficient and wild-type mice in the number of granulocytes, monocytes, and thrombocytes (data not shown). Pim mutant mice displayed a slight increase in the number of red blood cells; however, the hematocrit was not altered, most likely because PIM-deficient erythrocytes have a decreased mean cell volume (data not shown). This phenotype is comparable to the erythrocyte phenotype previously described for Pim1 KO mice (20), indicating that additional loss of Pim2 and Pim3 does not yield an additive effect.
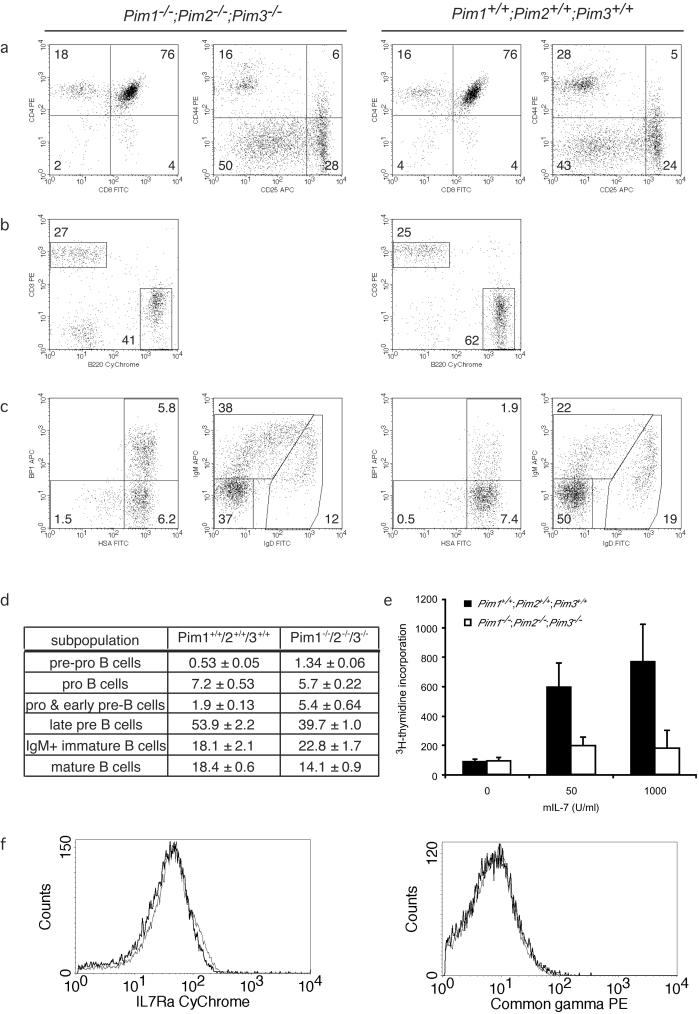
Next, we examined the thymic differentiation of T lymphocytes. No differences between Pim1 Pim2 Pim3 mutant and wild-type animals were observed in any of the subpopulations of differentiating T lymphocytes in the thymus (Fig. 3a). At the CD4/CD8 double-negative (DN) stage of T-cell differentiation, PIM-deficient mice displayed slightly higher levels of CD25 expression at DN2 and DN3 but no consistent differences were found in any of these fractions. Also, no differences between PIM-deficient and PIM-proficient animals were observed in the kinetics and levels of TCRβ expression in the transition from the CD4/CD8 DN to double-positive (DP) stage of development (data not shown). Pim mutant spleens have normal CD4+ and CD8+ T-cell numbers, displaying normal CD62L and CD44 profiles within the different populations (Fig. 3a and data not shown). Taken together, these data indicate that T-cell differentiation in and export from the thymus are unaffected by PIM deficiency.
FIG. 3.
B- but not T-lymphoid development is affected in Pim mutant mice. The first two columns represent PIM-deficient mice; the second series of two columns represents wild-type mice. FITC, fluorescein isothiocyanate; APC, antigen-presenting cells. (a) Flow cytometric analysis of wild-type and Pim mutant thymocytes by CD4/CD8 staining (panels 1 and 3) and CD25/CD44 staining on cells negative for CD4, CD8, B220, CD19, Mac1, Ter119, Gr-1, and NK1.1 (panels 2 and 4). (b) Mature B-cell and T-cell populations in Pim mutant and wild-type spleen. (c) Analysis of B-cell differentiation in the bone marrow. CD43-positive, B220-positive cells were gated and analyzed using BP1 and HSA markers to distinguish pre-pro-B, pro-B, and pro-B-early pre-B cells (panels 1 and 3). CD43-negative, B220-positive cells were gated and analyzed using IgM and IgD to distinguish late pre-B, IgM-positive immature B, and mature B cells (panels 2 and 4). (d) The number of cells belonging to distinct stages of B-cell development depicted as a percentage of total B220-positive cell numbers in the bone marrow of 6- to 8-week-old wild-type and Pim mutant mice. (e) Proliferation of Pim mutant bone marrow cells in response to IL-7. Bone marrow cells were cultured for 96 h in the presence of mIL-7. Proliferation was determined by [3H]thymidine incorporation. For the last 16 h, cells were cultured in the presence of [3H]thymidine. One representative of three independent experiments is shown. (f) Cell surface expression levels IL-7Rα and common-γ-chain on differentiating B cells in the bone marrow. The thin line represents wild-type cells; the thick line represents Pim-deficient cells.
In contrast to the T-cell results, the fraction of splenic B cells is reduced by approximately one-fourth in 8-week-old PIM-deficient mice compared to the results seen with age- and sex-matched wild types or Pim single mutants (Fig. 3b). The percentages of monocytes, granulocytes, and NK cells are comparable between Pim mutant and wild-type mice, whereas the erythroid progenitor fraction (Ter119+CD71+CD117+) and the population of mature denucleated erythroid cells (Ter119+CD71−) are increased in Pim mutant spleens (data not shown). At 20 weeks of age, no differences between PIM-deficient and PIM-proficient animals were observed in B-cell numbers in spleen (data not shown).
In view of the reduction of the splenic B-cell fraction and the transforming activity of overexpressed PIM in immature B lymphocytes (3, 45), we analyzed B-cell differentiation in Pim mutant bone marrow (for a review, see reference 15). Plots from representative PIM-deficient (left panel) and wild-type (right panel) animals are shown in Fig. 3c; averages for this analysis are shown in Fig. 3d. Within the subpopulation of CD43-positive B220+ B-cell progenitors, the early stages of B-cell development can be distinguished on the basis of the expression of HSA/CD24 and BP1. Pre-pro-B cells have not yet committed to the B-cell lineage and are characterized as HSAlo/BP1-negative cells. PIM-deficient bone marrow contains an increased fraction of these cells (Fig. 3c and d). HSAhi/BP1-negative cells are pro-B cells and seem to be present in slightly lower numbers in PIM-deficient bone marrow. Finally, HSAhi/BP1-positive cells contain both noncycling cells, which are in the process of rearranging their heavy chains (pro-B cells), and proliferating cells (early pre-B cells), which have successfully rearranged their heavy-chain locus. Although the proliferation of the early pre-B cells is pre-B-cell-receptor (BCR) initiated, it is propagated by IL-7 (16). In Pim mutant mice, HSAhi/BP1-positive cell levels are increased about threefold compared to wild-type control levels. After a certain number of cell divisions, differentiating B cells exit from the cell cycle, having lost expression of CD43, and initiate genomic rearrangements at the light-chain locus. These cells are characterized as CD43-negative, B220+, surface Ig-negative cells (late pre-B cells). In Pim mutant bone marrow, the number of late pre-B cells is substantially decreased (Fig. 3c and d). The increase in pre-pro-B cells (HSAlo/BP1 negative) and pro-B-early pre-B cells (HSAhi/BP1 positive), as well as the decrease in late pre-B-cells, was consistent between all experiments (a total of ∼20 animals of each genotype). Upon successful light-chain rearrangement, differentiating B cells, which now express a functional BCR complex at the cell surface, again enter a proliferative phase. PIM-deficient bone marrow contains an increased fraction of surface IgM-positive cells (Fig. 3c). However, the observed dissimilarity between wild-type and Pim mutant cells results differed between experiments. Finally, mature B cells, characterized by the expression of both IgD and IgM as well as by high levels of B220, recirculate into the bone marrow. In Pim mutant mice no reproducible differences were found for mature B cells. Additional analysis of the B-cell development in mice lacking only one Pim revealed that the major factor causing this defect is loss of PIM1 and that loss of PIM3 follows loss of PIM1 in importance, whereas PIM2 deficiency has almost no effect on B-cell development (data not shown).
These data indicate that Pim mutant bone marrow is relatively enriched for pre-pro-B cells and HSAhi/BP1-positive cells (pro-B and early pre-B cells). Pre-pro-B cells have not yet committed to the B-cell lineage. HSAhi/BP1-positive cells consist of cells in the process of rearranging their heavy-chain locus (pro-B cells) as well as cells that are actively cycling (early pre-B cells). The increase in these cells could, therefore, be caused by an impaired rearrangement at the heavy-chain locus (pro-B cells), by a prolonged proliferative expansion phase after successful rearrangement of the heavy-chain locus (early pre-B cells), or, in contrast, by less-efficient cell cycle progression in response to pre-BCR/IL-7 signaling causing a delay in the progression from early pre-B cells to late pre-B cells. Since the number of late pre-B cells, which are in the process of rearranging the light-chain locus, is decreased in PIM-deficient bone marrow, we hypothesized that the increase in the HSAhi/BP1-positive fraction (pro-B/late pre-B cells) reflects an effect on the proliferation rather than the rearrangement of differentiating B cells.
IL-7-mediated proliferation of late pre-B cells is decreased in the absence of PIM.
Since we used cell surface markers to delineate the functionally relevant subpopulation in bone marrow, we could not conclude whether an increase in the HSAhi/BP1-positive cell levels can be attributed to an enhanced proliferation upon heavy-chain rearrangement (resulting in more cells staining for these surface markers) or an attenuated cell cycle progression in response to pre-BCR/IL-7-mediated activation (resulting in a delayed exit from early pre-B to late pre-B, leading to an increased fraction of cells staining for the surface profile).
To distinguish between these possibilities, we grew IL-7-driven pre-B-cell cultures in vitro. Proliferation assays of B-cell progenitors demonstrated that the in vitro proliferative response to IL-7 is severely impaired in Pim mutant bone marrow cells (Fig. 3e), arguing against an increased proliferative response to IL-7 in vivo. To ensure that the diminished response to IL-7 was not the result of decreased expression of the IL-7 receptor complex, we analyzed the expression of the IL-7Rα and common-γ-chain on differentiating B cells in the bone marrow. IL-7Rα expression of immature B cells in Pim-deficient mice is marginally decreased compared to wild-type mouse results (Fig. 2f), whereas common-γ-chain expression levels were essentially the same (Fig. 2f). These data indicate that the differentiating Pim-deficient B cells might be slightly less sensitive to IL-7-induced signaling, but it seems unlikely that this difference in receptor expression is responsible for the observed differences in the IL-7-induced proliferative response.
In analyzing the contributions of the individual PIM family members to this phenotype, it can be shown that the extent of the observed in vivo defect (increased presence of the HSAhi/BP1-positive cells) correlated inversely with the ability to proliferate in response to the presence of IL-7 in vitro (see Fig. SA in the supplementary material). Although Pim1 and Pim3 are in part redundant, Pim1 is the most critical family member for modulating IL-7-driven proliferation of these immature B cells (see Fig. SA in the supplementary material). Taken together, these data suggest that the increased subpopulation of CD43+/B220+ cells that are HSAhi/BP1 positive in PIM-deficient bone marrow can be attributed, at least partially, to a decreased proliferative response to IL-7.
PIM affects cytokine-mediated cell growth and differentiation of bone marrow cells.
The induction of Pim expression is an early response to a variety of growth factors involved in hematopoiesis and is likely mediated by JAK/STAT signaling, as has been shown for IL-6-induced Pim1 expression (36). JAK-deficient as well as STAT-deficient hematopoietic progenitors show a reduction in their capacity to form colonies in response to distinctive growth factors (for a review, see reference 17). The growth of Pim1 mutant bone marrow-derived mast cells in response to the presence of IL-3 was previously shown to be diminished (10). Hence, to characterize the contribution of PIM proteins to myeloid and erythroid differentiation in response to growth factors, we performed colony formation assays of distinct Pim mutant hematopoietic progenitor populations (Table 1).
TABLE 1.
In vitro colony formation capacity of mutant PIM and wild-type bone marrow cells in response to distinct growth factors
| Growth factor | Colony type | Mean (±SD) colony formation capacity (n) for mice of Genotypea
|
||||
|---|---|---|---|---|---|---|
| Pim1+/+2+/+3+/+ | Pim1−/−2+/+3+/+ | Pim1+/+2−/−3+/+ | Pim1+/+2+/+3−/− | Pim1−/−2−/−3−/− | ||
| IL-3 | CFU-mix | 382 ± 43 (16) | 194 ± 53b (3) | 381 ± 14 (2) | 336 ± 55 (6) | 120 ± 37b (12) |
| SCF | CFU-mix | 119 ± 10 (16) | 82 ± 20 (3) | 126 ± 5 (2) | 95 ± 21b (5) | 64 ± 12b (12) |
| GM-CSF | CFU-GM | 358 ± 32 (18) | 402 ± 62 (2) | 365 ± 30 (2) | 385 ± 43 (3) | 357 ± 42 (12) |
| TPO | CFU-Meg | 17.9 ± 2.5 (9) | 0.26 ± 0.31 (3) | 19.6 ± 0.8 (2) | 18 ± 3 (6) | 0 ± 0b (9) |
| IL-5 | CFU-Eos | 19.6 ± 2.1 (10) | 8 ± 5.7 (2) | 20.4 ± 0.4 (2) | 18.6 ± 6.3 (3) | 2 ± 1b (9) |
Values represent numbers of colonies. n, number of mice. Pim1+/+2+/+3+/+, Pim1+/+ Pim2+/+ Pim3+/+.
P < 0.05 (t test analysis compared to the wild type).
IL-3 signaling induces Pim1 and Pim2 very efficiently and Pim3 to a lesser extent (Fig. 1d). The response of PIM-deficient bone marrow cells to the presence of IL-3 was strongly impaired, as indicated by a reduction in both colony number (approximately threefold) and colony size. Giemsa staining revealed that mixed hematopoietic colonies derived from PIM-deficient marrow contained cells predominantly of the erythroid lineage (see Fig. SB in the supplementary material), suggesting a substantial block in IL-3-dependent myeloid development. Although loss of PIM1 substantiates the largest effect, the additional loss of PIM2 and PIM3 does enhance the observed defect (P = 0.026). The disruption of all Pim genes decreases the granulocyte/macrophage content of IL-3-dependent CFU-mix colonies. However, the ability of Pim mutant bone marrow progenitors to form colonies in response to the presence of GM-CSF was unimpaired (Table 1) in spite of the fact that the GM-CSF receptor is a close family member of the IL-3 receptor and potently induces Pim expression (22). TPO stimulates the development of megakaryocytes. Pim mutant bone marrow cells were unable to differentiate into megakaryocytes in vitro, and again the loss of Pim1 displayed the largest effect. In in vivo experiments, we observed fewer and hypoproliferative megakaryocytes in spleen sections, as evidenced by the absence of mitotic cells (data not shown). Moreover, formation of eosinophilic granulocytes in vitro in response to IL-5 was impaired; it seems as if this had also been predominantly mediated by PIM1 loss, although once again the additional loss of PIM2 and PIM3 considerably enhanced the effect. Lastly, a reduction in the number (50% of wild-type numbers) and size of colonies was also seen in the response of Pim1 Pim2 Pim3 mutant bone marrow cells to the presence of SCF, a growth factor that signals via the KIT tyrosine kinase receptor and poorly induces Pim expression (3, 10). In contrast to IL-3-mediated colony formation, SCF showed an effect on growth and size of the colonies but not on the outgrowth of the different lineages within the colonies (data not shown).
PIM affects T-cell proliferation in response to TCR and IL-2 signaling.
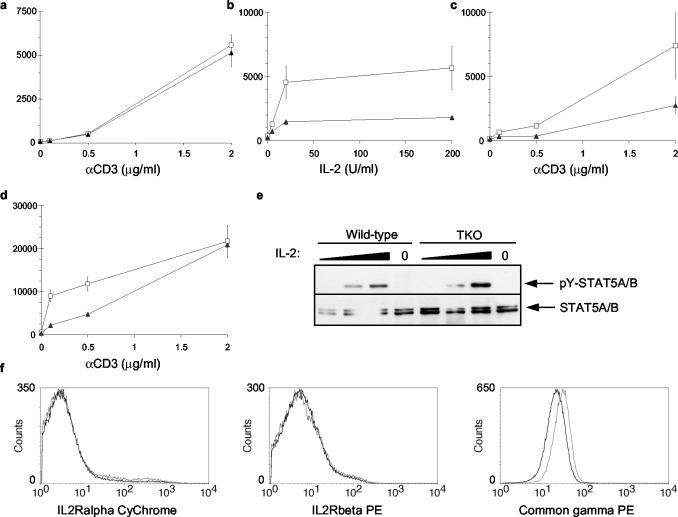
Since we observed no effect of PIM deficiency on T-lymphocyte differentiation in vivo, we assumed that peripheral T cells have been selected in a similar manner for the functionality of the TCR complex in both PIM-proficient and PIM-deficient mice. Therefore, we used peripheral T lymphocytes to study activation-induced proliferative responses of lymphocytes in the absence of PIM. IL-2 signaling efficiently induces Pim transcription (3, 8), and IL-2 signal transduction, like that of IL-7, is in part mediated by the common-γ-chain receptor. IL-2-induced signals display a strong synergy with TCR-mediated T-cell activation, which has also been shown to induce Pim1 expression (47). Therefore, we first analyzed the proliferation of Pim mutant peripheral T cells in response to CD3 cross-linking. Pim mutant T cells stimulated by CD3 cross-linking alone incorporated [3H]thymidine at rates similar to those seen with wild-type T cells (Fig. 4a). When T cells were activated by suboptimal levels of αCD3 and a dilution series of IL-2, however, PIM-deficient T cells exhibit a reduced proliferation, as indicated by a two- to fourfold reduction in [3H]thymidine incorporation (Fig. 4b). A significant reduction was also observed for Pim2 mutant T cells but not for cells from Pim1 or Pim3 single KOs (see Fig. SC in the supplementary material).
FIG. 4.
The loss of PIM proteins affects [3H]thymidine incorporation induced by synergistic TCR and IL-2 signaling. Splenic T lymphocytes derived from wild-type mice (open squares) or PIM-deficient mice (closed triangles) were cultured for 64 h in the presence of different concentrations of coated αCD3 (a), a suboptimal concentration of coated αCD3 (0.3 μg/ml) and increasing concentrations of hIL-2 (b), constant hIL-2 (5 U/ml) and increasing amounts of coated αCD3 (c), or constant hIL-2 (200 U/ml) and increasing amounts of αCD3 (d). The proliferation of T lymphocytes was assayed by measuring [3H]thymidine incorporation for the last 16 h of culture. Each panel represents one of three independent experiments with similar results (n = 3 for each genotype). (e) STAT5 phosphorylation in response to a dilution series of 0.02, 2.0, and 200 U of IL-2 of wild-type and PIM-deficient spleen cells. (f) Cell surface expression of components of the IL-2R complex on splenic T cells of wild-type mice (open squares) and TKO mice (solid triangles). PE, phycoerythrin.
To address whether the difference in proliferation is dependent on TCR activation, T cells were activated with different doses αCD3 in the presence of low (5 U/ml) or high (200 U/ml) levels of IL-2 (Fig. 4c and 4d). PIM-deficient T cells displayed a reduced proliferation at suboptimal levels of αCD3 irrespective of the IL-2 concentration, which confirms the data in Fig. 2b. However, this difference disappeared at high levels of αCD3 (2 μg/ml) in the presence of high levels of IL-2 (200 U/ml; Fig. 4d). These results indicate that PIM-deficient T cells display normal proliferative responses at optimal levels of αCD3 and hIL2 but that PIM proteins are required for proper proliferation upon suboptimal stimulation of either signaling pathway in vitro.
To determine whether expression levels of IL-2R complex components could be contributing to the observed decreased responses, we analyzed the expression levels of IL-2Rα, IL-2Rβ, and common-γ-chain on mature peripheral T cells (Fig. 4f). The expression levels of IL-2Rα and IL-2Rβ between wild-type and TKO mice were similar. However, Pim-deficient peripheral T cells expressed decreased levels of common-γ-chain on their cell surfaces. To elucidate whether the difference in common γ-chain expression also resulted in less-efficient STAT5 phosphorylation upon IL-2-induced signaling, we analyzed STAT5 phosphorylation in response to a dilution series of IL-2. We did not observe a difference between wild-type and TKO T cells in STAT5 phosphorylation upon IL-2 treatment (Fig. 4e). This indicates that the proliferative defect observed in PIM-deficient T cells is not caused by decreased STAT5 phosphorylation in response to the presence of IL-2.
PIM is required for efficient cycling of peripheral T cells.
The PIM proteins have been suggested to play an important role in proliferation (5, 28, 30, 31, 36); however, several of these reports and others have provided additional evidence for an antiapoptotic function of PIM (13, 28, 30, 31, 33, 36, 46). Since an apparent decrease in the incorporation of [3H]thymidine can be caused by decreased proliferation or decreased survival or a combination of the two, we investigated these processes in PIM-deficient peripheral T cells.
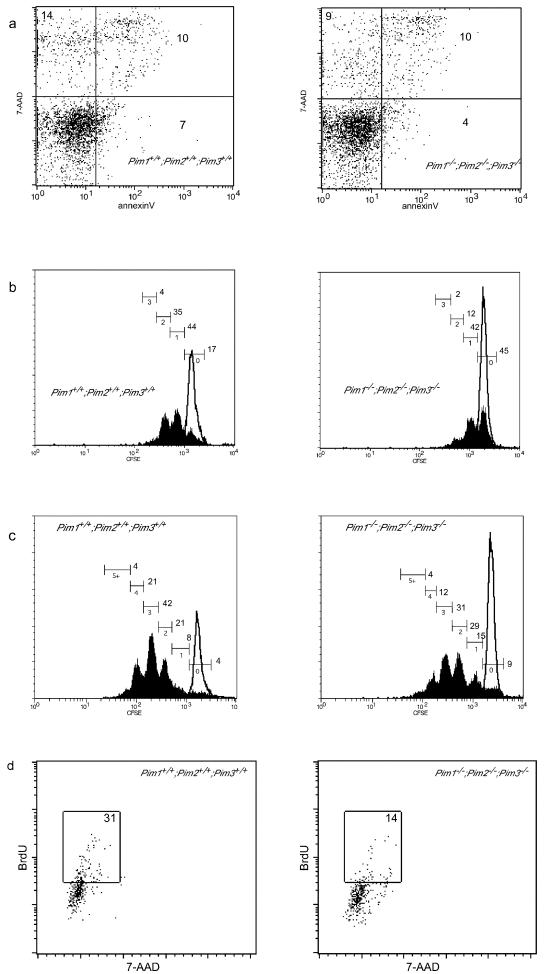
The apoptotic index and cell viability of peripheral T cells cultured in the presence of suboptimal levels of αCD3 and 200 U of hIL2 for 24 and 48 h were determined using Annexin V, 7-AAD, and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling staining. These analyses showed that the rate of apoptosis in PIM-deficient T-cells is not affected (Fig. 5a and data not shown).
FIG. 5.
PIM proteins appear to facilitate cell cycle entry in response to TCR and IL-2 signaling. Splenic T lymphocytes derived from wild-type or PIM-deficient mice were cultured in the presence of αCD3 (0.3 μg/ml) and hIL-2 (200 U/ml). For all analyses the T-cell population was characterized by flow cytometry using αTCRβ or αCD3 antibodies. (a) The proportions of apoptotic and dead cells after 48 h of culturing, as determined by Annexin V and 7-AAD staining. (b to c) The number of cell divisions was determined after 48 h (b) or 72 h (c) of culture growth, with CFSE used as a tracking dye (black lines correspond to unstimulated cells). (d) Analysis of DNA synthesis after 24 h of culture growth in the presence of hIL-2 (200 U/ml) and αCD3 (0.3 μg/ml).
Since the defect in [3H]thymidine incorporation was predominantly apparent in Pim2 single mutant and Pim compound KO T cells, we labeled wild-type and KO cells of both types with CFSE to track the number of cell divisions. After 48 h of culture in the presence of αCD3 and IL-2, 45% of the Pim mutant T cells had not yet divided compared to only 17% of the wild-type T cells. In addition, the proportion of PIM-deficient T cells that had divided more than once (14%) was markedly decreased compared to that of wild-type T cells (39%) (Fig. 5b). After 72 h, most of the PIM-deficient T-cells had divided at least once but they still lagged behind compared to their wild-type counterparts (Fig. 5c). Similar results were obtained by BrdU pulse labeling of the cells after 24 h of culturing under the same conditions (Fig. 5d). Also, for Pim2 mutant T cells the [3H]thymidine data correlated to a decrease in the number of cell divisions (see Fig. SC in the supplementary material).
Taken together, these data indicate that PIM-deficient T cells display a reduced proliferative response to suboptimal αCD3 activation in the presence of IL-2 due to a reduced capacity to undergo cell division under these conditions. No contribution of an enhanced induction of apoptosis was observed. Since the difference in the fractions of cells which had divided more than once did not increase in PIM-proficient T-cell cultures between 48 and 72 h compared to PIM-deficient T-cell culture results and since cells which had undergone multiple rounds of cell division were readily detected in PIM-deficient T-cell cultures, it seems that the observed diminished efficiency of cell cycle progression was specific for the first cell division of resting T cells.
DISCUSSION
In this study we analyzed the effect of targeted disruption of all PIM family members on murine development and hematopoietic differentiation. Surprisingly, the combined deficiency for all PIM family members resulted in a rather mild phenotype for the resultant compound Pim mutant mice. In light of the strong conservation of Pim genes between different species, this mild phenotype argues against a role for PIM family members in critical developmental processes. Apparently, life is compatible with the absence of PIM. On the other hand, the absence of an obvious phenotype in vivo under homeostatic conditions might in part be explained by the nature of Pim genes as early-response genes responding to growth factors and cytokines. In fact, critical functions of PIM family members might only be revealed under conditions of high stress, which severely disrupt the homeostatic situation.
We demonstrated that PIM proteins are important for body growth, since Pim1 Pim2 Pim3 compound KO mice display a profound reduction in body size at birth and throughout their entire live span. The decrease in body size was predominantly caused by a reduction in the number of cells rather than cell size, although a minor cell-size difference was consistently observed between PIM-proficient and -deficient animals. The reduction in body size (approximately 30%) observed at birth is, as such, reminiscent of a defect in insulin-like growth factor (IGF)-insulin signaling. Mice carrying defects in either of these pathways display a reduction in body size ranging from 20 to 60%, depending on the nature of the defect (for a review, see reference 1). However, it remains to be determined whether IGF-insulin signaling is indeed affected in Pim mutant mice.
The majority of hematopoietic growth factors are efficient inducers of Pim1, Pim2, and Pim3. In addition, a role for PIM1 in the proliferative response of hematopoietic cells to the presence of IL-3 and IL-7 was demonstrated in experiments with Pim1 KO bone marrow cells (9, 10). The generation of Pim1−/− Pim2−/− Pim3−/− compound KO mice and the subsequent isolation of PIM-deficient bone marrow cells demonstrated that PIM1, PIM2, and PIM3 can act redundantly in stimulating the colony formation of bone marrow cells in response to the presence of IL-3, IL-5, SCF, and TPO. It should, however, be noted that it seems that PIM1 is the most and PIM2 is the least crucial PIM protein in these responses. Since PIM2 is more efficiently induced than PIM3 by a number of these growth factors, PIM2 might have a role distinct from the functions of PIM1 and PIM3 in these settings. In contrast to results seen with respect to the response to IL-3, IL-5, SCF, and TPO, we did not observe an effect of PIM deficiency on the GM-CSF response. This is very remarkable, in particular because the receptor for GM-CSF is highly related to the IL-3 and IL-5 receptors, as they share the same common-β-chain, which acts as the signaling module. Thus, although GM-CSF signaling leads to transcriptional activation of Pim (22), the PIM proteins appear not to be necessary for efficient transduction of the GM-CSF signals. This is in contrast to the very similar IL-3 and IL-5 signaling, in which PIM kinases seem to play an important role.
In addition to the impaired colony formation of PIM-deficient bone marrow cells in response to a variety of growth factors, PIM-deficient early pre-B cells showed a reduced proliferative response to IL-7 in vitro. This was reflected in vivo by an attenuated progression of pro-B cells through the IL-7-driven proliferative stage that follows successful rearrangement of the heavy chain. Since differentiating B cells in the bone marrow of Pim-deficient animals show only marginally decreased IL-7Rα expression levels, it seems unlikely that the attenuated proliferative response to IL-7 can be attributed to lower IL-7R expression levels. In young animals, peripheral B-cell numbers were reduced, whereas in older animals, this was not the case. The observation that the number of B cells is unaltered in the periphery of older animals is likely explained by the continuous supplying of newly formed B cells from the bone marrow into the peripheral B-cell compartment and homeostatic proliferation of peripheral B cells (for a review, see reference 42).
The differentiation of T cells in the thymus is also characterized by a series of alternating phases of genomic recombination (resting cells) and IL-7-driven proliferation. We did not observe any effect of the absence of PIM on T-cell differentiation in the thymus. Although Pim mutant mice do not display abnormalities in thymic T-cell differentiation, the ability of peripheral T cells to proliferate in response to synergistic IL-2 and TCR activation was markedly impaired. The proliferative response of Pim-deficient and Pim-proficient T cells to a dilution series of CD3-cross-linking antibodies was identical. Likewise, serial dilutions of IL-2 on peripheral T cells elicited the same responses in terms of proliferation and STAT5 phosphorylation in both Pim-deficient and -proficient T cells. Yet when the two stimuli are combined, Pim-deficient T cells display a decreased response to the combined activities when either stimulus is present at suboptimal levels. PIM mutant peripheral T cells express lower levels of common-γ-chain but normal levels of the IL-2Rα and IL-2Rβ chains. This might indicate that common-γ-chain-transduced signals contribute to the decreased proliferation of PIM-deficient T cells. However, we did not find differences in STAT5 phosphorylation between PIM-proficient and PIM-deficient T cells. Moreover, it has been shown that the common-γ-chain is not required for antigen-driven proliferation of naïve T cells (21), indicating that additional mechanisms are likely to contribute to the decreased proliferation of peripheral T cells in response to the IL-2 and TCR activation. Further analysis of this proliferation defect demonstrates that PIM appears to be important for cell cycle entry of resting peripheral T cells at suboptimal levels of TCR signaling. PIM2 appeared to be the most critical PIM protein for this response, although PIM1 and PIM3 also contribute. These results indicate that the PIM proteins can augment the response to growth factor signals either directly or indirectly. The indirect pathway could involve signaling via the IGF receptors, as both IGF-I and -II can potentiate proliferation in B and T cells (14, 19). Such a role for PIM would also be consistent with previous results showing that overexpression of PIM1 can rescue the reduced thymic cellularity in common-γ-chain-deficient mice but only in the context of a functional CD3 complex (18).
We found no evidence that PIM deficiency compromises the survival of peripheral T cells. The results of other studies indicated that in vitro overexpression of PIM proteins protects cells against apoptosis (13, 28, 30, 31, 33, 36, 46). This apparent discrepancy emphasizes the notion that gene inactivation and overexpression studies do not necessarily reveal the same physiologically relevant functions of a protein. On the one hand, ectopic overexpression of proteins might evoke effects that are not representative of its function under physiological conditions. On the other hand, inactivation might elicit compensation by other proteins, obscuring or mitigating the normal role of the protein of interest. With respect to PIM proteins, which are usually expressed during a short time period due to the short half-lives of both mRNA and protein, overexpression studies using cells with enhanced PIM activity for prolonged periods are likely to yield data different from data obtained with studies using cells from which PIM activity had been deleted. The former situation might be more reflective of the role of PIM proteins during cellular transformation as observed in M-MuLV-induced tumors, whereas the latter could give more insight into the physiological role of PIM proteins in nontransformed cells. Therefore, the observation that an absence of PIM proteins does not give rise to cells which are more susceptible to apoptosis is not in contradiction with the notion that overexpression of PIM proteins might provide protection from apoptosis. The results from our study appear to be in agreement with Pim1- and Pim2-deficient mouse results from retroviral insertional mutagenesis screenings designed to identify genes that can substitute for PIM function in lymphomagenesis (25). The genes identified in this screen (e.g., Kit and Ccnd2) all regulate the G1 to S transition. Therefore, although we cannot exclude the possibility that rescue from apoptosis is indeed a function of PIM proteins in lymphomagenesis, it seems that regulation of cell cycle is at least as important.
Since Pim expression is directly regulated by the JAK/STAT pathway (36), the Pim mutant phenotypes described here might provide clues about the biological significance of the STAT proteins for transactivation of the Pim genes. In fact, Stat5a Stat5b compound KO bone marrow cells display a reduced ability to form colonies in response to the presence of IL-3, IL-5, IL-7, SCF, GM-CSF, and TPO (37). Thus, the inability of STAT-deficient cells to induce Pim expression might be responsible for some of the observed signaling defects. In particular, STAT-deficient T cells cannot proliferate in response to TCR/IL-2 signaling (27). Although this phenotype is much stronger than the defect observed in Pim mutant T cells, our data show that the induction of PIM proteins has a significant effect on TCR/IL-2-mediated proliferation.
In summary, the phenotypes of the Pim1 Pim2 Pim3 compound-KO mice illustrate a role for the PIM proteins in single-growth-factor signaling and in synergistic signaling pathways. The results with respect to synergistic growth factor signaling would suggest a view that PIM proteins might act as sensitizers of these pathways. This view would fit the induction of early transcription of the Pim genes by growth factors and the instability of the PIM mRNAs and proteins. Although such interplay between growth factor signals has been postulated before, the mediators have remained obscure. We show here that PIM proteins might fulfill such a role.
Supplementary Material
Acknowledgments
We thank P. Krimpenfort and K. van Veen for blastocyst injections, L. Tolkamp and H. Starreveld for animal care, and L. Oomen for confocal microscopy assistance.
This work was supported by the Dutch Cancer Society (H.M. and M.N.) and the Leukemia and Lymphoma Society of America (J.A.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1.Accili, D., J. Nakae, J. J. Kim, B. C. Park, and K. I. Rother. 1999. Targeted gene mutations define the roles of insulin and IGF-I receptors in mouse embryonic development. J. Pediatr. Endocrinol. Metab. 12:475-485. [DOI] [PubMed] [Google Scholar]
- 2.Allen, J. D., and A. Berns. 1996. Complementation tagging of cooperating oncogenes in knockout mice. Semin. Cancer Biol. 7:299-306. [DOI] [PubMed] [Google Scholar]
- 3.Allen, J. D., E. Verhoeven, J. Domen, M. van der Valk, and A. Berns. 1997. Pim-2 transgene induces lymphoid tumors, exhibiting potent synergy with c-myc. Oncogene 15:1133-1141. [DOI] [PubMed] [Google Scholar]
- 4.Baytel, D., S. Shalom, I. Madgar, R. Weissenberg, and J. Don. 1998. The human Pim-2 proto-oncogene and its testicular expression. Biochim. Biophys. Acta 1442:274-285. (Erratum, 1444:312-313, 1999.) [DOI] [PubMed] [Google Scholar]
- 5.Berns, A., H. Mikkers, P. Krimpenfort, J. Allen, B. Scheijen, and J. Jonkers. 1999. Identification and characterization of collaborating oncogenes in compound mutant mice. Cancer Res. 59:1773s-1777s. [PubMed] [Google Scholar]
- 6.Borg, K. E., M. Zhang, D. Hegge, R. L. Stephen, D. J. Buckley, N. S. Magnuson, and A. R. Buckley. 1999. Prolactin regulation of pim-1 expression: positive and negative promoter elements. Endocrinology 140:5659-5668. [DOI] [PubMed] [Google Scholar]
- 7.Cuypers, H. T., G. Selten, W. Quint, M. Zijlstra, E. R. Maandag, W. Boelens, P. van Wezenbeek, C. Melief, and A. Berns. 1984. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell 37:141-150. [DOI] [PubMed] [Google Scholar]
- 8.Dautry, F., D. Weil, J. Yu, and A. Dautry-Varsat. 1988. Regulation of pim and myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J. Biol. Chem. 263:17615-17620. [PubMed] [Google Scholar]
- 9.Domen, J., N. M. van der Lugt, D. Acton, P. W. Laird, K. Linders, and A. Berns. 1993. Pim-1 levels determine the size of early B lymphoid compartments in bone marrow. J. Exp. Med. 178:1665-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domen, J., N. M. van der Lugt, P. W. Laird, C. J. Saris, A. R. Clarke, M. L. Hooper, and A. Berns. 1993. Impaired interleukin-3 response in Pim-1-deficient bone marrow-derived mast cells. Blood 82:1445-1452. [PubMed] [Google Scholar]
- 11.Eichmann, A., L. Yuan, C. Breant, K. Alitalo, and P. J. Koskinen. 2000. Developmental expression of pim kinases suggests functions also outside of the hematopoietic system. Oncogene 19:1215-1224. [DOI] [PubMed] [Google Scholar]
- 12.Feldman, J. D., L. Vician, M. Crispino, G. Tocco, V. L. Marcheselli, N. G. Bazan, M. Baudry, and H. R. Herschman. 1998. KID-1, a protein kinase induced by depolarization in brain. J. Biol. Chem. 273:16535-16543. [DOI] [PubMed] [Google Scholar]
- 13.Fox, C. J., P. S. Hammerman, R. M. Cinalli, S. R. Master, L. A. Chodosh, and C. B. Thompson. 2003. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 17:1841-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, L. F., D. Piktel, and K. S. Landreth. 1993. Insulin-like growth factor-1 potentiates expansion of interleukin-7-dependent pro-B cells. Blood 82:3005-3011. [PubMed] [Google Scholar]
- 15.Hardy, R. R., and K. Hayakawa. 2001. B cell development pathways. Annu. Rev. Immunol. 19:595-621. [DOI] [PubMed] [Google Scholar]
- 16.Hess, J., A. Werner, T. Wirth, F. Melchers, H. M. Jack, and T. H. Winkler. 2001. Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc. Natl. Acad. Sci. USA 98:1745-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihle, J. N., W. Thierfelder, S. Teglund, D. Stravapodis, D. Wang, J. Feng, and E. Parganas. 1998. Signaling by the cytokine receptor superfamily. Ann. N. Y. Acad. Sci. 865:1-9. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, H., P. Krimpenfort, M. Haks, J. Allen, B. Blom, C. Demolliere, A. Kruisbeek, H. Spits, and A. Berns. 1999. PIM1 reconstitutes thymus cellularity in interleukin 7- and common gamma chain-mutant mice and permits thymocyte maturation in Rag- but not CD3γ-deficient mice. J. Exp. Med. 190:1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, E. W., L. A. Jones, and R. W. Kozak. 1992. Expression and function of insulin-like growth factor receptors on anti-CD3-activated human T lymphocytes. J. Immunol. 148:63-71. [PubMed] [Google Scholar]
- 20.Laird, P. W., N. M. van der Lugt, A. Clarke, J. Domen, K. Linders, J. McWhir, A. Berns, and M. Hooper. 1993. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 21:4750-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lantz, O., I. Grandjean, P. Matzinger, and J. P. Di Santo. 2000. Gamma chain required for naive CD4+ T-cell survival but not for antigen proliferation. Nat. Immunol. 1:54-58. [DOI] [PubMed] [Google Scholar]
- 22.Lilly, M., T. Le, P. Holland, and S. L. Hendrickson. 1992. Sustained expression of the pim-1 kinase is specifically induced in myeloid cells by cytokines whose receptors are structurally related. Oncogene 7:727-732. [PubMed] [Google Scholar]
- 23.Lilly, M., J. Sandholm, J. J. Cooper, P. J. Koskinen, and A. Kraft. 1999. The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 18:4022-4031. [DOI] [PubMed] [Google Scholar]
- 24.Matikainen, S., T. Sareneva, T. Ronni, A. Lehtonen, P. J. Koskinen, and I. Julkunen. 1999. Interferon-alpha activates multiple STAT proteins and upregulates proliferation-associated IL-2Rα, c-myc, and pim-1 genes in human T cells. Blood 93:1980-1991. [PubMed] [Google Scholar]
- 25.Mikkers, H., J. Allen, P. Knipscheer, L. Romeyn, A. Hart, E. Vink, and A. Berns. 2002. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 32:153-159. [DOI] [PubMed] [Google Scholar]
- 26.Miura, O., Y. Miura, N. Nakamura, F. W. Quelle, B. A. Witthuhn, J. N. Ihle, and N. Aoki. 1994. Induction of tyrosine phosphorylation of Vav and expression of Pim-1 correlates with Jak2-mediated growth signaling from the erythropoietin receptor. Blood 84:4135-4141. [PubMed] [Google Scholar]
- 27.Moriggl, R., D. J. Topham, S. Teglund, V. Sexl, C. McKay, D. Wang, A. Hoffmeyer, J. van Deursen, M. Y. Sangster, K. D. Bunting, G. C. Grosveld, and J. N. Ihle. 1999. Stat5 is required for IL-2-induced cell cycle progression of peripheral T cells. Immunity 10:249-259. [DOI] [PubMed] [Google Scholar]
- 28.Moroy, T., A. Grzeschiczek, S. Petzold, and K. U. Hartmann. 1993. Expression of a Pim-1 transgene accelerates lymphoproliferation and inhibits apoptosis in lpr/lpr mice. Proc. Natl. Acad. Sci. USA 90:10734-10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mountford, P., B. Zevnik, A. Duwel, J. Nichols, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieborowska-Skorska, M., G. Hoser, P. Kossev, M. A. Wasik, and T. Skorski. 2002. Complementary functions of the antiapoptotic protein A1 and serine/threonine kinase pim-1 in the BCR/ABL-mediated leukemogenesis. Blood 99:4531-4539. [DOI] [PubMed] [Google Scholar]
- 31.Nosaka, T., T. Kawashima, K. Misawa, K. Ikuta, A. L. Mui, and T. Kitamura. 1999. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 18:4754-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palaty, C. K., G. Kalmar, G. Tai, S. Oh, L. Amankawa, M. Affolter, R. Aebersold, and S. L. Pelech. 1997. Identification of the autophosphorylation sites of the Xenopus laevis Pim-1 proto-oncogene-encoded protein kinase. J. Biol. Chem. 272:10514-10521. [DOI] [PubMed] [Google Scholar]
- 33.Pircher, T. J., S. Zhao, J. N. Geiger, B. Joneja, and D. M. Wojchowski. 2000. Pim-1 kinase protects hematopoietic FDC cells from genotoxin-induced death. Oncogene 19:3684-3692. [DOI] [PubMed] [Google Scholar]
- 34.Saris, C. J., J. Domen, and A. Berns. 1991. The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG. EMBO J. 10:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selten, G., H. T. Cuypers, and A. Berns. 1985. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 4:1793-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirogane, T., T. Fukada, J. M. Muller, D. T. Shima, M. Hibi, and T. Hirano. 1999. Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 11:709-719. [DOI] [PubMed] [Google Scholar]
- 37.Sicinski, P., J. L. Donaher, S. B. Parker, T. Li, A. Fazeli, H. Gardner, S. Z. Haslam, R. T. Bronson, S. J. Elledge, and R. A. Weinberg. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621-630. [DOI] [PubMed] [Google Scholar]
- 38.Teglund, S., C. McKay, E. Schuetz, J. M. van Deursen, D. Stravopodis, D. Wang, M. Brown, S. Bodner, G. Grosveld, and J. N. Ihle. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93:841-850. [DOI] [PubMed] [Google Scholar]
- 39.Telerman, A., R. Amson, R. Zakut-Houri, and D. Givol. 1988. Identification of the human pim-1 gene product as a 33-kilodalton cytoplasmic protein with tyrosine kinase activity. Mol. Cell. Biol. 8:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Temple, R., E. Allen, J. Fordham, S. Phipps, H. C. Schneider, K. Lindauer, I. Hayes, J. Lockey, K. Pollock, and R. Jupp. 2001. Microarray analysis of eosinophils reveals a number of candidate survival and apoptosis genes. Am. J. Respir. Cell Mol. Biol. 25:425-433. [DOI] [PubMed] [Google Scholar]
- 41.te Riele, H., E. R. Maandag, A. Clarke, M. Hooper, and A. Berns. 1990. Consecutive inactivation of both alleles of the pim-1 proto-oncogene by homologous recombination in embryonic stem cells. Nature 348:649-651. [DOI] [PubMed] [Google Scholar]
- 42.Tough, D. F., and J. Sprent. 1995. Lifespan of lymphocytes. Immunol. Res. 14:1-12. [DOI] [PubMed] [Google Scholar]
- 43.Trumpp, A., Y. Refaeli, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 44.van der Lugt, N. M., J. Domen, E. Verhoeven, K. Linders, H. van der Gulden, J. Allen, and A. Berns. 1995. Proviral tagging in E mu-myc transgenic mice lacking the Pim-1 proto-oncogene leads to compensatory activation of Pim-2. EMBO J. 14:2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verbeek, S., M. van Lohuizen, M. van der Valk, J. Domen, G. Kraal, and A. Berns. 1991. Mice bearing the Eμ-myc and Eμ-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol. Cell. Biol. 11:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Z., N. Bhattacharya, M. K. Meyer, H. Seimiya, T. Tsuruo, J. A. Tonani, and N. S. Magnuson. 2001. Pim-1 negatively regulates the activity of PTP-U2S phosphatase and influences terminal differentiation and apoptosis of monoblastoid leukemia cells. Arch. Biochem. Biophys. 390:9-18. [DOI] [PubMed] [Google Scholar]
- 47.Wingett, D., A. Long, D. Kelleher, and N. S. Magnuson. 1996. pim-1 proto-oncogene expression in anti-CD3-mediated T cell activation is associated with protein kinase C activation and is independent of Raf-1. J. Immunol. 156:549-557. [PubMed] [Google Scholar]
- 48.Yip-Schneider, M. T., M. Horie, and H. E. Broxmeyer. 1995. Transcriptional induction of pim-1 protein kinase gene expression by interferon gamma and posttranscriptional effects on costimulation with steel factor. Blood 85:3494-3502. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.