Abstract
Following hybridization with embryonic stem (ES) cells, somatic genomes are epigenetically reprogrammed and acquire pluripotency. This results in the transcription of somatic genome-derived tissue-specific genes upon differentiation. During nuclear reprogramming, it is expected that DNA and chromatin modifications, believed to function in cell-type-specific epigenotype memory, should be significantly modified. Indeed, current evidence indicates that acetylation and methylation of histone H3 and H4 amino termini play a major role in the regulation of gene activity through the modulation of chromatin conformation. Here, we show that the reprogrammed somatic genome of ES hybrid cells becomes hyperacetylated at H3 and H4, while lysine 4 (K4) of H3 becomes globally hyper-di- and -tri-methylated. In the Oct4 promoter region, histones H3 and H4 are acetylated and H3-K4 is highly tri-methylated on both the ES and reprogrammed somatic genomes, which correlates with gene activation and DNA demethylation. However, H3-K4 is also di- and tri-methylated in the promoter regions of Neurofilament-M (Nfm), Nfl, and Thy-1, which are all silent in both ES and hybrid cells. Thus, H3-K4 di- and tri-methylation of reprogrammed somatic genomes is independent of gene activity and represents one of the major events that occurs during somatic genome reprogramming towards a transcriptional activation-permissive state.
Nuclear reprogramming of a somatic epigenotype to a totipotential epigenotype has been demonstrated by successful production of cloned animals via transplantation of somatic nuclei into enucleated oocytes (44). We have previously established an alternative experimental system that also induces the nuclear reprogramming of somatic nuclei, by in vitro cell fusion of mouse adult thymocytes with mouse embryonic stem (ES) cells. Reprogramming of the somatic nucleus to an undifferentiated state was demonstrated by reactivation of the pluripotential cell-specific marker gene Oct4-GFP and reactivation of the somatic cell-derived inactive X-chromosome. This correlated with reexpression of the undifferentiated state-specific Xist and Tsix genes (19, 48). The pluripotential competence of hybrid cells was shown through the formation of teratomas following subcutaneous injection into immunodeficient mice, through contribution of the hybrid cells to normal embryogenesis in chimeras and through the reprogrammed somatic genome-derived transcription of various tissue-specific mRNAs, both in teratomas redifferentiated in vivo and in mesencephalic dopaminergic neurons redifferentiated in vitro (46, 48). Therefore, pluripotential competence, represented by multilineage cell differentiation and transcription of tissue-specific genes, is conferred on the somatic genome by trans-acting factors present in undifferentiated ES cells.
One of the characteristic features of animal cloning experiments through nuclear transplantation of somatic cell nuclei has been the considerably low rate of survival to adulthood (44). This may be directly related to insufficient reprogramming of the somatic nuclei. This is supported by the observed expression of somatic nucleus-derived Oct4-GFP in a limited number of cloned blastocysts (4), aberrant reactivation of Oct4-related genes (5), and abnormal hypermethylation of histone H3-K9 associated with DNA hypermethylation in cloned preimplantation embryos (10, 40). Interestingly, in contrast with cloned embryos, reactivation of a somatic cell-derived Oct4-GFP transgene was found in nearly 100% of independently isolated ES hybrid clones (48). Furthermore, the reprogrammed somatic genome of these clones possessed pluripotential competence to redifferentiate into a variety of cell types (46). These findings indicate that ES hybrid cells in which the somatic genome has been sufficiently reprogrammed are able to survive selectively under appropriate culture conditions. Thus, the epigenetic profile of the reprogrammed somatic genome in the hybrid cells may reflect that of the fully reprogrammed cloned embryos, rather than that of embryos in which insufficient reprogramming has occurred.
The molecular mechanism(s) and factors involved in epigenotype reprogramming are largely unknown. DNA methylation and posttranslational acetylation, phosphorylation, and methylation on histone N termini function to regulate transcriptional activation or repression of genes (22). The histone modifications are thought to play certain key roles in regulating gene activity, most likely through modulation of chromatin structure, since in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Drosophila melanogaster appropriate gene regulation occurs in the absence of DNA methylation (26). To date at least eight acetylatable lysine positions are known in the N termini of histone H3 (K9, K14, K18, and K23) and H4 (K5, K8, K12, and K16) and six methylatable lysine positions exist in those of histone H3 (K4, K9, K27, K36, and K79) and H4 (K20). In general, acetylation of histone H3 and H4 correlates with gene activation, while deacetylation correlates with gene silencing (14). Methylation of H3-K4 also marks active chromatin, which contrasts with the modulation of inactive chromatin by methylation of H3-K9 (22). Methylation of H3-K27 is an epigenetic mark for recruitment of polycomb group (Pc-G) complexes (9) and is prominent in the inactivated X chromosome of female mammalian somatic cells (37, 43). The amino-terminal tail of histone H3 is subject to three distinctive methylation states: mono-, di-, and trimethylation. Pericentric heterochromatin is enriched for trimethylated H3-K9, while centromeric regions are enriched for the dimethylated state (22). At H3-K27, both di- and trimethylation are observed across several nucleosomes, and it is the trimethylated state that has been found to induce stable recruitment of Pc-G complexes (7). At H3-K4, fully activated promoters are enriched for the trimethylated state, while H3-K4 dimethylation correlates with the basal transcription-permissive state (41). Thus, it appears that dimethylation activity prepares histones for a trimethylating activity, which then propagates stably activated or silenced chromatin domains.
In this study, immunocytochemical and chromatin immunoprecipitation (ChIP) assays revealed that histone H3 and H4 amino termini are globally hypermethylated and hyperacetylated on the reprogrammed somatic genome in intersubspecific hybrid cells. For the Oct4, Neurofilament-M (Nfm), Nfl, Thy-1, and c-myc genes, histone H3-K4 is highly di- and trimethylated, and this is independent of the activity of these genes in the undifferentiated ES hybrid cells. Thus, reprogramming of the somatic genome is characterized by transcription activation-permissive chromatin. Decondensation of the reprogrammed chromatin, marked by H3-K4 di- and trimethylation, may be a prerequisite for erasing the somatic epigenotype prior to establishment of a pluripotential epigenotype.
MATERIALS AND METHODS
Cell hybridization.
Male Hm1 ES cells (Mus musculus domesticus) deficient for the X-linked Hprt gene were maintained in ES medium on inactivated primary embryonic fibroblasts (47). The ES cell line (at fewer than 10 passages) was hybridized with thymocytes collected from 6- to 8-week-old M. musculus molossinus mice. Cell hybridization was performed as previously described (47). Hybrid cells were selected with ES medium supplemented with HAT for 8 days. The ES hybrid cell clones were picked and subcultured every 2 days. ES hybrid cells at less than 15 passages were used for experiments.
Immunocytochemistry.
ES cells (104) and thymocytes (105) were pressed on an aminopropyl-triethoxysilane-coated glass slide (Matsunami) by spinning down at 200 × g for 6 min. The cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature. After three washes with 0.1% Triton X-100 in PBS (PBST), the cells were prehybridized with blocking buffer (1% bovine serum albumin in PBST) for 1 h and then incubated with anti-acetylated histone H3 (1:200 dilution; Upstate Biotechnology), anti-acetylated histone H4 (1:200 dilution; Upstate Biotechnology), anti-dimethylated H3-K4 (1:500 dilution; Upstate Biotechnology), anti-trimethylated H3-K4 (1:1,000 dilution; AbCam), anti-dimethylated H3-K9 (1:500 dilution; Upstate Biotechnology), anti-trimethylated H3-K9 (1:1,000 dilution; AbCam), or anti-trimethylated H3-K27 (1:1,000 dilution) (37) antibody in blocking buffer at 4°C overnight. Following three further washes with PBST, they were incubated with anti-rabbit immunoglobulin G conjugated with Alexa 546 (1:500 dilution; Molecular Probes) in blocking buffer for 1 h at room temperature. ES cells were visualized with mouse monoclonal antibody against Oct4 (1:200 dilution; Santa Cruz). The cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and were mounted with a SlowFade light antifade kit (Molecular Probes).
Western blot analysis.
Whole-cell extract was prepared and separated on sodium dodecyl sulfate-15% polyacrylamide gels. The amount of sample was prequantified with histone H3. After electrophoresis, the proteins were transferred onto Protran nitrocellulose membrane (Schleicher and Schuell). The membrane was probed with anti-histone H3 (monoclonal), anti-acetylated histone H3, anti-acetylated histone H4, anti-dimethylated H3-K4, and anti-dimethylated H3-K9 antibodies. Reactions with the first antibodies were performed according to the manufacturer's instruction. Specific bands were detected using alkaline phosphatase-conjugated secondary antibodies (Zymed). 5-Bromo-4-chloro-3-indolyl-phosphate and 4-nitro blue tetrazolium chloride were used as chromogenic substrate.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as described previously (13). Histone and DNA were cross-linked by incubation in 1% formaldehyde. The chromatin was then sonicated to an average DNA fragment length of 200 to 1,000 bp. Soluble chromatin reacted with and without anti-acetylated histone H3, anti-acetylated histone H4, anti-dimethylated H3-K4, anti-trimethylated H3-K4, anti-dimethylated H3-K9, anti-trimethylated H3-K9, or anti-trimethylated H3-K27 antibodies was purified and collected in elution buffer (0.1 M NaHCO3, 1% sodium dodecyl sulfate). Cross-linking was then reversed using elution buffer containing RNase A (0.03 mg/ml) and NaCl (0.3 M) by incubation for 4 h at 65°C. Supernatant obtained without antibody was used as the input control. Following treatment with proteinase K for 1 h at 45°C, the DNA was purified and analyzed by slot blot hybridization with specific probes and PCR using the following primer sets: Oct4-F, 5′-CTAGACGGGTGGGTAAGCAA-3′, and Oct4-R, 5′-CAGGAGGCCTTCATTTTCAA-3′; Nfm-F, 5′-GGGTGACAAGAGGTCTGGAA-3′, and Nfm-R, 5′-CAGCGTGTAGCTCATCTTGG-3′; Nfl-F, 5′-CAGGGAAGTTATGGGGGTCT-3′, and Nfl-R, 5′-AGAAGAACGGGGGAGAAGAG-3′; Thy-1-F, 5′-CTCCAAAGCCAAAACCTGTC-3′, and Thy-1-R, 5′-GCTGACTGGAGGTGTTCCAT-3′; and c-myc-F, 5′-GACGCTTGGCGGGAAAA-3′, and c-myc-R, 5′-CTCTGCACACACGGCTCTT-3′. The PCR products were digested with restriction enzymes or sequenced for DNA polymorphism analyses.
Slot blotting of DNA.
Immunoprecipitated DNA was blotted onto Hybond-N+ membrane (Amersham). The membrane was hybridized with probes of sonicated genomic DNA extracted from mouse primary embryonic fibroblasts and/or a B2 repeat cDNA probe and exposed to X-Omat AR X-ray film (Kodak). Band intensity was quantified using Lane analyzer 3.0 software (ATTO), and the value relative to the thymocyte was determined.
RT-PCR.
Total RNA was purified with Trizol (Invitrogen). Template cDNA synthesized using Superscript II reverse transcriptase (Invitrogen) with random hexamers was amplified with the following specific primer sets: Oct4/3′-F, 5′-AGAGAAGGATGTGGTTCGAG-3′, and Oct4/3′-R, 5′-ATGAGTGACAGACAGGCCAG-3′. The reverse transcription (RT)-PCR product of Oct4 was digested with MseI for polymorphism analysis.
Quantitative PCR analysis.
Immunoprecipitated DNA was analyzed by real-time PCR using SYBR GREEN Universal PCR Mix or TaqMan Universal PCR Master Mix, and Prism 7700 sequence detector (ABI). Quantitative PCR analysis was repeated at least three times in each case. Quantity of DNA was determined following the algebraic formula of 2−Ct (where Ct is the cycle threshold number). The relative amount of immunoprecipitated DNA to input DNA was calculated. Primer sets described above were used for real-time PCR with SYBR GREEN. For real-time PCR analysis of the Oct4 promoter B region (see Fig. 3B), we used the TaqMan probe, 5′-CCACAGCTCTGCTCCTCCACC-3′, and the following PCR primers: Oct4pp-F, 5′-CGCCTCAGTTTCTCCCACC-3′, and Oct4pp-R, 5′-AGCCTTGACCTCTGGCCC-3′.
FIG. 3.
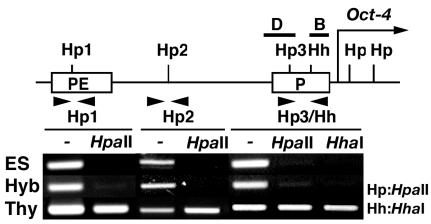
Reprogramming of DNA methylation at the Oct4 promoter region. The 5′ regulatory region of Oct4 is represented as a simple map. (B to D) The arrow indicates the transcription start site, and solid bars (B and D) indicate the regions analyzed by ChIP. Several sites recognized by the methylation-sensitive restriction enzymes HpaII (Hp) or HhaI (Hh) are shown as Hp1, Hp2, Hp3, and Hh. The promoter (P) and PE regions are boxed. Genomic DNA isolated from ES cells, hybrid cells, and thymocytes was digested with PvuII and HpaII or HhaI and then PCR-amplified with specific primer sets (arrowheads). PCR products were separated through 1% agarose and stained with ethidium bromide.
PCR-based methylation analysis.
PCR-based methylation analysis was performed as previously described (16). DNA (5 μg) digested with PvuII, and the methylation-sensitive restriction enzyme HpaII or HhaI was PCR amplified using the following primer sets: Hp1a, 5′-CCTCCTAATCCCGTCTCC-3′; Hp1b, 5′-CCAGCTCTCCACCTCTCC-3′; Hp2a, 5′-CCCAGTATTTCAGCCCATGTCC-3′; Hp2b, 5′-GTTAGAGCTGCCCCTCTG-3′; Hp3a, 5′-GGCACACGAACATTCAATGG-3′; Hp3b, 5′-GGAGAAACTGAGGCGAGCGC-3′; Hha, 5′-CTTTCGCCTACCCACTGCTC-3′; and Hhb, 5′-CCCTCCGAACGTGTACACAC-3′.
RESULTS
Global changes to histone modifications in reprogrammed somatic nuclei.
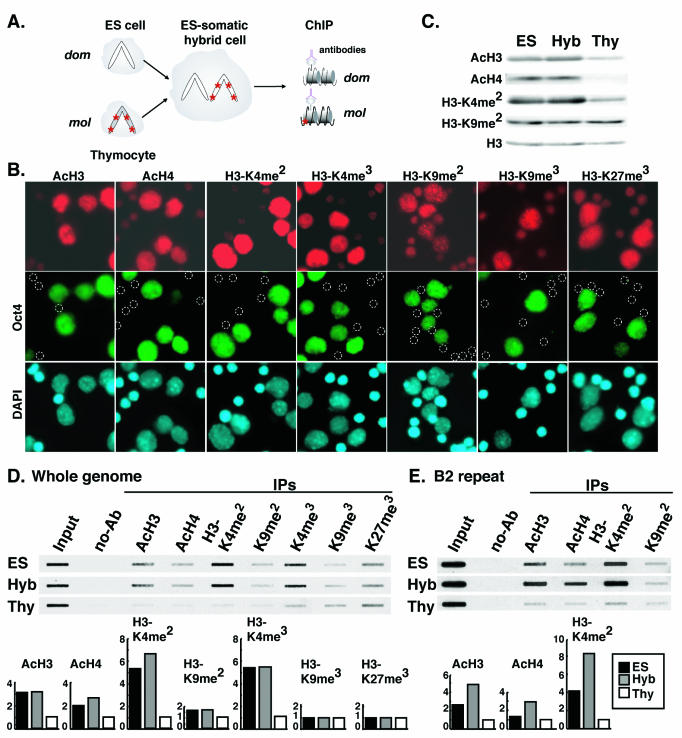
We have generated intersubspecific hybrid cells with M. musculus domesticus ES cells deficient for the X-linked Hprt gene and M. musculus molossinus thymocytes collected from adult mice (Fig. 1A). DNA sequence polymorphisms are frequent between M. musculus molossinus and M. musculus domesticus genomes, allowing us to monitor the origin of RNA and DNA in our hybrid clones. The mesodermal origin of the somatic cells was verified by lymphoid cell-specific DNA rearrangements of the Tcrb and IgH genes (data not shown) (48). First, the mixture of ES cells and thymocytes was immunostained with antibodies against histone H3 (K9 and K14) acetylation, histone H4 (K5, K8, K12, and K16) acetylation, histone H3-K4 dimethylation and trimethylation, histone H3-K9 di-methylation and trimethylation, and histone H3-K27 trimethylation (Fig. 1B). As for the hybrid cells, the ES cell nuclei were significantly hyperacetylated on histone H3 and H4, hyperdi- and -trimethylated on H3-K4, and hypertrimethylated on H3-K27 compared to thymocyte nuclei. No obvious difference in signal intensity of H3-K9 di- and trimethylation was found between the ES cell and thymocyte nuclei. H3-K9 trimethylation-positive signals were visualized as spots in nuclei. To verify the global histone acetylation and dimethylation level in the hybrid cells, we examined total histone protein extracted from thymocytes, ES cells, and hybrid cells by Western blot hybridization with antibodies against acetylation of histone H3 and H4, and dimethylation of H3-K4 and H3-K9 (Fig. 1C). Histone H3 and H4 were significantly more acetylated in ES and hybrid cells than in thymocytes. For histone H3, K4 dimethylation was also higher in ES and hybrid cells than in thymocytes. No difference was seen for histone H3 dimethylation at K9. To analyze the modifications on core histones, DNA extracted from immunoprecipitated chromatin was hybridized with mouse genomic DNA (Fig. 1D). The degree of histone H3 and H4 acetylation and H3-K4 di- and trimethylation was considerably higher in hybrid cells than that expected in a 1:1 mixture of ES cells and thymocytes, whereas no significant differences were seen in di- and trimethylation at H3-K9. Histone H3-K27 was also trimethylated to some extent but again little difference was shown between H3-K27 trimethylation was less different among ES cells, hybrid cells, and thymocytes. B2 repeat sequences, which are widely dispersed in the mouse genome, are abundantly expressed in early embryos through the cleavage stages and in undifferentiated ES cells. B2 repeat expression is much less common in differentiated somatic cells (52). ChIP analysis with a B2 repeat sequence probe revealed that H3-K4 was also more highly dimethylated at these sites in hybrid cells than would be expected from a 1:1 mixture of ES cells and thymocytes (Fig. 1E). Again, methylation at H3-K9 showed little difference. Furthermore, considerable acetylation of histone H3 and H4 was detected in the chromatin surrounded by B2 repeat DNA in hybrid cells. This increase in acetylation may be dependent on the up-regulation of B2 transcription in the hybrid cells.
FIG. 1.
Histone tail modifications in ES cells, hybrid cells, and thymocytes. (A) Experimental scheme with intersubspecific hybrid cells between M. musculus domesticus (dom) ES cells and M. musculus molossinus (mol) thymocytes. Asterisks represent DNA sequence-polymorphic sites. Histone modifications in ES cells, hybrid cells, and thymocytes were examined by Western blotting and ChIP assays with antibodies specific to acetylated H3 (AcH3), acetylated H4 (AcH4), dimethylated H3 lysine 4 (H3-K4me2), trimethylated H3-K4 (H3-K4me3), dimethylated H3 lysine 9 (H3-K9me2), trimethylated H3-K9me3, and trimethylated H3 lysine 27 (H3-K27me3). (B) Immunocytochemical analysis of histone modifications in ES cells and thymocytes. The cells were immunostained with antibodies (red) against AcH3, AcH4, H3-K4me2, H3-K4me3, H3-K9me2, H3-K9me3, and H3-K27me3. ES cell nuclei were immunostained with antibody against OCT4 (green). Nuclei were counterstained with DAPI (blue). Circles indicate thymocytes. (C) Western blotting analysis of histone modifications in ES cells, hybrid cells, and thymocytes. Total protein extracted from the cells was immobilized onto nitrocellulose membrane and hybridized with antibodies against AcH3, AcH4, H3-K4me2, and H3-K9me2. Total H3 was used as loading control. (D) Slot blot hybridization analysis of histone modifications with a sonicated genomic DNA probe from mouse embryonic fibroblasts. DNA immunoprecipitated with antibodies against AcH3, AcH4, H3-K4me2, H3-K4me3, H3-K9me2, H3-K9me3, and H3-K27me3 was slot blotted and hybridized. The relative levels of the histone modifications detected in ES cells, hybrid cells, and thymocytes are summarized in histogram form. (E) Slot blot hybridization analysis of histone modifications with a B2 repeat probe. DNA immunoprecipitated with antibodies against AcH3, AcH4, H3-K4me2, and H3-K9me2 was analyzed as detected in panel D.
Chromatin reprogramming of a pluripotential cell-specific gene.
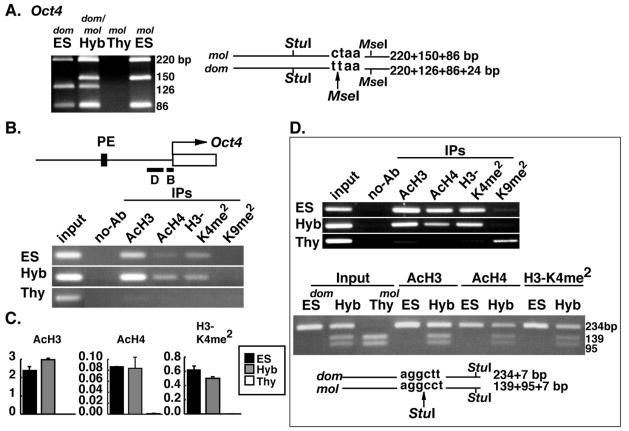
Expression of the Oct4 gene, which is required for maintaining a pluripotential state, is restricted to undifferentiated embryonic cells, including ES cells. Oct4 is silenced in all somatic cells, including thymocytes (32). Following cell hybridization, the thymocyte-derived endogenous Oct4 gene was reactivated, and this was verified by sensitivity of RT-PCR product to digestion with MseI (Fig. 2A). Roughly equal amounts of Oct4 were transcribed from the ES and thymocyte genomes. In the region immediately upstream of the transcription start site (region B in Fig. 2B), histone H3 and H4 were hyperacetylated and H3-K4 was hyperdimethylated in the hybrid cells. These histone modifications were not detected in thymocytes (Fig. 2B). Quantitative PCR analysis showed that the degree of acetylation and dimethylation in the hybrid cells was significantly higher than that expected from a 1:1 mixture of ES cells and somatic cells (Fig. 2C). Acetylation and di-methylation levels were roughly equal in ES and hybrid cells. A similar profile of histone tail modification was detected in the region 186 bp upstream of the transcription start site (region D in Fig. 2B). Histone H3 and H4 were acetylated and H3-K4 (but not H3-K9) was dimethylated in hybrid cells (Fig. 2D). Interestingly, dimethylation at H3-K9 that does exist in region D in thymocytes was not present in hybrid cells (Fig. 2D). A polymorphic StuI site that we identified in this region specific to the thymocyte-derived DNA (M. musculus molossinus) was used to determine the origin of histone H3 and H4 acetylation and H3-K4 dimethylation in the hybrid cells (Fig. 2D). PCR products derived from the immunoprecipitated chromatin showed M. musculus molossinus-specific StuI digestion in all cases, demonstrating that each of the modifications had occurred on the thymocyte-derived chromatin in addition to the ES cell-derived chromatin. To address whether the observed changes to histone tail modifications are linked to loss of CpG methylation from the thymocyte-derived genome, three HpaII sites (Hp1, Hp2, and Hp3) and one HhaI (Hh) site were examined using a methylation-sensitive PCR-based assay (Fig. 3). The Hp1 site is located in the proximal enhancer (PE) region, and the Hp3 site is in region D (Fig. 2B) of the Oct4 promoter, respectively. All of the CpG sites examined were unmethylated in ES and hybrid cells, whereas they were methylated in thymocytes. Demethylation of these CpG sites is therefore linked to with reactivation of thymocyte-derived Oct4 in the hybrid cells.
FIG. 2.
Reprogramming of histone modifications in the somatic cell-derived Oct4 promoter region. (A) RT-PCR analysis of Oct4 expression in hybrid cells. RT-PCR products were digested with StuI and MseI. Transcripts from the ES cell-derived genome (dom) generate a 126-bp fragment, and those from the thymocyte-derived genome (mol) generate a 150-bp fragment. (B) Histone modifications at the Oct4 promoter region. DNA prepared from chromatin immunoprecipitated with antibodies against AcH3, AcH4, H3-K4 dimethylation (H3-K4me2), and H3-K9 dimethylation (H3-K9me2) was PCR amplified with a primer set specific to the Oct4 promoter region B (black bar). (C) Real-time quantitative PCR analysis. Histograms show the relative amount of immunoprecipitated DNA to input DNA for AcH3, AcH4, and H3-K4me2. (D) Determination of the origin of immunoprecipitated DNA. DNA prepared from chromatin immunoprecipitated with antibodies to AcH3, AcH4, H3-K4me2, and H3-K9me2 was PCR amplified with a primer set specific to region D (black bar indicated in panel B). The origin of PCR products was determined by sensitivity to StuI digestion. The thymocyte-derived products (mol) are digested into three fragments, whereas the ES cell-derived products (dom) produce only two fragments.
Chromatin reprogramming of somatic cell-specific genes.
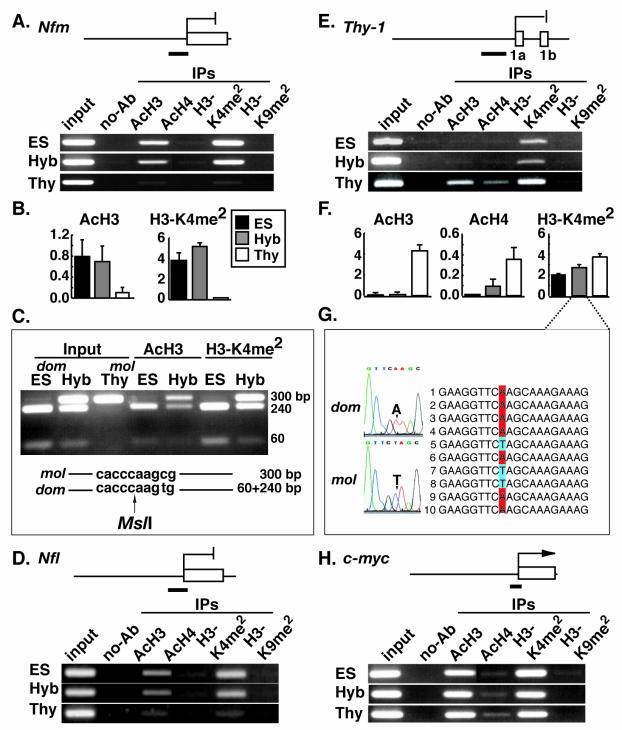
Next, we analyzed changes to histone tail modifications at the neuronal cell-specific genes Nfm and Nfl, which are repressed in both thymocytes and ES cells and also in hybrid cells (39). In the promoter region of Nfm, ES cell chromatin showed both histone H3 acetylation and H3-K4 dimethylation, but not histone H4 acetylation and H3-K9 dimethylation (Fig. 4A). Thymocyte chromatin was unmodified at all sites tested. The pattern of chromatin modifications seen in the hybrid cells was similar to that of ES cells. Quantitative PCR analysis on the immunoprecipitated chromatin revealed that the thymocyte genome had acquired remarkably high levels of histone H3 acetylation and H3-K4 dimethylation in the hybrid cells (Fig. 4B). Polymorphic restriction enzyme digestion analysis with MslI demonstrated that the ES cell genome-specific PCR product (M. musculus domesticus) was sensitive to digestion resulting in bands at 240 and 60 bp, while the thymocyte-derived product (M. musculus molossinus) was not (Fig. 4C). In the hybrid cells, both ES- and thymocyte-specific products were immunoprecipitated in similar amounts, demonstrating that the thymocyte and ES cell-derived histone H3 were equivalently acetylated and equivalently dimethylated at K4. Similar results were observed for the promoter region of Nfl. Histone H3 was hyperacetylated and H3-K4 was hyperdimethylated in the hybrid cells but not in thymocytes (Fig. 4D). Importantly, these specific modification changes to the thymocyte-derived chromatin occurred in the absence of transcriptional activation.
FIG. 4.
Reprogramming of histone modifications in the promoter regions of several somatic tissue-specific genes. (A) Histone modifications at the Nfm promoter region (black bar). DNA prepared from chromatin immunoprecipitated with antibodies against AcH3, AcH4, H3-K4 dimethylation (H3-K4me2), and H3-K9 dimethylation (H3-K9me2) was PCR amplified with a primer set specific to the Nfm promoter region. (B) Real-time quantitative PCR analysis of Nfm. Histograms show the relative amount of immunoprecipitated DNA to input DNA for AcH3 and H3-K4me2. (C) Determination of the origin of immunoprecipitated DNA. The origin of PCR products was determined by sensitivity to MslI digestion. The thymocyte-derived products (mol) remain undigested (300 bp), whereas the ES cell-derived products (dom) are cut into two fragments (240 and 60 bp). (D) Histone modifications at the Nfl promoter region (black bar). DNA prepared from chromatin immunoprecipitated with antibodies to AcH3, AcH4, H3-K4me2, and H3-K9me2 was PCR amplified with a primer set specific to the Nfl promoter region. (E) Histone modifications at the Thy-1 promoter region (black bar). DNA prepared from chromatin immunoprecipitated with antibodies to AcH3, AcH4, H3-K4me2, and H3-K9me2 was PCR amplified with a primer set specific to the Thy-1 promoter region. (F) Real-time quantitative PCR analysis of Thy-1. Histograms show the relative amount of immunoprecipitated DNA to input DNA for AcH3, AcH4, and H3-K4me2. (G) Determination of the origin of immunoprecipitated DNA. DNA prepared from chromatin immunoprecipitated with antibodies to H3-K4me2 was PCR amplified with a primer set specific to the Thy-1 promoter region. The origin of PCR products was determined by sequencing of DNA extracted from independent clones. An adenine residue in the ES cell-derived genome (dom) is replaced with a thymine residue in the thymocyte-derived genome (mol). (H) Histone modifications at the c-myc promoter region (black bar). DNA prepared from chromatin immunoprecipitated with antibodies to AcH3, AcH4, H3-K4me2, and H3-K9me2 was PCR amplified with a primer set specific to the c-myc promoter region.
We next looked at modifications at the Thy-1 gene, which is expressed in thymocytes and repressed in ES and hybrid cells (12). The transcriptionally active thymocyte-derived promoter region was characterized by hyperacetylation of histone H3 and H4 and hyperdimethylation of H3-K4 but not H3-K9 (Fig. 4E). Conversely, the transcriptionally inactive ES cell-derived Thy-1 promoter region only showed dimethylation at H3-K4. In the hybrid cells, histone H3 and H4 acetylation was dramatically reduced, which correlated with transcriptional repression, whereas H3-K4 dimethylation was maintained at a certain level. Using quantitative PCR, we found that the degree of H3-K4 dimethylation in hybrid cells corresponded well with the expected levels in a 1:1 mixture of ES cells and thymocytes (Fig. 4F). Indeed, H3-K4 was dimethylated both in the thymocyte and ES cell-derived chromatin again demonstrated using a sequence polymorphism present within the PCR products (Fig. 4G). In the promoter region of c-myc, a constitutively active gene, the chromatin was characterized by histone H3 hyperacetylation, H3-K4 hyperdimethylation, and lack of H3-K9 dimethylation in the hybrid cells, ES cells and thymocytes (Fig. 4H).
Chromatin reprogramming via histone H3 trimethylation of tissue-specific genes.
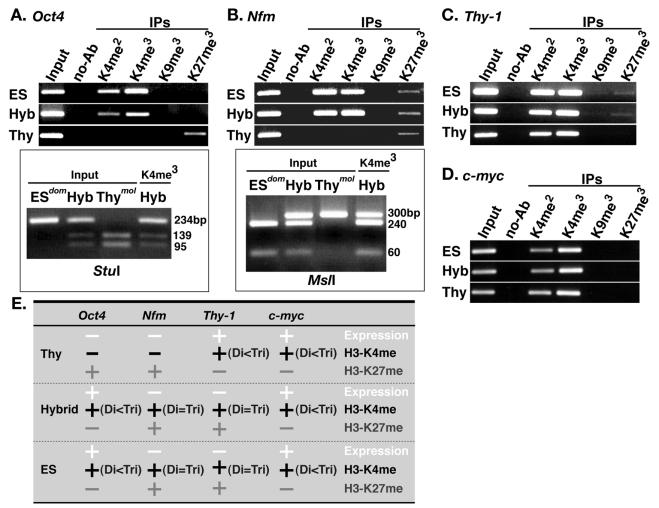
In yeast, histone H3-K4 dimethylation is associated with both active and repressed gene states, whereas histone H3-K4 trimethylation is generally only observed under active conditions (41). To address whether trimethylation of histone H3 on the mammalian reprogrammed somatic nucleus is tightly linked with gene activity, di- and trimethylation of H3-K4, H3-K9, and H3-K27 were analyzed by ChIP in the intersubspecific ES hybrid cells.
In thymocytes, neither H3-K4 di- nor trimethylation could be detected at the repressed Oct4 and Nfm genes, while both di- and trimethylation were prominent in the expressed Thy-1 and c-myc genes (Fig. 5A to E). Thus, H3-K4 hyperdi- and -trimethylation correlates with activity of the these genes. However, in pluripotential ES and ES hybrid cells, considerable levels of both di- and trimethylation were detected even at the repressed genes Nfm and Thy-1 (Fig. 5B, C, and E). For the expressed genes Oct4 and c-myc, trimethylation was more prominent than dimethylation (Fig. 5A, D, and E). To identify the origin of ChIP-mediated PCR product, the trimethylation-specific products were digested with StuI (for Oct4) and with MslI (for Nfm). At each gene, both thymocyte-derived (M. musculus molossinus) and ES cell-derived (M. musculus domesticus) products both in Oct4 and Nfm were detected in hybrid cells (Fig. 5A and B), demonstrating that thymocyte-derived genomes become trimethylated following cell hybridization with ES cell genomes. These findings suggest that, in pluripotential stem cells, H3-K4 trimethylation is a mark for open chromatin structure but is not directly associated with gene activity. Thus, the transition from dimethylation to trimethylation occurs irrespective of gene activity.
FIG. 5.
Reprogramming of histone H3 trimethylation at K4, K9, and K27 in the promoter regions of Oct4, Nfm, Thy-1, and c-myc. (A) Histone modifications at in the Oct4 promoter region (region D in Fig. 2B). DNA prepared from chromatin immunoprecipitated with antibodies against H3-K4 dimethylation (H3-K4me2) and trimethylation (H3-K4me3) or H3-K9 trimethylation (H3-K9me3) and H3-K27 trimethylation (H3-K27me3) was PCR amplified with a primer set specific to the Oct4 promoter region. The origin of PCR products of H3-K4me3 was determined by StuI digestion. The thymocyte-derived products (mol) were digested into three fragments, whereas the ES cell-derived products (dom) produced only two fragments. (B) Histone modification at the Nfm promoter region. DNA prepared from chromatin immunoprecipitated as described for panel A was PCR amplified with a primer set specific to the Nfm promoter region. The origin of PCR products of H3-K4me3 was determined by MslI digestion. The thymocyte-derived products (mol) are undigested, whereas the ES cell-derived products (dom) are digested into two fragments. (C) Histone modification at the Thy-1 promoter region. Immunoprecipitated DNA prepared as described for panel A was PCR amplified with a primer set specific to the Thy-1 promoter region. (D) Histone modification at the c-myc promoter region. DNA prepared as described for panel A was PCR amplified with a primer set specific to the c-myc promoter region. (E) Summary of histone H3 trimethylation at K4, K9, and K27 in the Oct4, Nfm, Thy-1, and c-myc promoter regions.
For H3-K27, trimethylation was directly associated with gene repression (Oct4 and Nfm in thymocytes and Nfm and Thy-1 in ES and hybrid cells) and not with gene activation (Fig. 5A to E). Thus, high level of H3-K4 trimethylation is linked to absence of H3-K27 trimethylation, implying that H3-K4 tri-methylation and H3-K27 trimethylation are mutually exclusive.
As for H3-K9 trimethylation, signals could not be detected at any of the promoter regions of the genes examined (Fig. 5A to E), as similar to H3-K9 dimethylation (Fig. 4). This is consistent with findings by immunocytochemical analysis (Fig. 1B) where H3-K9 trimethylation was predominantly localized to centromeric regions.
DISCUSSION
Cloned animals and ES cells from adult cells are produced by nuclear reprogramming of the somatic epigenotype to a pluripotential epigenotype. Acetylation and methylation of histone H3 and H4 amino termini are epigenetic modifications involved in regulating gene activity by modulating chromatin conformation. We have demonstrated that the acquisition of pluripotential competence by a reprogrammed somatic genome is accompanied, independent of gene activity, by global decondensation of the somatic cell-derived chromatin. This is most clearly marked by hyperdimethylation at histone H3-K4. We suggest that this erasure of somatic cell-specific histone modifications is a crucial step in the induction of successful nuclear reprogramming.
The pluripotential state-specific Oct4 gene is a member of the mammalian POU family of transcriptional factors, which is essential for maintenance of pluripotency in embryonic germ cells, pregastrulation embryos, and ES cells (30, 32). The unique expression pattern of Oct4 is achieved by two distinct elements; the distal enhancer, which is required for expression specific to preimplantation embryos and embryonic germ cells; and the PE, which is specific to the epiblast (55). Basal levels of Oct4 transcription are directed by the region immediately upstream of the transcription start site, encompassing the promoter and PE elements (3, 16). We have demonstrated that in the thymocyte genome, where Oct4 is silent, the promoter region is dimethylated at H3-K9 but not at H3-K4. This is associated with histone H3 and H4 hypoacetylation. In the reprogrammed thymocyte genome, where Oct4 is reactivated, this region is dimethylated at H3-K4 but not at H3-K9, and hyperacetylated at H3 and H4 (Fig. 3A to D). It has been previously shown that H3-K4 methylation is preferentially associated with histone H3 acetylation in yeast, chicken, and HeLa cells (25, 33, 34). Furthermore, reciprocal methylation of H3-K4 and H3-K9 is known to define active and silent chromosomal domains, respectively, from yeast to human (25, 33, 45). We have found that, in the process of nuclear reprogramming of Oct4, the transcriptionally inactive state (marked by dimethylation of H3-K9 and hypoacetylation of H3 and H4), is converted to a transcriptionally active state. This is marked by dimethylation of H3-K4 and hyperacetylation of histone H3 and H4. This suggests that the methylation of histone H3, acetylation of histone H3 and H4, and gene activity are mechanistically linked in this region. It has been shown that fully activated promoters appear to be enriched in trimethylation of H3-K4, while basal transcription correlates with a dimethylated state (41). Additional ChIP analysis indicates that the Oct4 promoter region in the reprogrammed somatic genome is highly trimethylated at H3-K4.
It has been suggested that in Neurospora crassa (49) and Arabidopsis thaliana (17), DNA methylation and histone H3-K9 methylation are mechanistically linked, and that H3-K9 methylation is a prerequisite for DNA methylation. In mammals, ES cells deficient for a histone H3-K9 methyltransferase, Suv39h, display a DNA hypomethylation profile at pericentric major satellite repeats (24). Furthermore, failure of DNA demethylation is associated with genome-wide H3-K9 hypermethylation in cloned bovine preimplantation embryos, created by somatic nuclear transfer (10, 40). Closely associated with its expression status, DNA at the Oct4 promoter region is unmethylated during preimplantation and early postimplantation stages but undergoes de novo methylation at 6.5 days after fertilization. The hypermethylated state is maintained in all adult somatic tissues (16). Here, we have demonstrated that DNA at the Oct4 promoter region was demethylated in the reprogrammed somatic genome, in contrast with its hypermethylated state before hybridization with ES cells. Thus, for the Oct4 promoter region, epigenetic reprogramming to hypomethylation of DNA, hypermethylation of H3-K4, hyperacetylation of histone H3 and hypomethylation of H3-K9 are closely associated with reactivation of Oct4 transcription. Orchestrated regulation of these epigenetic modifications to DNA and histones may be required for inducing appropriate reexpression of Oct4, which is essential for reprogramming to a pluripotential state (4).
The mammalian homologues of Drosophila Pc-G gene Enhancer of zeste [E(z)] are known as Ezh1, which is expressed in adult tissues, and Ezh2, which is expressed in early embryonic development (23). Ezh2 interacts with the embryonic ectoderm development gene, Eed, and is essential for mouse early embryonic development and derivation of ES cells (35). Ezh2, which contains a conserved SET (suppressor of variegation, enhancer of zeste and trithorax) domain, has histone methyltransferase (HMTase) activity preferential for H3-K27. However, it may also be involved in methylating H3-K9 through the formation of a Ezh/Eed complex (9, 11, 21). Maternal or zygotic EZH2 is found in pluripotential mouse preimplantation-embryonic cells, including blastocyst inner cell mass (ICM) cells and derived ES cells. Interestingly, ICM and ES cells also stain intensely with antibodies against dimethylation and trimethylation of H3-K27 (11). Thus, H3-K27 methylation may be involved in promoting stable recruitment of Pc-G complexes and controlling developmental processes through formation of inactive chromatin. Importantly, we have observed intensive di- and trimethylation of H3-K4 in mouse ICM and ES cells through immunostaining with specific antibodies. In general, H3-K4 methylation marks active chromatin, in contrast with H3-K27 methylation, which is specific to inactive chromatin. Our ChIP analysis data suggest that high level of H3-K4 trimethylation may be incompatible with H3-K27 tri-methylation. It remains unclear whether methylation of H3-K4 and H3-K27 coexist on the same nucleosome. However, full silencing of some homeotic genes by regional formation of H3-K27 methylation-associated inactive chromatin is likely to be required for stable maintenance of a pluripotential state. Other chromatin regions in pluripotential nuclei are characterized by decondensed active chromatin, marked by H3-K4 methylation. These data suggest that prior to formation of an inactive chromatin state, a decondensed active chromatin state is required to facilitate the binding of inactive chromatin-related factors.
The dynamic reprogramming of histone tail modifications is induced by position-specific de novo HMTases. To date, four H3-K4 HMTases—Set7/Set9 (31, 51), MLL (27, 29), Ash1 (2), and Set1 (54)—have been identified. MLL and Ash1 are trithorax (trx-G) group proteins that, in contrast to PcG proteins, act to perpetuate a transcriptionally active state. In Drosophila, histone methylation generated by Ash1 prevents binding of the repressive PcG and HP1 proteins and recruits a Brahma (Brm)-containing ATP-dependent chromatin remodeling factor (2). Drosophila Brm and human SWI/SNF are part of a large complex containing several proteins with similarity to subunits of the yeast SWI/SNF subfamily (20, 36). Another member of the SWI2/SNF2 superfamily is the ISWI ATPase (6, 50). Both SWI/SNF and ISWI are ATP-dependent chromatin remodeling factors but are functionally distinct (18). Remarkably, mammalian SWI/SNF and Brm-associated factor complexes contain conserved subunits (1, 53), implying that H3-K4 methylation may function as an epigenetic mark to induce recruitment of the Brm-associated SWI/SNF chromatin remodeling factors. In fact, it has been recently shown that H3-K4 di- and trimethylation and Isw1p ATPase activity are tightly linked in transcription regulation of certain genes in yeast (42). This is consistent with decondensed chromatin in the reprogrammed somatic nuclei. Our own RT-PCR analysis has found that transcription of Set7/Set9, MLL, and Ash1 occurs not only in ES cells but also in thymocytes. Thus, accessibility of these H3-K4 HMTase complexes may be controlled by an unknown key cofactor, which play a crucial role in the initiation of somatic nuclear reprogramming.
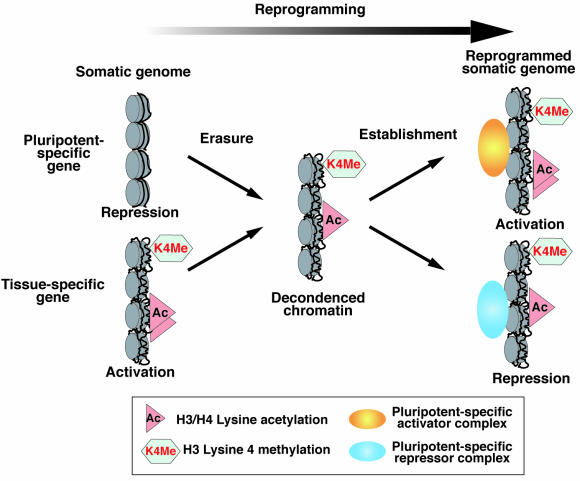
Transcriptional activity-independent de novo histone di-methylation at H3-K4 in the reprogrammed somatic genome suggests that the basal transcription activation-permissive chromatin may be the default state following erasure of the somatic cell-specific epigenotype (Fig. 6). Subsequently, the pluripotential cell-specific epigenotype that regulates activation and/or repression of genes involved in maintenance of pluripotency may be reestablished under the control of key genes such as Oct4, Ezh2, and Nanog (8, 15, 28, 35). We propose that the nuclear reprogramming of somatic cells involves at least two distinct events (Fig. 6). We previously demonstrated that reactivation of a somatic cell-derived Oct4-GFP takes approximately 36 to 48 h to occur, following cell hybridization with ES cells (48). This suggests that up to 2 days is required for completing erasure of the somatic epigenotype. The erasure mechanism may serve to prevent the inheritance of unnecessary epigenetic memory to the next generation in germ cells and in preimplantation development. In nuclear transfer experiments, the comparatively high success rate for production of cloned mice with ES cell-derived nuclei (up to 20%) compared to somatic nuclei (2 to 3%) may be due to the reduced requirement for initial erasure of the somatic cell-specific epigenotype (38). The main cause of the death of cloned animals may be due to insufficient reestablishment of a pluripotential cell-specific epigenotype following erasure of the somatic cell-specific epigenotype. Consistent with this, somatic nucleus-derived Oct4-GFP was found to be reexpressed in only 10% of cloned mouse blastocysts (4). We predict that expression of other key genes that are also required for maintenance of pluripotency in mouse ES cells, including Ezh2 and Nanog, would be restricted to the limited number of Oct4-positive cloned blastocysts. In total, only the 2 to 3% of cloned embryos that are sufficiently reprogrammed and express all of the key genes, may have the capacity to develop to adulthood.
FIG. 6.
A two-step model for somatic cell nuclear reprogramming. Step 1 (erasure step): the somatic cell-specific epigenotype is globally erased by the activity of reprogramming factors. Consequently, histone H3 and H4 become hyperacetylated and histone H3 lysine 4 becomes hyperdi- and -trimethylated. This leads to chromatin decondensation, which is required to form the transcription-permissive or repressive state. Step 2 (reestablishment step): an epigenotype required for acquisition of pluripotential competence is reestablished as a result of the activation of pluripotential cell-specific genes and the repression of somatic cell-specific genes.
Acknowledgments
We thank the Genetic Stock Research Center, National Institute of Genetics, Mishima, Japan, for kindly providing JF1 mice; Yi Zhang for providing antibody to H3-K27 trimethylation; Emiko Moribe and Yoshimitsu Toyoda for DNA sequencing; and Justin F. X. Ainscough for critical reading of the manuscript.
REFERENCES
- 1.Armstrong, J. A., and B. M. Emerson. 1998. Transcription of chromatin: these are complex times. Curr. Opin. Genet. Dev. 8:165-172. [DOI] [PubMed] [Google Scholar]
- 2.Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857-862. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Shushan, E., E. Pikarsky, A. Klar, and Y. Bergman. 1993. Extinction of Oct-3/4 gene expression in embryonal carcinoma × fibroblast somatic cell hybrids is accompanied by changes in the methylation status, chromatin structure, and transcriptional activity of the Oct-3/4 upstream region. Mol. Cell. Biol. 13:891-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boiani, M., S. Eckardt, H. R. Scholer, and K. J. McLaughlin. 2002. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 16:1209-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bortvin, A., K. Eggan, H. Skaletsky, H. Akutsu, D. L. Berry, R. Yanagimachi, D. C. Page, and R. Jaenisch. 2003. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development 130:1673-1680. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R. 1998. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem. Sci. 23:20-25. [DOI] [PubMed] [Google Scholar]
- 7.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. [DOI] [PubMed] [Google Scholar]
- 9.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrota. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 10.Dean, W., F. Santos, M. Stojkovic, V. Zakhartchenko, J. Walter, E. Wolf, and W. Reik. 2001. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA 98:13734-13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erhardt, S., I. H. Su, R. Schneider, S. Barton, L. Perez-Burgos, T. Jenuwein, T. Kouzarides, A. Tarakhovsky, and M. A. Surani. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130:4235-4248. [DOI] [PubMed] [Google Scholar]
- 12.Forejt, J., S. Gregorova, K. Dohnal, and J. Nosek. 1984. Gene expression of differentiated parent in teratocarcinoma cell hybrids. Repression or reprogramming? Cell Differ. 15:229-234. [DOI] [PubMed] [Google Scholar]
- 13.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry, C. J., and C. L. Peterson. 2001. Chromatin remodeling enzymes: who's on first? Curr. Biol. 11:R185-197. [DOI] [PubMed] [Google Scholar]
- 15.Fujikura, J., E. Yamato, S. Yonemura, K. Hosoda, S. Masui, K. Nakano, J. Miyazaki, and H. Niwa. 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16:784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gidekel, S., and Y. Bergman. 2002. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 277:34521-34530. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 18.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, H., M. Tada, S. Hatano, M. Miyazaki, N. Nakatsuji, and T. Tada. 2002. Chromatin reprogramming of male somatic cell-derived XIST and TSIX in ES hybrid cells. Cytogenet. Genome Res. 99:106-114. [DOI] [PubMed] [Google Scholar]
- 20.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lachner, M., R. J. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117-2124. [DOI] [PubMed] [Google Scholar]
- 23.Laible, G., A. Wolf, R. Dorn, G. Reuter, C. Nislow, A. Lebersorger, D. Popkin, L. Pillus, and T. Jenuwein. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehnertz, B., Y. Ueda, A. A. Derick, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 25.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 26.Lyko, F. 2001. DNA methylation learns to fly. Trends Genet. 17:169-172. [DOI] [PubMed] [Google Scholar]
- 27.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107-1117. [DOI] [PubMed] [Google Scholar]
- 28.Mitsui, K., Y. Tokuzawa, H. Itoh, K. Segawa, M. Murakami, K. Takahashi, M. Maruyama, M. Maeda, and S. Yamanaka. 2003. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113:631-642. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 30.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 31.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromerge, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 33.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 34.Noma, K., and S. I. Grewal. 2002. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16438-16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Carroll, D., S. Erhardt, M. Pagani, S. C. Barton, M. A. Surani, and T. Jenuwein. 2001. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955-3966. [DOI] [PubMed] [Google Scholar]
- 37.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 38.Rideout, W. M., 3rd, T. Wakayama, A. Wutz, K. Eggan, L. Jackson-Grusby, J. Dausman, R. Yanagimachi, and R. Jaenisch. 2000. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat. Genet. 24:109-110. [DOI] [PubMed] [Google Scholar]
- 39.Rohwedel, J., T. Kleppisch, U. Pich, K. Guan, S. Jin, W. Zuschratter, C. Hopf, W. Hoch, J. Hescheler, V. Witzemann, and A. M. Wobus. 1998. Formation of postsynaptic-like membranes during differentiation of embryonic stem cells in vitro. Exp. Cell Res. 239:214-225. [DOI] [PubMed] [Google Scholar]
- 40.Santos, F., V. Zakhartchenko, M. Stojkovic, A. Peters, T. Jenuwein, E. Wolf, W. Reik, and W. Dean. 2003. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr. Biol. 13:1116-11121. [DOI] [PubMed] [Google Scholar]
- 41.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are trimethylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 42.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 43.Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff. 2003. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4:481-495. [DOI] [PubMed] [Google Scholar]
- 44.Solter, D. 2000. Mammalian cloning: advance and limitations. Nat. Rev. Genet. 1:199-207. [DOI] [PubMed] [Google Scholar]
- 45.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tada, M., A. Morizane, H. Kimura, H. Kawasaki, J. F. Ainscough, Y. Sasai, N. Nakatsuji, and T. Tada. 2003. Pluripotency of reprogrammed somatic genomes in embryonic stem hybrid cells. Dev. Dyn. 227:504-510. [DOI] [PubMed] [Google Scholar]
- 47.Tada, M., T. Tada, L. Lefebvre, S. C. Barton, and M. A. Surani. 1997. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16:6510-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tada, M., Y. Takahama, K. Abe, N. Nakatsuji, and T. Tada. 2001. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11:1553-1558. [DOI] [PubMed] [Google Scholar]
- 49.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 50.Varga-Weisz, P. D., and P. B. Becker. 1998. Chromatin-remodeling factors: machines that regulate? Curr. Opin. Cell Biol. 10:346-353. [DOI] [PubMed] [Google Scholar]
- 51.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromerge, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 52.Whitelaw, E., and D. I. Martin. 2001. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 27:361-365. [DOI] [PubMed] [Google Scholar]
- 53.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 54.Wysocka, J., M. P., Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Hum. Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeom, Y. I., G. Fuhrmann, C. E. Ovitt, A. Brehm, K. Ohbo, M. Gross, K. Hubner, and H. R. Scholer. 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122:881-894. [DOI] [PubMed] [Google Scholar]