Summary
Transmitochondrial cybrids and multiple OMICs approaches were used to understand mitochondrial reprogramming and mitochondria-regulated cancer pathways in triple negative breast cancer (TNBC). Analysis of cybrids and established breast cancer (BC) cell lines showed that metastatic TNBC maintains high levels of ATP through fatty acid β-oxidation (FAO) and activates Src oncoprotein through autophosphorylation at Y419. Manipulation of FAO including the knocking down of carnitine palmitoyltransferase-1 (CPT1) and 2 (CPT2), the rate-limiting proteins of FAO, and analysis of patient-derived xenograft models, confirmed the role of mitochondrial FAO in Src activation and metastasis. Analysis of TCGA and other independent BC clinical data further reaffirmed the role of mitochondrial FAO and CPT genes in Src regulation and their significance in BC metastasis.
Introduction
While the Warburg effect has been validated by numerous studies, there has also been tremendous advancements towards the understanding of many aspects of cancer metabolism, including the roles of glycolysis, glutaminolysis, fatty acid (FA) synthesis, and most recently, fatty acid β oxidation (FAO) (Carracedo et al., 2013; Ward and Thompson, 2012). Multiple reports have suggested that despite enhanced glycolysis, cancer cells can produce a significant fraction of their ATP via mitochondrial respiration (Caino et al., 2015; LeBleu et al., 2014; Lu et al., 2015; Maiuri and Kroemer, 2015; Tan et al., 2015; Viale et al., 2015; Ward and Thompson, 2012; Xu et al., 2015). In a growing tumor, adaptive metabolic reprogramming, precipitated in part by oncogenic transformation, gives cancer cells the advantage of active proliferation, functional motility, and metastasis (Basak and Banerjee, 2015; Caino et al., 2015; LeBleu et al., 2014). A recent study by Tan et al. has described that when mitochondrial DNA (mtDNA)-depleted tumor cells (ρ0 cells) were injected into mice, they enhanced their tumor growth property by acquisition of mtDNA from the host mouse cells and reassembling a mitochondrial electron transport chain complex (ETC) and respiratory function (Tan et al., 2015). These observations suggest that, at least in selected subgroups of cancers, mitochondrial biogenesis is important for their oncogenesis and tumor progression.
Based on the differential metabolic preferences of a tumor cell compared to a normal cell, targeting tumor cell-specific metabolic characteristics is increasingly becoming a more attractive potential therapeutic strategy (Caino et al., 2015; Ghosh et al., 2015; Ward and Thompson, 2012). To better evaluate therapeutic potentials, it is important to elucidate how these metabolic programs couple with or converge into oncogenic signals such as those leading to unbridled growth, reduced apoptosis, and metastatic potential. The extensive crosstalk between the mitochondria and the nucleus known as mitochondrial retrograde regulation (MRR) is triggered by mitochondrial dysfunction/reprogramming and is not a simple on-off switch, but rather responds in a continuous manner to the changing metabolic needs of the cell (Erol, 2005).
Triple negative breast cancer (TNBC) are negative for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) amplification. TNBC suffers a poor prognosis compared to other cancer subtypes, caused by significant heterogeneity and limited understanding of the driver signaling pathways. Thus, for TNBC, clinical benefit from currently available targeted therapies is limited, and new therapeutic strategies are urgently needed. Most of the conventional chemotherapeutic agents, the current clinical standard for TNBC treatment, generally kill cells by activating mitochondrial apoptosis (Costantini et al., 2000; Hail, 2005). Thus, understanding MRR and the mitochondria-mediated oncogenic signature is critical to improve understanding of the currently limited known etiology and treatment resistance of TNBC.
Mitochondrial studies using whole cell approaches make it difficult to distinguish mitochondria-specific effects from those contributed by the nucleus. We overcome this gap by using transmitochondrial cybrid (cybrid) models for mitochondria function and pathway discovery (Ishikawa et al., 2008; Kaipparettu et al., 2013; Kaipparettu et al., 2010; King and Attardi, 1989; Vithayathil et al., 2012). The cybrid system is an excellent tool to compare different mitochondria on a common defined nuclear background to understand mitochondria-specific effects on cellular properties. We have used the cybrid approach to discover mitochondria-regulated energy and cancer pathways in TNBC. These initial findings were then further validated in established breast cancer (BC) cell lines, patient-derived xenograft (PDX) models, and BC patient data.
c-Src is a proto-oncogene involved in signaling that culminates in the control of multiple biological functions. Like most protein kinases, Src family members require phosphorylation within a segment of the kinase domain termed the activation loop for full catalytic activity. The chief phosphorylation sites of human Src include an activating autophosphorylation of Y419 in the kinase domain and an inhibitory phosphorylation of Y530 in the regulatory tail. While phosphorylation of Y530 inactivates Src through the folding of Src into a closed, inaccessible bundle, the full activation of the Src signature depends on autophosphorylation at Y419 that allows access of the substrate (Aleshin and Finn, 2010; Roskoski, 2015; Zhang and Yu, 2012). Src Y530 phosphorylation results from the action of other protein-tyrosine kinases including Csk and Chk. Importantly, the doubly phosphorylated enzyme is active, indicating that Y419 autophosphorylation overrides inhibition produced by Y530 phosphorylation (Roskoski, 2015; Zhang and Yu, 2012). Aberrant Src activation plays prominent roles in cancer formation and progression (Aleshin and Finn, 2010; Finn, 2008; Mayer and Krop, 2010). The Src pathway is one of the most commonly upregulated pathways in TNBC (Anbalagan et al., 2012; Tryfonopoulos et al., 2011). While Src inhibitors hold promise in treating metastatic TNBC (Pal and Mortimer, 2009; Tryfonopoulos et al., 2011), initial clinical studies using Src inhibitor monotherapy in unselected patients with advanced BC showed only minor response (Finn et al., 2011; Gucalp et al., 2011; Herold et al., 2011; Mayer and Krop, 2010). Since the discovery of Src approximately 40 years ago, much effort has been invested in understanding the role of Src in cell regulation and Src signaling pathways in cancer. While structural biology, molecular and cell biology, and genetic studies have provided crucial information, the complete picture of molecular mechanisms and tumor biology of Src signaling remain elusive (Oneyama and Okada, 2015; Roskoski, 2015; Sirvent et al., 2015; Zhang and Yu, 2012). Therefore, elucidating how aberrant Src activation occurs in the cancer cells and which mechanisms fuel this process would be of utmost interest in the cancer field.
Here we use cybrid technology, BC cell line, PDX models, and BC patient data to understand the role of energy reprogramming in metastatic TNBC and its significance in the regulation of tumor properties via the activation of Src cancer pathway. This study reports the significance of mitochondrial energy reprogramming in the regulation of a major cancer pathway by posttranslational modification.
Results
Characteristics of Mitochondrial Origin Determine Oncogenic Properties of the Cybrids
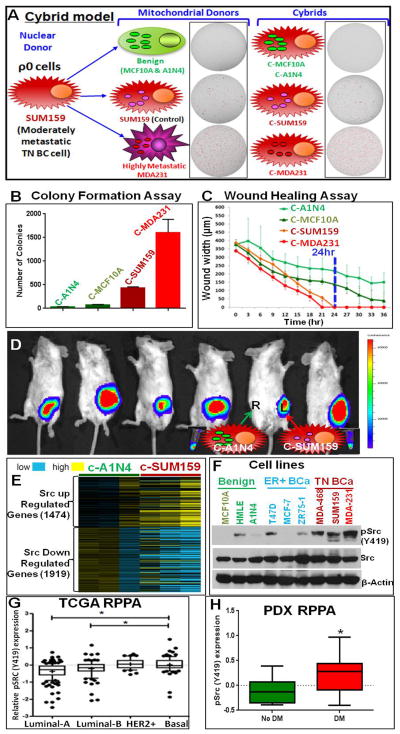
In order to understand the mitochondria-regulated energy and cancer pathways in TNBC, we have generated cybrid models with the moderately metastatic TNBC cell line SUM159 as the nuclear background and mitochondria from TN benign breast cells MCF-10A and A1N4, moderately metastatic SUM159, and highly metastatic MDA-MB-231 (MDA231) TNBC cells (Figure 1A). mtDNAs from original cell lines used for the generation of cybrids were sequenced (Table S1). Cybrids were confirmed by analyzing their nuclear DNA and mtDNA variations. mtDNA copy numbers were quantified from each cybrid clone and only clones with comparable copy numbers were used for further analysis.
Figure 1.
(A) SUM159 TNBC cybrid model with mitochondria from benign (MCF10A and A1N4), moderately metastatic (SUM159), and highly metastatic (MDA231) TN cells under a defined nuclear background of SUM159 mitochondrial DNA depleted ρ0 cells. The cybrids are named as C-MCF10A, C-A1N4, C-SUM159, and C-MDA231 respectively. Images of soft agar colony formation assay of mitochondrial donor cells and cybrids are shown with the cartoon. Colonies are red marked using GelCount™ software. (B) Quantification of soft agar colonies of cybrids. (C) Analysis of wound healing assays performed in an IncuCyte ZOOM® kinetic imaging system with live-cell imaging every three hours. While cybrids with mitochondria from cancer cells completely healed (100% relative wound density) within 24 hours, cybrids with mitochondria from benign cells could not heal the wound even after 36 hours. (D) Bioluminescence imaging of tumor growth in cybrids. Mice injected in the mammary fat pads with cybrids C-A1N4 [right (R); cartooned with green mitochondria] and C-SUM159 [left (L); cartooned with pink mitochondria]. Benign mitochondria (A1N4) abolished the in vivo tumorigenicity of SUM159 cells. Pellets show the luminescence property of both cybrids. (E) Microarray analysis of cybrids suggest that Src transcriptional signature (Creighton, 2008) is abolished in C-A1N4 with benign mitochondria. c-Src upregulated genes are down and c-Src downregulated genes are up in C-A1N4 compared to C-SUM159. Yellow shows higher and blue shows lower expression. (F) pSrc (Y419) analysis in benign, ER positive, and TNBC cell lines showing increased Src Y419 phosphorylation in TNBC cells compared to benign or ER+ cells. (G) The RPPA data from TCGA BC patient tumors suggest an increased pSrc (Y419) expression in basal subtype compared to hormone positive subtypes. The * represents significant increase in the relative pSrc (Y419) expression in basal subtype compared to luminal-A and B subtypes. (H) Comparison of RPPA data from TNBC PDX models with distant metastatic (DM) and non-distant metastatic (No DM) potential, suggesting pSrc (Y419) level is significantly upregulated in PDXs with DM potential.
See also Figure S1.
These cybrids showed in vitro and in vivo tumor-like properties according to their mitochondrial origin. Mitochondria derived from benign cells almost completely abolished the in vitro and in vivo tumor properties of SUM159 cells (Figures 1A, 1B, 1C, and 1D) resulting in significantly lower number of colonies on soft agar (Figure 1B), like their mitochondrial donor cells (Figure S1), and abolished cell migration potential in wound healing assays (Figure. 1C). However, mitochondria from metastatic cells significantly induced tumor properties. In vivo tumor formation assay after mammary gland transplantation suggested that introduction of benign mitochondria resulted in complete inhibition of the in vivo tumor formation potential of SUM159 cells (Figures 1D and S1).
Src Autophosphorylation Depending on Mitochondrial ETC
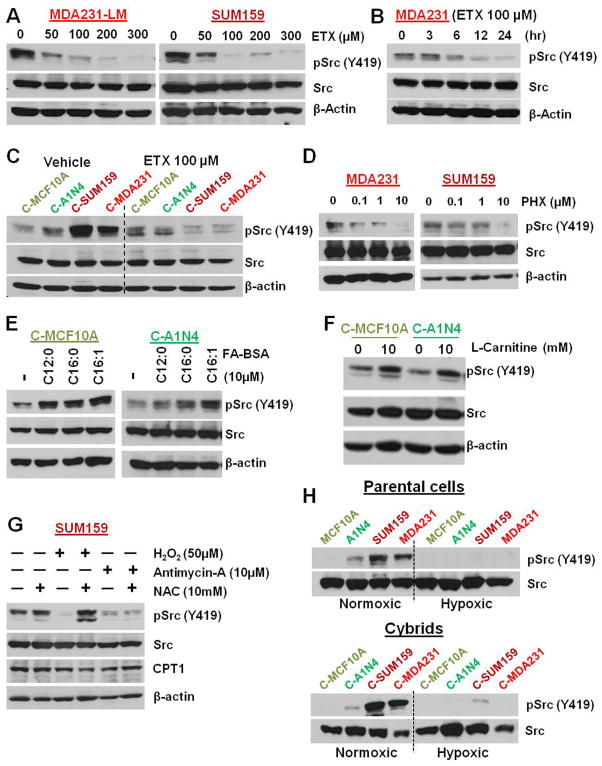
We performed microarray analysis in the cybrid models to identify pathways that are altered according to the mitochondrial regulation. One of the major pathways altered was the Src oncogenic pathway (Figure 1E). Analysis of multiple BC cell lines suggested that pSrc (Y419) is increased in TNBC cell lines compared to benign or ER+ BC cell lines (Figure 1F). The Cancer Genome Atlas (TCGA) data analysis confirmed that Src (Y419) phosphorylation is significantly increased in the basal subtype of BC compared to hormone positive luminal A and B subtypes of BC (Figure 1G). Reverse phase protein array (RPPA) data was analysed from PDX models with and without distant metastatic potential. These PDX models are derived directly from BC patients without intervening culture in vitro, and are maintained by serial transplantation from one animal to another, so that they have never been exposed to in vitro culture conditions. As a result, these xenografts maintain histological and molecular features of the tumor of origin, including their characteristic expression levels (Zhang et al., 2013). Multiple PDX lines were included from some of the patients (Figures S1C and S1D). Analysis suggested significantly increased pSrc (Y419) levels in TNBC PDXs with distant metastasis (Figures 1H and S1C), while, no significant difference was observed with pSrc (Y530) (Figures S1D and S1E).
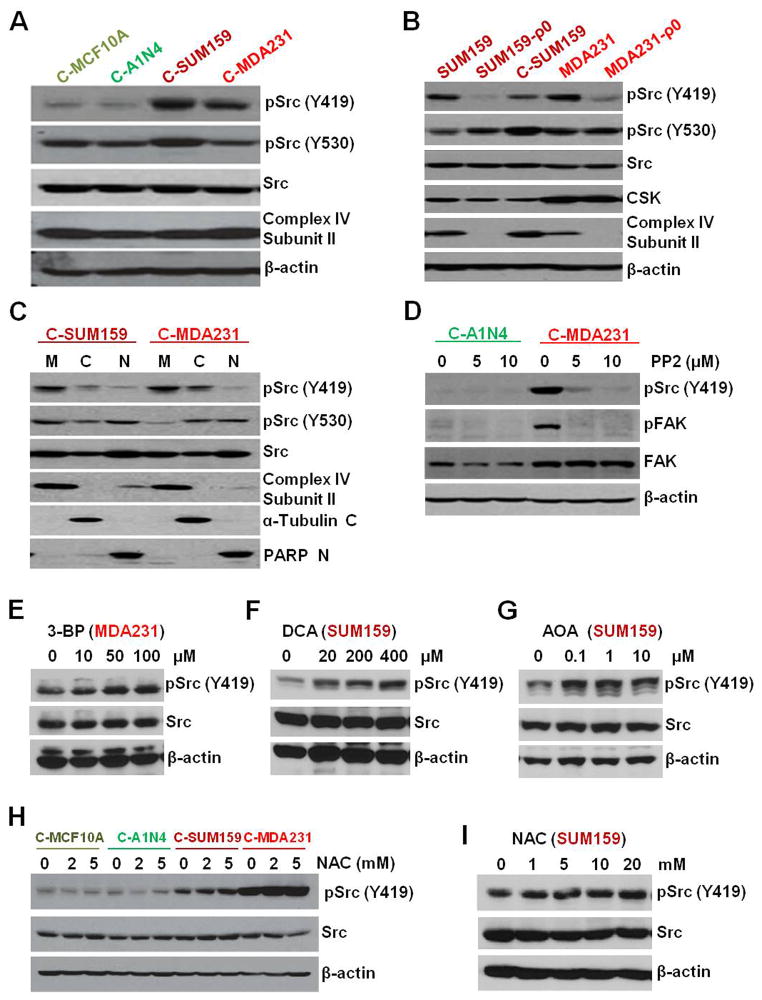
Extensive evaluation of cybrid models and parental cells confirmed that Src (Y419) is activated by metastatic TNBC mitochondria without any major effect at Y530 (Figures 2A and 2B). Cellular fractionation (Figure 2C) confirmed the localization of Src and pSrc (Y419) in TNBC mitochondria. Src (Y419) autophosphorylation (Figure 2D and S2A) of cybrids and cancer potential (Figures S2) of cybrids and parental cells were decreased by treatment with the Src inhibitors PP2 and dasatinib, suggesting that their tumorigenic properties depend on the Src pathway.
Figure 2.
(A) Western Blot showing decreased Src (Y419) phosphorylation in cybrids with mitochondria from benign cells compared to TNBC mitochondria. However, there was no major difference in the total Src levels or Src (Y530) phosphorylation status. Complex-IV subunit II and β-actin were used as mitochondrial and nuclear loading controls. (B) Depletion of mitochondria (ρ0 cells) abolished pSrc (Y419) in parental cells but reappeared in cybrids with TNBC mitochondria with no major effect on total Src, pSrc (Y530), and CSK. (C) Cell fractional analysis suggests localization of pSrc in mitochondrial fraction (M=mitochondria, C= Cytoplasmic, and N= Nuclear fraction). (D) pSrc (Y419) and its target pFAK in cybrids are abolished after treatment with a selective inhibitor for Src-family kinases, PP2. (E and F) TNBC cells treated with glycolysis inhibitors 3-BP (E) and DCA (F), showing no considerable decrease in Src phosphorylation status. (G) Treatment with glutamine pathway inhibitor AOA did not show major reduction in Src (Y419). (H and I) Treatment with ROS scavenger NAC in cybrids (H) and parental cell SUM159 (I) did not abolished the increased Src (Y419) phosphorylation in TNBC.
See also Figure S2 and S3.
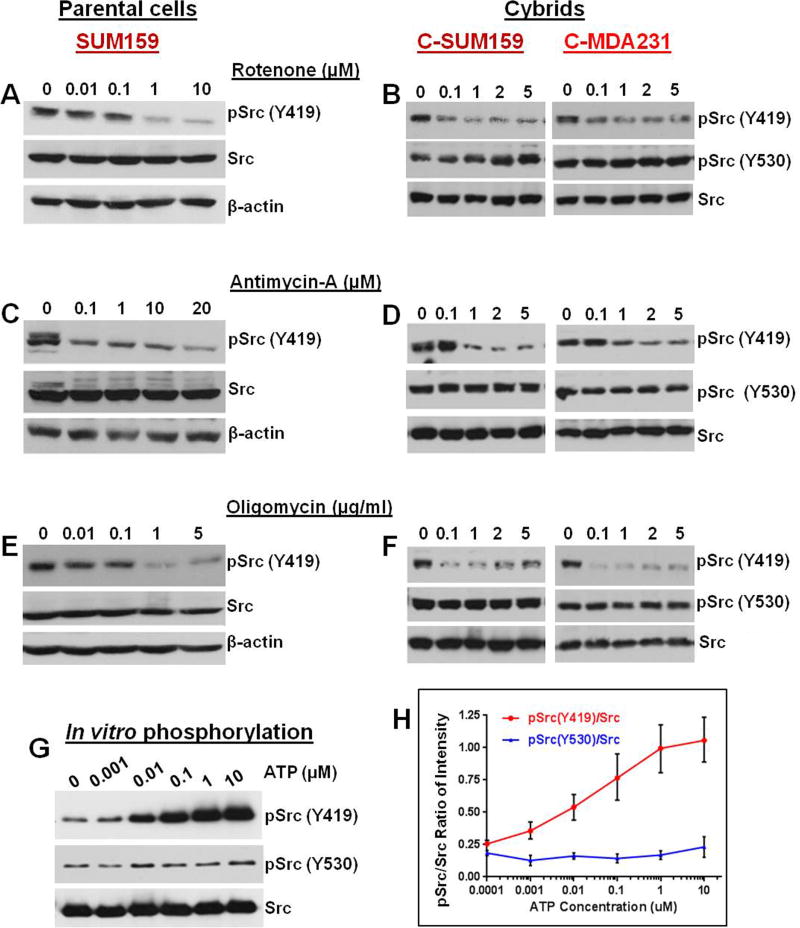
Inhibition of glycolysis by 3-bromopyruvate (3-BP) or dichloroacetate (DCA) did not decrease the increased pSrc (Y419) in TNBC cells (Figures 2E and 2F). Inhibiting the glutamine pathway using aminooxyacetic acid (AOA) (Figure 2G) or scavenging basal reactive oxygen species (ROS) using N-acetylcysteine (NAC) (Figures 2H and 2I) also did not inhibit the induced Src (Y419) autophosphorylation in TNBC cell lines or cybrids. We then analyzed the role of mitochondrial ETC complexes that are responsible for ATP generation inside mitochondria in the regulation of Src autophosphorylation. Interestingly treatment with agents that target ETC including the complex-1 inhibitor rotenone (Figures 3A and 3B), the complex III inhibitor antimycin-A (Figures 3C and 3D), and the ATP synthase inhibitor oligomycin (Figures 3E and 3F) dose-dependently reduced Src (Y419) autophosphorylation both in parental TNBC cells and in cybrids. However, ETC inhibition did not show any major effect on the phosphorylation of Src (Y530) (Figures 3B, 3D, and 3F). In vitro phosphorylation experiments suggested that Src (Y419) autophosphorylation but not pSrc (Y530) is directly dependent on the ATP concentration (Figures 3G and 3H). Though other known mitochondria-localizing Src family kinases (SFKs) (Acin-Perez et al., 2014; Salvi et al., 2002; Vahedi et al., 2015) are also present in the TNBC cells we used (Figure S3A), as observed with pSrc (Y530) their phosphorylation status did not show major alterations according to the mitochondrial character of the cybrid (Figure S3B and S3C)
Figure 3.
Src (Y419) phosphorylation depends on ATP from ETC. (A–F) Cells were treated with the mitochondrial ETC complex I inhibitor Rotenone (A and B), the complex III inhibitor Antimycin-A (C and D), or the complex V inhibitor Oligomycin (E and F). Inhibitors dose-dependently inhibit Src (Y419) autophosphorylation in parental cells (A, C, and E) and cybrids (B, D, and F) respectively, suggesting a critical role of ATP from mitochondrial ETC in Src Y419 autophosphorylation. (G) Western blot of in vitro phosphorylation assay of purified Src protein using varying concentrations of ATP. Src (Y419) autophosphorylation is increased with the ATP concentration. However, no ATP dose dependency is observed in pSrc (Y530). (H) Quantification of the average pSrc/Src ratio from three independent in vitro phosphorylation experiments. The error bars represent S.E.M.
Metastatic TNBC Cells Demonstrate Energy Dependency on Mitochondrial FAO
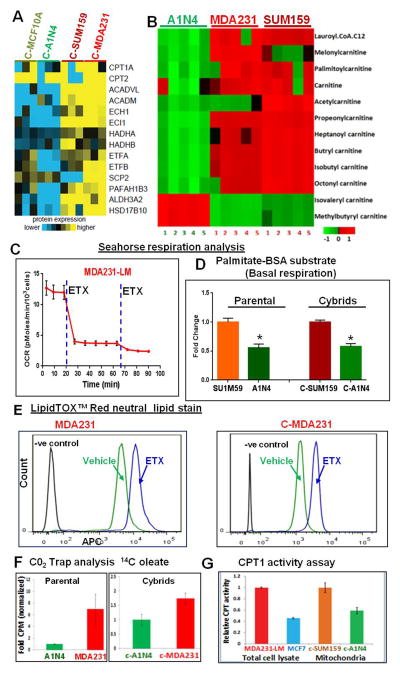
Proteomic analysis suggested that several proteins related to mitochondrial FAO are upregulated in cybrids with mitochondria derived from metastatic TNBC (Figures 4A, S4A, and S4B). Mass spectrometry analysis of carnitines in parental cells showed increased levels of several carnitines in metastatic TNBC cells (Figure 4B). Oxygen consumption rate (OCR) analysis suggested a sharp decrease in OCR after treatment with etomoxir (ETX), which suppresses mitochondrial FAO rate-limiting enzyme, carnitine palmitoyltransferase-1 (CPT1)(Figures 4C and S4C). While ETX treatment decreased the OCR, it also simultaneously increased the glycolysis, as seen by the high extracellular acidification rate (ECAR) (Figure S4D and S4E). Medium containing palmitate conjugated with bovine serum albumin (BSA) significantly increased OCR and basal respiration of TNBC cells and cybrids with mitochondria from TNBC compared to benign cells and cybrids containing mitochondria from benign cells respectively (Figure 4D). Neutral lipid staining suggested increased lipid formation after ETX treatment in TNBC metastatic cells and cybrids with TNBC mitochondria but not in ER+ BC cells (Figures 4E and S4F). A CO2 trap assay using 14C radiolabelled FA oleate-BSA further confirmed increased FAO in metastatic TNBC cells and cybrids (Figure 4F). Moreover, CPT1 activity is upregulated in metastatic TNBC cells and cybrids with TNBC mitochondria (Figure 4G).
Figure 4.
Metastatic TNBC shows energy dependence on mitochondrial FAO. (A) Shotgun Jet Stream Proteomics analysis of cybrids on a UHPLC/AJS-iFunnel Q-TOF suggested increased (yellow) expression of mitochondrial FAO proteins in cybrids with cancer mitochondria. (B) Mass spectrometric analysis of carnitines in benign and metastatic TNBC cells showing increased carnitine levels in TNBC cells (MDA231 and SUM159) compared to benign cells (A1N4). (C) Seahorse XF analysis suggesting that addition of FAO inhibitor almost completely inhibited the increased respiration of lung metastatic TNBC cells (MDA231-LM). (D) Seahorse XF analysis using medium containing palmitate-BSA. Metastatic cells and cybrids with mitochondria from metastatic cells showed increased basal respiration compared to benign cells and cybrids with mitochondria from benign cells respectively. (E) Flowcytometry analysis using the allophycocyanin (APC) channel after lipidTOX neutral lipid staining. Treatment with the ETX enhanced the APC florescence in a TNBC cell line (MDA231) and C-MDA231 cybrids. (F) CO2 trap assays using 14C-labeled oleate in parental cells (left) and cybrids (right) confirm significantly higher FAO in metastatic TNBC cells and its cybrids compared to benign cells and its cybrids. (G) CPT1 activity assay in whole cell lysate from TNBC cell MDA231-LM and ER+ cell MCF7 showing increased CPT1 activity in metastatic TNBC cells (left). Isolated mitochondria from cybrids with mitochondria from cancer cells (C-SUM159) and from benign cells (C-A1N4) show increased CPT1 activity in C-SUM159 cybrid (right).
See also Figure S4.
Mitochondrial FAO Inhibitors or Knockdown of CPT1 and 2 Genes Abolish Src Autophosphorylation
Inhibition of FAO by ETX (Figures 5A–5C) or perhexiline (PHX) (Figure 5D) dose-dependently abolished Src activation in parental cells and cybrids. In reverse experiments, we activated FAO by treatment with FA conjugated with BSA (Figure 5E) or L-carnitine (Figure 5F) as substrates of CPT1 to understand the induction of pSrc (Y419) in cybrids with benign mitochondria. As expected, activation of FAO by FA and L-carnitine induced pSrc (Y419) in cybrids with benign mitochondria (Figures 5E and 5F). ROS induction is known to decrease CPT1 functional activity (Setoyama et al., 2013). Antimycin-A, which inhibits Src autophosphorylation (Figures 3C and 3D), also significantly induces mitochondrial ROS (Figures S5A). To further understand the role of ROS in Src (Y419) autophosphorylation, cells were treated with H2O2, antimycin-A, and/or ROS scavenger NAC (Figures S5). As expected, treatment with H2O2 and antimycin-A inhibited pSrc (Y419) in the parental cell line (Figure 5G) and the cybrid (Figure S5C). The H2O2-mediated inhibition was reversed by the addition of NAC in H2O2-treated cells but not in antimycin-A-treated cells (Figure 5G). This suggests that the ETC activity is important in the activation of pSrc (Y419). Another important question is what happens to Src (Y419) autophosphorylation in conditions where tumor cells are under hypoxia, where ETC function is diminished? Parental cells were analysed under normoxic and hypoxic conditions. As expected, Src (Y419) phosphorylation was abolished under hypoxia, confirming its dependence on ATP generation from mitochondrial oxidative phosphorylation (Figure 5H).
Figure 5.
Inhibition of FAO abolishes Src (Y419) phosphorylation. (A) Treatment with ETX dose-dependently decreased Src (Y419) autophosphorylation in MDA231-LM cells (left) and SUM159 cells (right). (B) Time dependency for ETX-mediated Src dephosphorylation in MDA231 cells. (C) Treatment with ETX depletes pSrc (Y419) phosphorylation in cybrids with mitochondria from cancer cells but not in cybrids with benign mitochondria. (D) Treatment with another CPT1 inhibitor PHX also dose-dependently depleted pSrc (Y419) in MDA231 (left) and SUM159 (right) cells. (E) Stimulation of FAO by treatment with BSA-conjugated FA increased pSrc (Y419) levels in cybrids with benign mitochondria. (F) Addition of known FAO enhancer L-carnitine also increased pSrc (Y419) in cybrids with benign mitochondria. (G) Stimulation of ROS with H2O2 and antimycin-A inhibits pSrc (Y419) in SUM159 cells. Addition of NAC reverses H2O2-mediated but not antimycin-A-mediated inhibition of pSrc (Y419). (H) pSrc (Y419) phosphorylation is depleted in parental cells (upper panel) and cybrids (lower panel) cultured under hypoxic condition with 1% O2.
See also Figure S5.
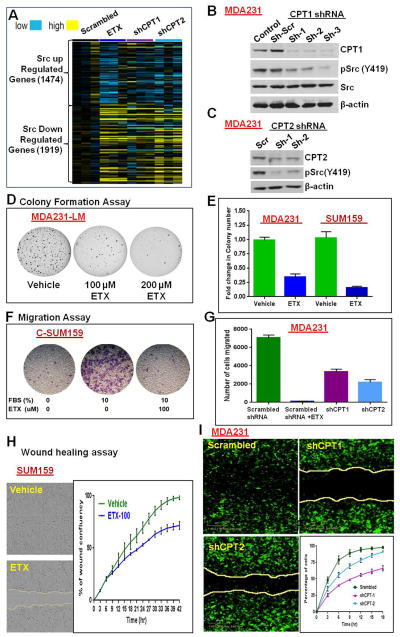
Gene expression data from MDA231 cells confirm that inhibition of FAO by ETX or CPT shRNA reverse the published Src-regulated gene pattern (Creighton, 2008) (Figure 6A). The known Src up-regulated genes are down-regulated and Src down-regulated genes are up-regulated in FAO-inhibited cells. Knockdown of CPT1 (Figure 6B) or CPT2 (Figure 6C) also inhibited Src (Y419) autophosphorylation as observed with ETX (Figure 5). However, FAO inhibition did not show major variation in the expression of Src-related proteins (Figure S6A) or phosphorylation of other analyzed SFKs (Figures S6A–S6D).
Figure 6.
Mitochondrial FAO inhibition represses in vitro tumor properties. (A) Src gene signature from microarray analysis in MDA231 cells. Inhibition of FAO by ETX or CPT shRNA broadly reversed the Src-regulated gene signature compared to scrambled shRNA-transfected cells. (B and C) Knockdown of CPT1 (B) and CPT2 (C) by shRNA down-regulated Src (Y419) phosphorylation. (D) ETX inhibited colony formation in MDA231-LM cells. (E) Two days pretreatment with ETX significantly reduced the soft agar colony formation potential of MDA231 and SUM159 cells. ETX was not added in the soft agar medium. (F and G) Transwell migration assay. ETX treatment significantly decreased the migration potential of cybrids (F) and parental cells (G). (H and I) Images of wound healing assay in SUM159 cells (36 hours) and MDA231 cells (15 hours). Mean percentage of wound confluence analyzed from live-cell imaging every three hours shows significantly reduced wound healing potential after ETX treatment (H) or knockdown of CPT genes by shRNA (I).
See also Figure S6.
FAO Inhibition Abolishes Tumor and Metastatic Properties
In vitro analysis of tumor properties showed the critical role of FAO in TNBC cancer progression (Figures 6D–6I). Colony formation assays of TNBC cell lines using agar medium with ETX suggested dose-dependent decrease of colony formation (Figure 6D). Pretreatment of cells with ETX also significantly decreased the colony formation potential of TNBC cell lines (Figure 6E). Similarly, ETX treatment significantly decreased the migration potential of cybrids and parental cells (Figures 6F and 6G). Knockdown of CPT1 or CPT2 by shRNA also significantly inhibited the transwell migration potential of MDA231 cells (Figure 6G). ETX treatment (Figure 6H) or knockdown of CPT genes (Figure 6I) also significantly inhibited the wound healing potential. However, ETX-mediated inhibition was significantly lower in CPT knockdown cells compared to control cells (Figure S6E).
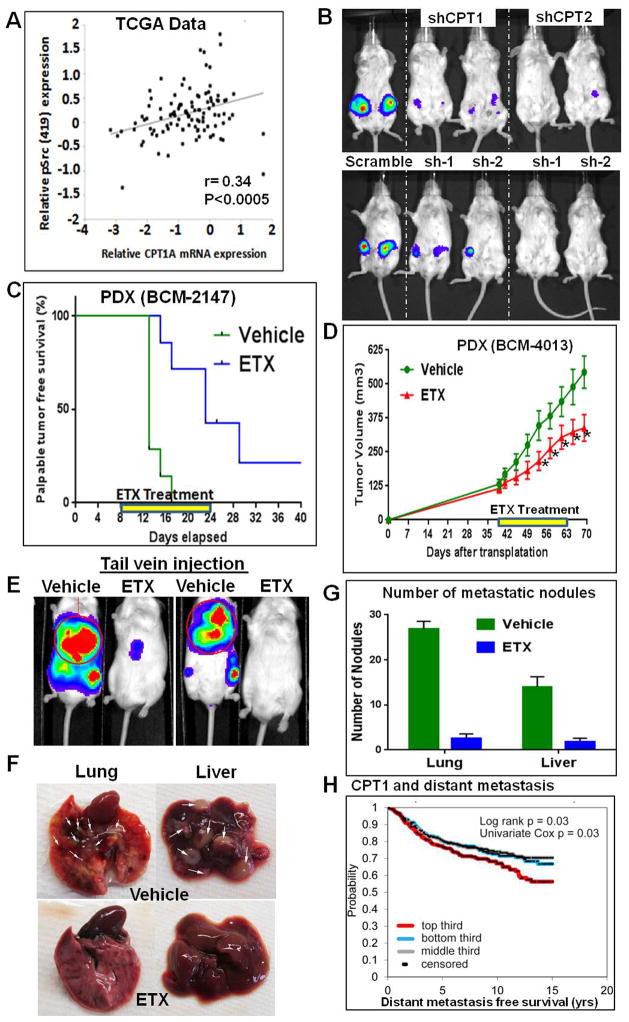
RPPA and gene expression data from TCGA patient tumor analysis suggested a significant positive correlation between CPT1A mRNA expression and Src (Y419) phosphorylation status in basal BC subtypes (Figure 7A). However, no such correlation was observed in other BC subgroups (Figure S7A). In addition, no significant correlation was observed with Src (Y530) phosphorylation status (Figure S7B). CPT1 and CPT2 shRNA also significantly decreased the tumor growth of MDA231 cells compared to scrambled shRNA transfected control cells (Figure 7B). Cell line data validated using PDX models (Zhang et al., 2013). One week after PDX transplantation to the mammary gland, the mice were treated with ETX for two weeks. Short-term ETX treatment significantly delayed the palpable tumor appearance (Figure 7C) and tumor growth of the PDX (BCM-2147, Figure S7C). In mice transplanted with another PDX model (BCM-4013), ETX was administered after the tumor size reached around 150 mm3. As observed above, ETX treatment significantly decreased the tumor growth (Figure 7D). For distant metastasis analysis, MDA231 cells were pretreated for two days with ETX before tail vein injection. The ETX treatment continued for one more week in mice. After three weeks, bioluminescence imaging showed considerably lower metastatic signals in ETX-treated mice (Figure 7E). Also significantly low number of metastatic nodules observed in ETX-treated mice, confirming the role of FAO in distant metastatic potential (Figures 7F and 7G). Similar results were observed even without ETX pre-treatment (Figures. S7D and S7E). Finally, an independent unselected BC patient data set (n=1302) with long-term clinical follow-up data suggested that high CPT1A mRNA expression in the tumor promotes distant metastasis (Kessler et al., 2012) (Figure 7H).
Figure 7.
Mitochondrial FAO regulates in vivo tumor properties. (A) RPPA and gene expression data from the TCGA patient database (n=105) show significant positive correlation between Src (Y419) status and relative CPT1 mRNA expression (r=0.34, p<0.0005, Pearson’s correlation). (B) Bioluminescence images of mice showing that knockdown of CPT1 or CPT2 in MDA231 cells inhibits in vivo tumor growth potential. (C and D) ETX treatment in PDX. PDX BCM-2147 and BCM-4013 were transplanted to the 4th mammary glands of mice. BCM-2147-transplanted mice were treated one week after the transplantation. BCM-4013-transplanted mice were treated after the tumors reached around 150 mm3 (treatment period illustrated with the yellow lines). ETX treatment significantly delayed the palpable tumor formation of BCM-2147 (C) and decreased the tumor growth of BCM-4013 (D) (* represent p<0.05 in one-tailed t-test). (E–G) Significance of FAO in distant metastasis. Control and ETX-pretreated (2 days) MDA231 cells (1.5 × 105) were injected in the tail vein of NOD SCID Gamma mice. ETX treatment was continued in the mice for one more week. Bioluminescence imaging after three weeks showed decreased metastasis in ETX treated mice (E). Lung and liver images (nodules indicated with white arrows) (F) and nodule count (G) confirm significantly decreased metastasis in ETX-treated mice compared to control-treated mice. (H) Gene expression data from independent BC data sets (n=1302) show significantly increased risk of distant metastasis with high CPT1 mRNA expression. P-value by log-rank statistic. See also Figure S7.
Discussion
Cancer cells essentially undergo a metabolic rewiring program to meet the demands of a cancer phenotype (Cantor and Sabatini, 2012). This concept of metabolic reshuffling emanates from Warburg’s hypothesis stating that cancer cell mitochondria are defective and hence switch to energy inefficient pathways (glycolysis). Our data suggest that mitochondrial energy reprogramming to FAO is critical in the activation of Src signaling in TNBC. Several recent reports questioned the conventional concept that in cancer cells the energy is produced only through glycolysis without major contribution from mitochondrial respiration (Biswas et al., 2012; Caino et al., 2015; Carracedo et al., 2013; LeBleu et al., 2014; Liu et al., 2015; Maiuri and Kroemer, 2015; Tan et al., 2015; Villanueva, 2015; Ward and Thompson, 2012; Xu et al., 2015). However, the vast majority of the research into cancer metabolism has been limited to a handful of metabolic pathways, while other pathways have remained in the dark (Carracedo et al., 2013). In the past decade, researchers have revisited the Warburg theory and reached a better understanding of the ‘metabolic switch’ in cancer cells, including the intimate and causal relationship between cancer genes and metabolic alterations, and their potential to be targeted for cancer treatment (Biswas et al., 2012; Carracedo et al., 2013; Ward and Thompson, 2012). Recently alternative energy pathways like FAO have been getting increasing attention in cancer research (Balaban et al., 2015; Harjes et al., 2015). Association of FAO with apoptotic machinery has been reported in cancer (Balaban et al., 2015; Samudio et al., 2010), suggesting that FA metabolism provides a survival advantage to the cancer cells and closely communicates with the cancer signaling pathways.
The alterations in malignant mitochondria or the metabolic pathways governing mitochondrial function vary by cancer type and level of disease progression (Constance and Lim, 2012). Drugs that target mitochondria and exert anti-cancer activity have become a focus of recent research due to their great clinical potential (Neuzil et al., 2013). Thus, mitochondrial energy reprogramming from regulated oxidative phosphorylation and glycolysis to alternative pathways like mitochondrial FAO will have a major influence on tumor response to currently available anticancer drugs. Sensitizing mitochondria (Ni Chonghaile et al., 2011) may be an ideal way to overcome currently non-targetable tumors like TNBC. Mitochondria are known to harbor variations in mtDNA that have been linked to BC development (Ishikawa et al., 2008; Ohsawa et al., 2012). However, here mtDNA sequencing did not show any common variation that could contribute to the energy reprogramming in the analyzed cell models. Thus, the study focused on overall functional characteristics of the mitochondria rather than individual mtDNA variations.
Src is one of the most commonly upregulated pathways in TNBC (Tryfonopoulos et al., 2011). However, even with promising preclinical studies on the significance of the Src pathway in TNBC, single-agent like dasatinib studies in patients with metastatic TNBC did not show a major clinical advantage in unselected populations (Finn et al., 2011; Mayer and Krop, 2010; Tryfonopoulos et al., 2011). From the current results, it should be noted that these patients with advanced tumors might have larger tumors in which several tumor areas are hypoxic and cannot depend on mitochondrial respiration for their survival. Our data shows that those tumors without active mitochondrial oxidative phosphorylation, as in a hypoxic situation, may not maintain Src activity via its autophosphorylation (Figure 5H). Thus, those tumors or tumor areas may be depending on alternative cancer pathways and require combination therapies with other mitochondrial targets or chemotherapeutic agents. Identifying which patients should be selected for Src-directed therapies will be important to the clinical success of these agents.
Mitochondrial protein phosphorylation is an important mechanism for the modulation of mitochondrial function (Acin-Perez et al., 2009; O’Rourke et al., 2011). Previous reports including immunoelectron microscopy studies have shown that Src is located inside mitochondria, and Src activity is also important for the regulation of mitochondrial functions and cell viability (Hebert-Chatelain, 2013; Miyazaki et al., 2003; Salvi et al., 2002; Tibaldi et al., 2008). Src can phosphorylate several proteins involved in the mitochondrial ETC. Cells with high Src activity are also known to maintain high ETC activity, as several mitochondrial proteins were shown to be substrates of Src kinases (Hebert-Chatelain, 2013). For example, Y193 at NADH dehydrogenase [ubiquinone] flavoprotein 2 of respiratory complex I, which is indispensable for NADH dehydrogenase (complex I) activity and ATP production, Y215 at succinate dehydrogenase A of complex II, as well as direct phosphorylation of cytochrome c oxidase subunit-II of complex-IV are some of the mitochondrial phosphorylation targets for Src (Miyazaki et al., 2003; Ogura et al., 2012). Together with these findings, our data propose that increased FAO in TNBC cells activates Src autophosphorylation, and activated Src phosphorylates mitochondrial ETC proteins to maintain its activated status.
Studies have also shown that Src regulates ROS production in localized subcellular districts including mitochondria. Mitochondrial Src can phosphorylate flotillin-1 at Y56 and Y149, which are critical for its interaction with Complex-II and the prevention of ROS production (Ogura et al., 2014). Increased ROS can inhibit CPT1 enzymatic activity (Setoyama et al., 2013). Moreover, L-carnitine is considered as a potential anti-oxidant (Mescka et al., 2011; Ribas et al., 2014) and L-carnitine can protect the H2O2-induced cytotoxity by enhancing CPT1 activity (Li et al., 2012). All these confirm the role of ROS in the down-regulation of FAO via inhibition of CPT1. Our results also support these findings in TNBC. Treatment with H2O2 or antimycin-A (which increases mitochondrial ROS) abolishes Src autophosphorylation (Fig 5G). Interestingly, while ROS scavengers rescued the H2O2-mediated pSrc (Y419) inhibition, it could not reverse antimycin-A-mediated pSrc (Y419) inhibition. However, in addition to increasing ROS, antimycin-A is a strong inhibitor of complex-III function and ETC activity. This further confirms the importance of ETC activity in the autophosphorylation of Src.
Large prospective studies have shown that dietary fat intake is associated with the risk of postmenopausal invasive BC. The Women’s Intervention Nutrition Study (WINS) trial reported that low-fat diet and corresponding weight loss has a favorable effect on BC recurrence, especially in postmenopausal women with ER-negative cancer (Hoy et al., 2009). The recent WINS report highlighted that deaths of women with TNBC were reduced by up to 54% when they followed a program to reduce their dietary fat intake (Chlebowski and Blackburn, 2015). This highlights the significance of fat in TNBC progression. Considering our important finding on the role of fat metabolism in the activation of the Src signature in TNBC, it is also possible that due to a high fat environment, mitochondria will reprogram to a FA dependent energy pathway in obese TNBC patients. Obese women also have a greater risk for TNBC than non-obese women, where menopause status may be a mitigating factor (Pierobon and Frankenfeld, 2013). In pre-menopausal women, a significant association between obesity and TNBC was observed in both case–case and case–control studies. Clinical data reported significantly shorter overall survival and disease-free survival in obese patients with TNBC (Turkoz et al., 2013). Meta analysis on obesity and TNBC reported that compared to over-weight and normal-weight individuals, pre-menopausal women with BMI equal to or greater than 30 have up to a 42% increased risk of developing TNBC (Pierobon and Frankenfeld, 2013). Post-menopausal women often present with less aggressive phenotypes and with estrogen-dependent lesions most likely driven by the production of steroidal hormones from the adipocytes (Lorincz and Sukumar, 2006; Roberts et al., 2010). Since TNBC lacks expression of hormone receptors, distinct molecular mechanisms must link obesity to the TNBC subtype compared to ER+ tumors. Given the worldwide epidemic of overweight and obesity, as well as the aggressiveness associated with TNBC, it is critical to understand the impact of obesity and fat metabolism in FAO-regulated Src-driven TNBC. This will help in identifying and targeting a higher risk group of patients with this aggressive form of BC.
Altogether, these show the role of mitochondrial energy reprogramming to FAO in TNBC and its significance in regulating the driving protein of a major cancer pathway via its post-translational modification. Further studies are in progress to understand the role of obesity in the activation of Src in TNBC, and to seek suitable combination therapies with mitochondrial targets to manage the currently non-targetable TNBC.
Experimental Procedures
Cells, Cybrids and PDX
Cybrids were generated according to the previously published protocol (Vithayathil et al., 2012). Parental cells and cybrids were used and maintained in culture as described in the Supplemental Information. TNBC PDX models (Zhang et al., 2013) were serially transplanted in SCID/Beige female mice (Harlan Laboratories).
In vivo Tumor Studies
For mammary transplantation assay, 2 ×105 cells and cybrids with Firefly luciferase or small tumor pieces from PDXs were transplanted into the fourth mammary fat pads of 4–5 week old SCID/Beige female mice (Zhang et al., 2013). For tail vein injection, 1.5 × 105 MDA231 cells were injected into the tail veins of 4–5 week old NOD SCID Gamma mice. Details are described in the Supplemental Information.
Src Gene Signature Analysis
The mRNA expression profile was performed using U133plus 2.0 array, and the Src transcriptional signature was analyzed as previously described (Creighton, 2008). Details are described in the Supplemental Information.
Supplementary Material
Acknowledgments
Dr. Hostetler provided FA-BSA, Drs. Zhang and Insun supported animal experiments and Mr. Sederstrom supported flowcytometry.
Grant Support
Funding to BAK: NIH (R21CA179720, R21CA173150, R21CA179720S1, R21CA179720S2 & U54-CA149196 Pilot Project), DOD (DAMD W81XWH-11-1-0292), and DLDCC Pilot Projects. Other supports: BCM Cancer Center Grant (P30CA125123), DEF (NIH-R01CA184208), MTL (P50CA50183, R01CA112305, U54 CA149196, P01CA30195), AS (R21CA185516 & U01CA179674), NP (ACS 127430-RSG-15-105-01-CNE), CPRIT Proteomics and Metabolomics Core Facility (RP120092, & Mass spectrometry Center of Excellence by Agilent) and Cytometry and Cell Sorting Core (NIAID P30AI036211, & NCRR S10RR024574).
Footnotes
Accession Numbers
The accession number for the microarray data submitted for cybrid models is GEO: GSE72319. The accession number for the microarray data submitted for MDA231 CPT knockdown is GSE72320.
Authors’ Contributions
Conception and design: BAK, LJW, and JHP; Methodology: BAK, LJW, CJC, MTL, AS, NP, PY, DEF, JHP, SV, BHG, LED, KA, and SK; Data analysis: JHP, SK, SV, PS, VP, VG, KA, DW, ET, SB, TRD, VB, YZ, ET, SM, TRD, CC, CJC, NP and BAK; Writing, review, editing: BAK, LJW, CJC, MTL, AS, JHP, LED, NP, AS, and MTL: Supervision: BAK and LJW
References
- Acin-Perez R, Carrascoso I, Baixauli F, Roche-Molina M, Latorre-Pellicer A, Fernandez-Silva P, Mittelbrunn M, Sanchez-Madrid F, Perez-Martos A, Lowell CA, et al. ROS-triggered phosphorylation of complex II by Fgr kinase regulates cellular adaptation to fuel use. Cell Metab. 2014;19:1020–1033. doi: 10.1016/j.cmet.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R, Salazar E, Brosel S, Yang H, Schon EA, Manfredi G. Modulation of mitochondrial protein phosphorylation by soluble adenylyl cyclase ameliorates cytochrome oxidase defects. EMBO Mol Med. 2009;1:392–406. doi: 10.1002/emmm.200900046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleshin A, Finn RS. SRC: a century of science brought to the clinic. Neoplasia. 2010;12:599–607. doi: 10.1593/neo.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anbalagan M, Moroz K, Ali A, Carrier L, Glodowski S, Rowan BG. Subcellular localization of total and activated Src kinase in African American and Caucasian breast cancer. PLoS One. 2012;7:e33017. doi: 10.1371/journal.pone.0033017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban S, Lee LS, Schreuder M, Hoy AJ. Obesity and cancer progression: is there a role of fatty acid metabolism? Biomed Res Int. 2015;2015:274585. doi: 10.1155/2015/274585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak NP, Banerjee S. Mitochondrial dependency in progression of acute myeloid leukemia. Mitochondrion. 2015;21:41–48. doi: 10.1016/j.mito.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Biswas S, Lunec J, Bartlett K. Non-glucose metabolism in cancer cells--is it all in the fat? Cancer Metastasis Rev. 2012;31:689–698. doi: 10.1007/s10555-012-9384-6. [DOI] [PubMed] [Google Scholar]
- Caino MC, Ghosh JC, Chae YC, Vaira V, Rivadeneira DB, Faversani A, Rampini P, Kossenkov AV, Aird KM, Zhang R, et al. PI3K therapy reprograms mitochondrial trafficking to fuel tumor cell invasion. Proc Natl Acad Sci U S A. 2015;112:8638–8643. doi: 10.1073/pnas.1500722112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A, Cantley LC, Pandolfi PP. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT, Blackburn GL. Abstract S5-08: Final survival analysis from the randomized Women’s Intervention Nutrition Study (WINS) evaluating dietary intervention as adjuvant breast cancer therapy. Cancer Res. 2015;75:S5-08–S05-08. [Google Scholar]
- Constance JE, Lim CS. Targeting malignant mitochondria with therapeutic peptides. Ther Deliv. 2012;3:961–979. doi: 10.4155/tde.12.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini P, Jacotot E, Decaudin D, Kroemer G. Mitochondrion as a novel target of anticancer chemotherapy. J Natl Cancer Inst. 2000;92:1042–1053. doi: 10.1093/jnci/92.13.1042. [DOI] [PubMed] [Google Scholar]
- Creighton CJ. Multiple oncogenic pathway signatures show coordinate expression patterns in human prostate tumors. PLoS One. 2008;3:e1816. doi: 10.1371/journal.pone.0001816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erol A. Retrograde regulation due to mitochondrial dysfunction may be an important mechanism for carcinogenesis. Med Hypotheses. 2005;65:525–529. doi: 10.1016/j.mehy.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Finn RS. Targeting Src in breast cancer. Ann Oncol. 2008;19:1379–1386. doi: 10.1093/annonc/mdn291. [DOI] [PubMed] [Google Scholar]
- Finn RS, Bengala C, Ibrahim N, Roche H, Sparano J, Strauss LC, Fairchild J, Sy O, Goldstein LJ. Dasatinib as a single agent in triple-negative breast cancer: results of an open-label phase 2 study. Clin Cancer Res. 2011;17:6905–6913. doi: 10.1158/1078-0432.CCR-11-0288. [DOI] [PubMed] [Google Scholar]
- Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, Lisanti S, Rampini P, Giroda M, Caino MC, et al. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gucalp A, Sparano JA, Caravelli J, Santamauro J, Patil S, Abbruzzi A, Pellegrino C, Bromberg J, Dang C, Theodoulou M, et al. Phase II trial of saracatinib (AZD0530), an oral SRC-inhibitor for the treatment of patients with hormone receptor-negative metastatic breast cancer. Clin Breast Cancer. 2011;11:306–311. doi: 10.1016/j.clbc.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hail N., Jr Mitochondria: A novel target for the chemoprevention of cancer. Apoptosis. 2005;10:687–705. doi: 10.1007/s10495-005-0792-8. [DOI] [PubMed] [Google Scholar]
- Harjes U, Kalucka J, Carmeliet P. Targeting fatty acid metabolism in cancer and endothelial cells. Crit Rev Oncol Hematol. 2015;97:15–21. doi: 10.1016/j.critrevonc.2015.10.011. [DOI] [PubMed] [Google Scholar]
- Hebert-Chatelain E. Src kinases are important regulators of mitochondrial functions. Int J Biochem Cell Biol. 2013;45:90–98. doi: 10.1016/j.biocel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Herold CI, Chadaram V, Peterson BL, Marcom PK, Hopkins J, Kimmick GG, Favaro J, Hamilton E, Welch RA, Bacus S, et al. Phase II trial of dasatinib in patients with metastatic breast cancer using real-time pharmacodynamic tissue biomarkers of Src inhibition to escalate dosing. Clin Cancer Res. 2011;17:6061–6070. doi: 10.1158/1078-0432.CCR-11-1071. [DOI] [PubMed] [Google Scholar]
- Hoy MK, Winters BL, Chlebowski RT, Papoutsakis C, Shapiro A, Lubin MP, Thomson CA, Grosvenor MB, Copeland T, Falk E, et al. Implementing a low-fat eating plan in the Women’s Intervention Nutrition Study. J Am Diet Assoc. 2009;109:688–696. doi: 10.1016/j.jada.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Kaipparettu BA, Ma Y, Park JH, Lee TL, Zhang Y, Yotnda P, Creighton CJ, Chan WY, Wong LJ. Crosstalk from non-cancerous mitochondria can inhibit tumor properties of metastatic cells by suppressing oncogenic pathways. PLoS One. 2013;8:e61747. doi: 10.1371/journal.pone.0061747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaipparettu BA, Ma Y, Wong LJ. Functional effects of cancer mitochondria on energy metabolism and tumorigenesis: utility of transmitochondrial cybrids. Ann N Y Acad Sci. 2010;1201:137–146. doi: 10.1111/j.1749-6632.2010.05621.x. [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. 1001–1015. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Wang QY, Luan HY, Kang ZC, Wang CB. Effects of L-carnitine against oxidative stress in human hepatocytes: involvement of peroxisome proliferator-activated receptor alpha. J Biomed Sci. 2012;19:32. doi: 10.1186/1423-0127-19-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cho SN, Akkanti B, Jin N, Mao J, Long W, Chen T, Zhang Y, Tang X, Wistub, et al. ErbB2 Pathway Activation upon Smad4 Loss Promotes Lung Tumor Growth and Metastasis. Cell Rep. 2015;S2211-1247(15):142–4. doi: 10.1016/j.celrep.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz AM, Sukumar S. Molecular links between obesity and breast cancer. Endocr Relat Cancer. 2006;13:279–292. doi: 10.1677/erc.1.00729. [DOI] [PubMed] [Google Scholar]
- Lu CL, Qin L, Liu HC, Candas D, Fan M, Li JJ. Tumor cells switch to mitochondrial oxidative phosphorylation under radiation via mTOR-mediated hexokinase II inhibition--a Warburg-reversing effect. PLoS One. 2015;10:e0121046. doi: 10.1371/journal.pone.0121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Kroemer G. Essential role for oxidative phosphorylation in cancer progression. Cell Metab. 2015;21:11–12. doi: 10.1016/j.cmet.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Mayer EL, Krop IE. Advances in targeting SRC in the treatment of breast cancer and other solid malignancies. Clin Cancer Res. 2010;16:3526–3532. doi: 10.1158/1078-0432.CCR-09-1834. [DOI] [PubMed] [Google Scholar]
- Mescka C, Moraes T, Rosa A, Mazzola P, Piccoli B, Jacques C, Dalazen G, Coelho J, Cortes M, Terra M, et al. In vivo neuroprotective effect of L-carnitine against oxidative stress in maple syrup urine disease. Metab Brain Dis. 2011;26:21–28. doi: 10.1007/s11011-011-9238-x. [DOI] [PubMed] [Google Scholar]
- Miyazaki T, Neff L, Tanaka S, Horne WC, Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J Cell Biol. 2003;160:709–718. doi: 10.1083/jcb.200209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil J, Dong LF, Rohlena J, Truksa J, Ralph SJ. Classification of mitocans, anti-cancer drugs acting on mitochondria. Mitochondrion. 2013;13:199–208. doi: 10.1016/j.mito.2012.07.112. [DOI] [PubMed] [Google Scholar]
- Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, del Moore GV, Deng J, Anderson KC, Richardson P, Tai YT, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke B, Van Eyk JE, Foster DB. Mitochondrial protein phosphorylation as a regulatory modality: implications for mitochondrial dysfunction in heart failure. Congest Heart Fail. 2011;17:269–282. doi: 10.1111/j.1751-7133.2011.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Yamaki J, Homma MK, Homma Y. Mitochondrial c-Src regulates cell survival through phosphorylation of respiratory chain components. Biochem J. 2012;447:281–289. doi: 10.1042/BJ20120509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura M, Yamaki J, Homma MK, Homma Y. Phosphorylation of flotillin-1 by mitochondrial c-Src is required to prevent the production of reactive oxygen species. FEBS Lett. 2014;588:2837–2843. doi: 10.1016/j.febslet.2014.06.044. [DOI] [PubMed] [Google Scholar]
- Ohsawa S, Sato Y, Enomoto M, Nakamura M, Betsumiya A, Igaki T. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012;490:547–551. doi: 10.1038/nature11452. [DOI] [PubMed] [Google Scholar]
- Oneyama C, Okada M. MicroRNAs as the fine-tuners of Src oncogenic signalling. J Biochem. 2015;157:431–438. doi: 10.1093/jb/mvv036. [DOI] [PubMed] [Google Scholar]
- Pal SK, Mortimer J. Triple-negative breast cancer: novel therapies and new directions. Maturitas. 2009;63:269–274. doi: 10.1016/j.maturitas.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Pierobon M, Frankenfeld CL. Obesity as a risk factor for triple-negative breast cancers: a systematic review and meta-analysis. Breast Cancer Res Treat. 2013;137:307–314. doi: 10.1007/s10549-012-2339-3. [DOI] [PubMed] [Google Scholar]
- Ribas GS, Vargas CR, Wajner M. L-carnitine supplementation as a potential antioxidant therapy for inherited neurometabolic disorders. Gene. 2014;533:469–476. doi: 10.1016/j.gene.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Roberts DL, Dive C, Renehan AG. Biological mechanisms linking obesity and cancer risk: new perspectives. Annu Rev Med. 2010;61:301–316. doi: 10.1146/annurev.med.080708.082713. [DOI] [PubMed] [Google Scholar]
- Roskoski R., Jr Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015;94:9–25. doi: 10.1016/j.phrs.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Salvi M, Brunati AM, Bordin L, La Rocca N, Clari G, Toninello A. Characterization and location of Src-dependent tyrosine phosphorylation in rat brain mitochondria. Biochim Biophys Acta. 2002;1589:181–195. doi: 10.1016/s0167-4889(02)00174-x. [DOI] [PubMed] [Google Scholar]
- Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest. 2010;120:142–156. doi: 10.1172/JCI38942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setoyama D, Fujimura Y, Miura D. Metabolomics reveals that carnitine palmitoyltransferase-1 is a novel target for oxidative inactivation in human cells. Genes Cells. 2013;18:1107–1119. doi: 10.1111/gtc.12098. [DOI] [PubMed] [Google Scholar]
- Sirvent A, Urbach S, Roche S. Contribution of phosphoproteomics in understanding SRC signaling in normal and tumor cells. Proteomics. 2015;15:232–244. doi: 10.1002/pmic.201400162. [DOI] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 2015;21:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Tibaldi E, Brunati AM, Massimino ML, Stringaro A, Colone M, Agostinelli E, Arancia G, Toninello A. Src-Tyrosine kinases are major agents in mitochondrial tyrosine phosphorylation. J Cell Biochem. 2008;104:840–849. doi: 10.1002/jcb.21670. [DOI] [PubMed] [Google Scholar]
- Tryfonopoulos D, Walsh S, Collins DM, Flanagan L, Quinn C, Corkery B, McDermott EW, Evoy D, Pierce A, O’Donovan N, et al. Src: a potential target for the treatment of triple-negative breast cancer. Ann Oncol. 2011;22:2234–2240. doi: 10.1093/annonc/mdq757. [DOI] [PubMed] [Google Scholar]
- Turkoz FP, Solak M, Petekkaya I, Keskin O, Kertmen N, Sarici F, Arik Z, Babacan T, Ozisik Y, Altundag K. The prognostic impact of obesity on molecular subtypes of breast cancer in premenopausal women. J BUON. 2013;18:335–341. [PubMed] [Google Scholar]
- Vahedi S, Chueh FY, Chandran B, Yu CL. Lymphocyte-specific protein tyrosine kinase (Lck) interacts with CR6-interacting factor 1 (CRIF1) in mitochondria to repress oxidative phosphorylation. BMC Cancer. 2015;15:551. doi: 10.1186/s12885-015-1520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Corti D, Draetta GF. Tumors and Mitochondrial Respiration: A Neglected Connection. Cancer Res. 2015;75(18):3685–6. doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- Villanueva MT. Metabolism: The mitochondria thief. Nat Rev Cancer. 2015;15:70–71. doi: 10.1038/nrc3901. [DOI] [PubMed] [Google Scholar]
- Vithayathil SA, Ma Y, Kaipparettu BA. Transmitochondrial cybrids: tools for functional studies of mutant mitochondria. Methods Mol Biol. 2012;837:219–230. doi: 10.1007/978-1-61779-504-6_15. [DOI] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Biener-Ramanujan E, Yang W, Ramanujan VK. Targeting metabolic plasticity in breast cancer cells via mitochondrial complex I modulation. Breast Cancer Res Treat. 2015;150:43–56. doi: 10.1007/s10549-015-3304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yu D. Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends Pharmacol Sci. 2012;33:122–128. doi: 10.1016/j.tips.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Claerhout S, Prat A, Dobrolecki LE, Petrovic I, Lai Q, Landis MD, Wiechmann L, Schiff R, Giuliano M, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.