Abstract
Igf2 and H19 are reciprocally imprinted genes on mouse distal chromosome 7. They share several regulatory elements, including a differentially methylated region (DMR) upstream of H19 that is paternally methylated throughout development. The cis-acting sequence requirements for targeting DNA methylation to the DMR remain unknown; however, it has been suggested that direct tandem repeats near DMRs could be involved. Previous studies of the imprinted Rasgrf1 locus demonstrate indeed that a direct repeat element adjacent to a DMR is responsible for establishing paternal allele-specific methylation at the DMR and therefore allelic expression of the Rasgrf1 transcript. We identified a prominent and conserved direct tandem repeat 1 kb upstream of the H19 DMR and proposed that it played a similar role in imprinted regulation of H19. To test our hypothesis, we generated mice harboring a 1.7-kb targeted deletion of the direct repeat element and analyzed fetal growth, allelic expression, and methylation within the Igf2-H19 region. Surprisingly the deletion had no effect on imprinting. These results together with deletions of other repeats close to imprinted genes suggest that direct repeats may not be important for the targeting of methylation at the majority of imprinted loci and that the Rasgrf1 locus may be an exception to this rule.
Imprinting is an epigenetic mechanism that determines expression of genes according to their parental origin. Many imprinted genes have associated differentially methylated regions (DMRs), and loss of DNA methylation at these DMRs usually results in loss of imprinted expression (6, 18, 25). It is not yet clear what sequences are involved in the establishment and maintenance of the allele-specific methylation at DMRs, but recent evidence suggests that, in some cases, direct tandem repeats may play a role in this process. Repetitive DNA sequences are found at a high frequency in the vicinity of heavily methylated, silenced regions of the genome. These repetitive sequences are often parasitic in origin, and it has been proposed that they become methylated and silenced as part of a host defense mechanism (3, 27, 45). Recent evidence from plants and fungi suggests that such a mechanism may involve the RNA interference pathway, leading to histone methylation, which, in turn, recruits DNA methylation (1, 35, 44). It now seems likely that this pathway also operates in mammals (22). Direct tandem repeats may be particularly susceptible to RNA interference-mediated heterochromatization (26).
Direct tandem repeats have been found in close vicinity to several imprinted genes in mice and humans, and this has led to the prominent suggestion in the field that they are important in targeting methylation to DMRs (27), although it is not yet clear to what extent this is exclusive to imprinted loci. There are indeed some examples of genes that are imprinted in one species having an associated repeat region, while their nonimprinted homologues in other species have no direct repeat. This is true for the U2afbp-rs1 gene, the Impact gene, and the Rasgrf1 gene (28, 31, 32). However, paradoxically, deletion of the repeat region from U2afbp-rs1 in mice did not abolish imprinting (37). There are also examples of DMRs that possess repeats in one species but not another, but which are nevertheless imprinted in both species (2). Yoon et al. (46) provided the first functional evidence demonstrating the importance of repeats at imprinted loci by deleting the direct repeat downstream of the paternally methylated mouse Rasgrf1 DMR. Paternal transmission of this deletion resulted in loss of methylation at the DMR and loss of gene expression, but this only occurred in some strains of mice.
Here we investigate the role of a direct repeat region upstream of the paternally methylated H19 DMR. The maternally expressed mouse H19 gene lies 90 kb downstream of the reciprocally imprinted Igf2 gene. The two genes share several regulatory elements, including enhancers lying 3′ of H19 and a DMR 5′ of H19 (23, 24, 42, 43). The H19 DMR is essential for imprinted regulation of the locus. On the unmethylated maternal chromosome, it acts as a CCCTC-binding factor (CTCF)-dependent boundary element preventing expression of maternal Igf2 (5, 17, 20, 38, 40). On the paternal chromosome, methylation inhibits binding of the CTCF insulator protein, allowing the downstream enhancers to activate paternal Igf2 expression.
Sequence elements that target DNA methylation to the H19 DMR have so far not been identified. Downstream of the DMR, there is a G-rich direct repeat element; this has been deleted in two previous studies but does not appear to be involved in the regulation of imprinting (34, 41). Several transgenic studies have been used to define the minimal region upstream of H19 required to establish the methylation imprint at the H19 DMR. At the outset, transgenes included only 4 kb of 5′ flanking sequence (4, 13, 33). However, these transgenes only showed imprinted expression when integrated in high copy numbers. A more recent study using a transgene containing 5.5 kb of sequence upstream of the H19 transcriptional start site found that single-copy integrants also showed imprinted expression (10). In addition, an insertion of the region from kb −10 to −0.8 downstream of the H19 gene exhibited the endogenous methylation pattern (19). Thus, it seems possible that an element between kb −5.5 and −4 upstream of H19 is necessary for the regulation of imprinting at this locus as discussed by Cranston et al. (10).
We have identified a 1-kb direct repeat unit (the K repeat) in the region kb −5 to −6 upstream of H19. Positional and sequence conservation of this repeat coupled with evidence from the transgenic experiments described above led us to propose that the K repeat may be involved in regulation of methylation at the H19 DMR. Upon targeted deletion of the paternally inherited copy of the repeat, we expected that the paternal DMR would become hypomethylated, leading to reactivation of the boundary element and loss of Igf2 expression.
MATERIALS AND METHODS
Generation of H19ΔK mice.
To generate the targeting construct pΔK1.7DTA, an XbaI-SacI fragment containing a neomycin resistance cassette flanked by two loxP sites was ligated into litmus 28. The 3.8-kb SacI-SacI 3′ homologous region from cDHI (provided by H. Sasaki) was cloned into the SacI site of plitmus, and the 7.8-kb XbaI-HincII 5′ homologous region fragment from BAC189M11 (provided by H Sasaki) was cloned into the SpeI and EcoRV sites. This plasmid was then digested with SnaBI, and a blunt-ended fragment from pgkdtabpa (provided by D. Adams) containing the diphtheria toxin A (DTA) negative selection cassette was cloned into the site. The Babraham Institute Gene Targeting Facility then electroporated pΔK1.7DTA, linearized with SwaI, into embryonic stem (ES) cells as follows. DNA (3 × 50 μg) was electroporated into passage 9, embryonic development day 14 (E14) 129ola male ES cells (10,000,000 cells for each of the three electroporations) at 250 V and 950 μF. After 24 h in nonselective medium, cells were incubated for 8 days with G418 medium (200 μg/μl) to select for neomycin resistance. The 380 surviving colonies were picked and grown up in four 96-well plates. DNA was extracted and digested with AccIII and StuI to screen for correct integration of the 5′ and 3′ ends by Southern blotting as described below. The 5′ probe consists of a 2.8-kb PCR fragment (90437 to 93289 from AP03184) and the 1.3-kb AvaI-AvaI H19 cDNA 3′ probe is previously described (15). The neomycin resistance cassette (neo) was removed by transiently transfecting Cre recombinase. Correctly excised clones were verified by digestion with StuI, injected into C57BL/6J blastocysts, and transferred into pseudopregnant C57BL/6J females. Male chimeras produced by these injections were paired with C57BL/6J females, and germ line pups (as evidenced by coat color) were screened for the targeted allele by using the StuI digest as described above and subsequently the following PCR assay. PCR amplification was performed with Roche Taq DNA polymerase and buffer with 2.25 mM (50 μl) of MgCl2, using the primers ΔKf (ACACTTCCTCTGAACAAGG) and ΔKr (TACAATGAGGGCAGTAAGC) and a program of 94°C for 2 min followed by 10 cycles consisting of 94°C for 15 s, 55°C for 30 s, and 68°C for 3 min, and then 20 cycles consisting of 94°C for 10 s, 55°C for 30 s, and 68°C for 3 min plus 20 extra s for each cycle at 68°C for 5 min.
Southern blot analysis.
Genomic DNA was extracted from ES cells or tissues as previously described (21). Twenty micrograms of DNA was digested with 65 U of each restriction enzyme, loaded onto a 0.8 to 1% agarose gel, transferred to Hybond N+ membrane (Amersham Pharmacia) under alkaline conditions, and hybridized with specific radiolabeled probes (8, 16).
Growth analysis.
Wet weights of placenta, whole fetuses, and newborn mice were recorded. Data were analyzed by means of two-way analyses of variance with litters and genotype as the two factors.
RNA analysis.
Total RNA was isolated from mouse tissues with the RNeasy mini kit (QIAGEN). For expression analysis by Northern blotting, 10 μg of RNA was separated in low-percentage formaldehyde gels and transferred to Hybond N+ membrane (Amersham Pharmacia) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Radiolabeling of probes and hybridization of filters were as described for DNA analysis. Hybridization was carried out with a 0.9-kb KpnI-BamHI probe specific for Igf2, a 1.3-kb AvaI-AvaI probe specific for H19, and a 0.25-kb HindIII-PstI probe specific for Gapdh as described by Feil et al. (15). Quantifications of expression levels were performed with a phosphorimager (Fujifilm FLA-3000 and AIDA software), and a t test was carried out with the Microsoft Excel statistical function. The reverse transcriptase PCR (RT-PCR) assays used to detect allele-specific expression of Igf2 and H19 were performed as described previously (12).
Bisulfite sequencing.
DNA was prepared from mouse tissues and sperm, extracted with phenol-chloroform, precipitated with 100% ethanol, and digested with BamHI. Digested DNA was denatured and treated with sodium bisulfite solution as previously described (29). PCR amplification was carried out on single agarose beads with nested primers specific for bisulfite-treated DNA. Amplifications were performed in 100 μl of reaction mixture using Roche Taq polymerase and 2.5 mM MgCl2, using the primers OF-GAAAGAAAAAGGTTGGTGAGAAAT and OR-CATAAACCCCTAACCTCATAAAACC at 94°C for 2 min followed by 10 cycles consisting of 94°C for 30 s, 50°C for 2 min, and 72°C for 1 min and then 25 cycles consisting of 94°C for 30 s, 51°C for 1 min 30 s, and 72°C for 2 min plus 5 extra s for each cycle at 72°C for 5 min. Two microliters of the first amplification products was used in a second reaction with the nested primers IF-AGGTTGGTGAGAAAATAGAGATT and IR-CAACCCTAATCTTTACACACAAAAA.
RESULTS
Identification of a conserved repeat region upstream of the H19 DMR.
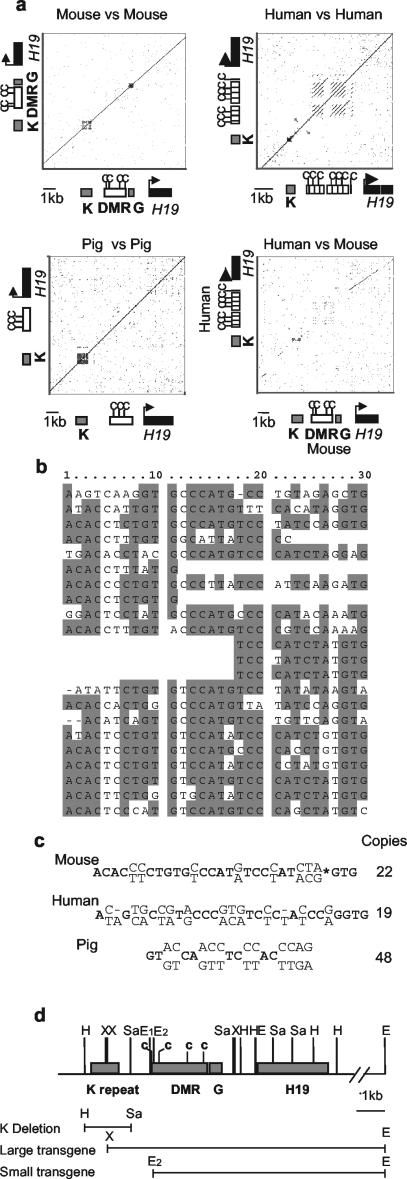
Sequence analysis of the region 5′ of the mouse H19 DMR revealed a 1-kb direct tandem repeat unit (Fig. 1a and b). We have termed this repeat the K repeat. Similar analysis showed that this region also contains a repeat in humans and pigs. A comparison with the human H19 locus (using the Omiga sequence analysis package) showed positional conservation of the K repeat and some conservation of the sequence (Fig. 1a and c) but no conservation of the downstream G-rich repeat. The human CTCF-dependent boundary element contains additional repeat elements as previously described (39). There are also two smaller regions of conservation upstream of the repeat, one of which contains a retroviral long terminal repeat element. The K repeat consists of a 30-bp unit repeated 22 times in mice and a 31-bp unit repeated 19 times in humans. In pigs, the repeat consists of a 19-bp unit repeated 48 times, but there is no significant sequence conservation with the mouse or human K repeat (Fig. 1c) Further comparisons show that the sequence and position of this repeat are highly conserved in rats (data not shown). Figure 1d shows the position of the K repeat and the deleted region described below with respect to the transgenic studies discussed (4, 10, 13). The large transgene, which imprints as a single-copy integrant, contains 400 bp of the K repeat, whereas the short transgene, which only imprints in high-copy-number integrants, does not contain the K repeat. These transgenic studies, coupled with the highly conserved position of this repeat and the sequence conservation across species, support the hypothesis that the K repeat is involved in the regulation of methylation and/or imprinting at the H19 DMR.
FIG. 1.
Position and structure of the H19 DMR K repeat. (a) Dot plots carried out using the Omiga sequence (Oxford Molecular) analysis package. The window size is set at 30, and the threshold sequence identity level is set at 65%. The sequence elements corresponding to the dot plot are shown adjacent to the relevant axes. The mouse H19 DMR is shown by an open rectangle, and the repeat regions are marked by gray rectangles. The human and pig equivalent to the DMR region is shown, but neither has an obvious homologue of the G repeat. The CTCF binding sites are marked with c. The mouse-versus-mouse dot plot shows the position of the two repeat regions (K and G) relative to the DMR and the H19 gene. The human-versus-human plot shows the K repeat and the repeats within the CTCF-dependent boundary element. The pig-versus-pig plot shows that the K repeat is the only repeat in the region. The mouse-versus-human dot plot clearly shows some conservation of the K repeat but very little of the G repeat region, although there may be some conservation of repeats within the DMR region, probably including the CTCF binding sites. Upstream regions contain some conserved interspersed repetitive elements. (b) The 22 repeat units within the mouse K repeat. (c) The 30- and 31-bp consensus mouse and consensus human sequences are shown, with the highly conserved residues in boldface and the variable residues shown above and below the consensus. The 19-bp consensus of the pig is also shown. Accession numbers: mouse, AF049091 and AP003184; human, AF087017 and AF043430; and pig, AY044827. (d) Scale diagram showing the position of the K repeat and deletion relative to the large and small transgenes (4, 10, 13, 33). The EcoRI, HincII, SacI, and XbaI restriction sites are labeled E, H, Sa, and X, respectively.
Targeted deletion of the K repeat in the mouse.
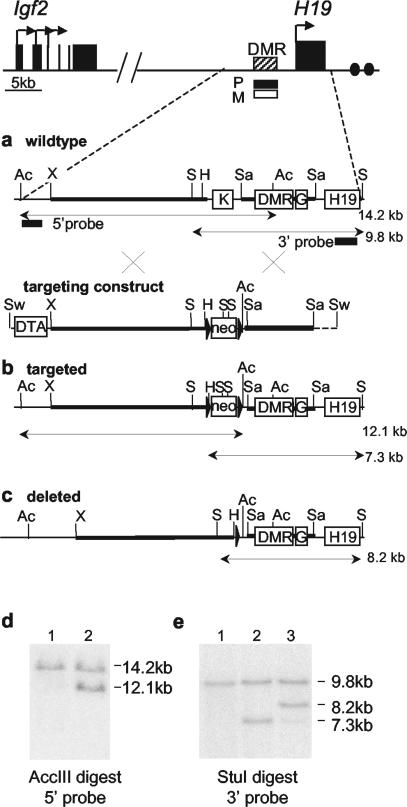
We deleted a 1.7-kb region (from kb −4.7 to −6.4) spanning the K repeat in mice by homologous recombination in ES cells using the strategy shown in Fig. 2a, b, and c. The targeting construct (Fig. 2b) included a neomycin resistance cassette (neo) and also a cassette containing the DTA gene designed to enrich for homologous recombination. Recombinant clones were screened by Southern blot (Fig. 2d and e). The neo cassette was deleted from a correctly targeted clone, and mice carrying the deletion (H19ΔK) were generated. Heterozygous mice were then mated to C57BL/6J mice for analysis of growth and gene expression levels and homozygous SD7 (a Mus musculus domesticus strain containing the distal portion of chromosome 7 of Mus spretus origin) mice for analysis of allelic expression and allele-specific DNA methylation levels.
FIG. 2.
Targeting strategy for deletion of the K repeat. (a) Map of the Igf2-H19 region with a large-scale restriction map of the wild-type H19 upstream region and targeting vector. The DMR at kb −2 to −4 from the transcription start site of H19 is methylated on the paternal allele and unmethylated on the maternal allele. The K repeat and the G repeat lie 1 kb upstream and directly downstream of the DMR, respectively. Black rectangles show the positions of the 5′ and 3′ probes used for screening of ES cells by Southern blot. The targeting construct contains a floxed neomycin resistance cassette (neo) replacing a 1.7-kb region containing the K repeat and a DTA cassette to enrich for homologous recombination. Heavy black lines show regions of homology, and dotted lines indicate the vector sequence. The crosses represent homologous recombination events. The restriction fragments used for Southern blot screening are indicated. (b) Restriction map of the correctly targeted locus. (c) The targeted locus after deletion of the neomycin resistance cassette by Cre recombination in ES cells. (d) Screening for integration of the 5′ end of the targeting construct by Southern blot. The wild-type allele AccIII digest gives a 14.2-kb band, and the targeted allele gives a 12.1-kb band. Lanes 1: wild type; 2 H19ΔKneo+/−. (e) Screening for integration of the 3′ end of the targeting construct by Southern blot. DNA was digested with StuIand probed with the 3′ probe. The wild-type allele digest gives a 9.8-kb band, and the targeted allele gives a 7.3-kb band and an 8.2-kb band after Cre deletion. Lanes: 1, wild type; 2, H19ΔKneo+/−; 3, H19ΔK+/−. The AccIII, ApaI, HincII, SacI, StuI, SwaI, and XbaI restriction sites are labeled Ac, A, H, Sa, S, Sw, and X, respectively.
Deletion of the K repeat has no effect on fetal growth.
We hypothesized that paternal transmission of the deletion would cause a reduction in paternal allele-specific DNA methylation at the H19 DMR resulting in reactivation of the CTCF boundary element, a decrease in Igf2 expression, and therefore a reduction in fetal growth. In order to test this prediction, 6 litters of H19ΔK+/− mice and their wild-type littermates were weighed at E18 and at birth (data not shown). An analysis of variance test showed no significant difference between the mutant and wild-type embryos and newborns. In addition, no difference was observed in placental weight at E18 after paternal transmission of the deletion. Similarly, maternal transmission of the H19ΔK deletion had no significant effect on growth (data not shown).
Deletion of the K repeat has no effect on expression of H19 or Igf2.
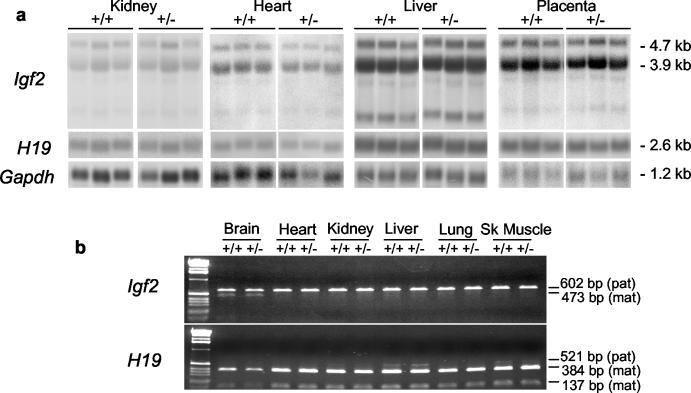
Substantial changes in the levels of Igf2 expression do not always result in comparable changes in fetal growth (9, 40). We therefore analyzed the expression levels of Igf2 and H19 by Northern blot after paternal transmission of the deletion (Fig. 3a). No significant differences were seen between mutant and wild-type levels in newborn tissues or E18 placenta. Allelic expression in the locus was also unchanged, with both wild-type and H19ΔK+/− animals expressing Igf2 from only the paternal chromosome (except for some regions of the brain) and H19 only from the maternal chromosome (except for low levels of paternal expression in skeletal muscle and liver) (Fig. 3b). Again, no changes were observed after maternal transmission of the deleted allele (data not shown).
FIG. 3.
Analysis of Igf2 and H19 expression levels and imprinting after paternal transmission of H19ΔK deletion. (a) Heterozygous H19ΔK males were mated to C57BL/6J females, and the expression levels of Igf2 and H19 in the newborn H19ΔK (+/−) offspring and their wild-type (+/+) littermates were analyzed by Northern blot along with E18 H19ΔK and wild-type placentas. RNA was electrophoresed under denaturing conditions and hybridized with probes specific for Igf2, H19, and Gapdh. Six samples of each genotype from each tissue were quantified with a phosphorimager (Fujifilm FLA-3000 and AIDA software) (three representative samples are shown). The total Igf2 and H19 transcripts were normalized against Gapdh levels. A t test was carried out and showed no significant difference in Igf2 and H19 expression levels between wild-type and H19ΔK tissues. (b) Heterozygous H19ΔK males were mated to homozygous SD7 females, and allele-specific expression of Igf2 and H19 was analyzed in tissues from newborn H19ΔK (+/−) offspring and their wild-type (+/+) littermates by RT-PCR. For Igf2, an SD7-specific BsaAI polymorphism was used to distinguish between parental alleles (602-bp band for C57BL/6 [paternal] and 473-bp band for SD7 [maternal]). For H19, an SD7-specific BglI polymorphism was used to distinguish between parental alleles (521-bp band for C57BL/6 [paternal] and 384- and 137-bp bands for SD7 [maternal]). Two samples of each tissue were analyzed. No change in imprinted expression of Igf2 or H19 was detected.
Deletion of the K repeat has no effect on differential methylation.
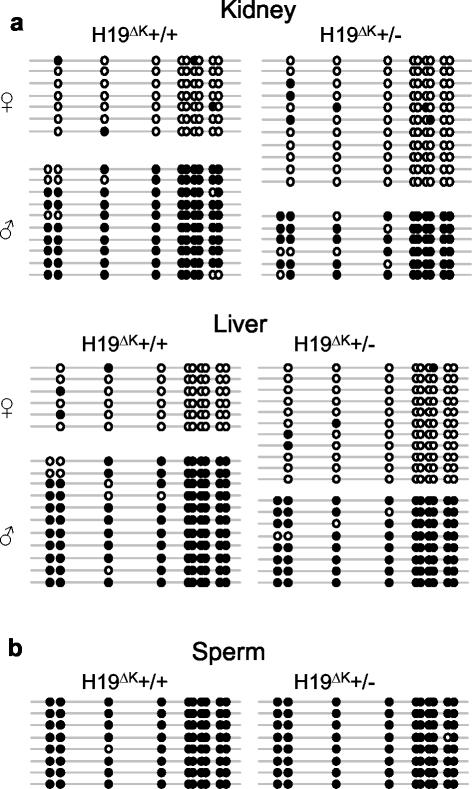
Methylation at the H19 DMR was analyzed in tissues from newborn mice by bisulfite sequencing (Fig. 4a) and Southern blot (data not shown) after paternal transmission of the H19ΔK allele. No differences were seen in paternal or maternal methylation levels. Paternal allele-specific methylation levels remained high even at the 5′ end of the DMR, which lies only 590 bp downstream of the deletion. In addition, sperm DNA was also found to be highly methylated in H19ΔK+/− adult males by bisulfite sequencing (Fig. 4b) and Southern blot (data not shown). Methylation analysis by Southern blot also revealed no methylation changes after maternal transmission of the deletion (data not shown).
FIG. 4.
Analysis of methylation levels at the H19 DMR after paternal transmission of H19ΔK deletion. (a) Heterozygous H19ΔK males were mated to homozygous SD7 females. Methylation of the H19 DMR was analyzed in tissues from newborn H19ΔK (+/−) offspring and their wild-type (+/+) littermates by bisulfite sequencing. Each line represents a single template molecule; solid circles represent methylated CpGs. Single nucleotide polymorphisms were used to distinguish the parental origin of the sequences. Two samples of each tissue were analyzed. No change in methylation was detected. (b) Sperm was collected from the caudal epididymus of adult heterozygous H19ΔK (+/−) males and their wild-type (+/+) littermates. Methylation of the H19 DMR was analyzed by bisulfite sequencing. No change in methylation was detected.
DISCUSSION
We have identified a direct repeat element 1 kb upstream of the mouse H19 DMR, which is conserved in humans, rats, and pigs. We proposed that this repeat may be involved in regulation of methylation in the DMR, but targeted deletion did not affect imprinting or methylation in the Igf2-H19 locus.
Given the dominant hypothesis in the imprinting field that associates tandem repeats with targeting of methylation to DMRs (27), the considerable evidence for a role for tandem repeats in heterochromatization, and the conservation of the K repeat across several species, this result was unexpected. However similar deletions of the G repeat upstream of the H19 DMR and the direct repeat in the U2afbp-rs1 locus also have no effect on imprinting (34, 37, 41). To date, only one repeat deletion in an imprinted region has given rise to aberrant DNA methylation and imprinting: the repeat downstream of the Rasgrf1 DMR (47). (It should be noted that in mice carrying this deletion, the loss of methylation varies in a strain-dependent manner and in some cases methylation is at wild-type levels.) It is therefore possible that Rasgrf1 is the exception rather than the rule. In support of this, a conditional deletion of the de novo methylase gene Dnmt3a in the male germ line causes a reduction in methylation at paternally methylated DMRs upstream of H19 and Gtl2 but has no effect on methylation at Rasgrf1, indicating that a different enzymatic pathway may be responsible for methylation of the Rasgrf1 DMR in the male germ line (H. Sasaki, personal communication). Perhaps the pathway used at Rasgrf1 is partly dependent on the presence of a direct repeat, whereas the Dnmt3a pathway used at H19 and Gtl2 require a different methylation targeting signal. Taken together, these results suggest that direct tandem repeats are not universal signals for methylation of DMRs.
The transgenic experiments described above indicate that an extra 1.5 kb of sequence upstream of the DMR enables transgenes to imprint as single-copy insertions. This extra sequence includes 400 kb (12 repeat units) of the K repeat, which led us to hypothesize that the repeat was important for imprinting. However, it must be noted that the original short transgenes used a slightly truncated form of the DMR lacking about 80 bp of the DMR as defined by methylation studies (4, 13, 33, 42). Though relatively short in length, this sequence contains one of the four CTCF binding sites. We now know that that CTCF binding not only is important for boundary function but also is involved in protecting against de novo methylation on the maternal H19 allele (14, 30, 36). Perhaps it is this small sequence element that enables the long transgenes to imprint as single-copy insertions rather than an incomplete copy of the K repeat.
Although the upstream region of H19 is essential for imprinting, there are also downstream elements involved in the establishment of differential methylation (7, 11). Maternal transmission of the Mnt mutation, a chromosomal inversion beginning 25 kb downstream of H19, causes hypermethylation of the maternal H19 DMR. This indicates that a number of cis elements are required to establish and maintain differential methylation and imprinting at H19. An interesting possibility is that there may be redundancy between some of these elements with the presence of one or two being sufficient to establish the methylation imprint at the endogenous DMR even if others are deleted.
In conclusion, our results taken together with others now suggests that the simple (and appealing) hypothesis that tandem repeats are necessary for targeting of methylation to DMRs in imprinted genes is not generally correct.
Acknowledgments
We thank The Babraham Institute Gene Targeting Facility for their help with generating the H19ΔK mice, H. Sasaki for providing the cDH1 cosmid and the 189M11 Bac, and D. Adams for providing the DTA-containing plasmid. We thank G. Kelsey for comments on the manuscript and E. Walters for help with statistical analysis.
This work was supported by BBSRC and MRC.
Footnotes
Present address: Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202-5251.
REFERENCES
- 1.Aravin, A. A., N. M. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozovsky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. [DOI] [PubMed] [Google Scholar]
- 2.Arnaud, P., D. Monk, M. Hitchins, E. Gordon, W. Dean, C. V. Beechey, J. Peters, W. Craigen, M. Preece, P. Stanier, G. E. Moore, and G. Kelsey. 2003. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum. Mol. Genet. 12:1005-1019. [DOI] [PubMed] [Google Scholar]
- 3.Barlow, D. P. 1993. Methylation and imprinting: from host defense to gene regulation? Science 260:309-310. [DOI] [PubMed] [Google Scholar]
- 4.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 6.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 7.Cerrato, F., W. Dean, K. Davies, K. Kagotani, K. Mitsuya, K. Okumura, A. Riccio, and W. Reik. 2003. Paternal imprints can be established on the maternal Igf2-H19 locus without altering replication timing of DNA. Hum. Mol. Genet. 12:3123-3132. [DOI] [PubMed] [Google Scholar]
- 8.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constancia, M., W. Dean, S. Lopes, T. Moore, G. Kelsey, and W. Reik. 2000. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 26:203-206. [DOI] [PubMed] [Google Scholar]
- 10.Cranston, M. J., T. L. Spinka, D. A. Elson, and M. S. Bartolomei. 2001. Elucidation of the minimal sequence required to imprint H19 transgenes. Genomics 73:98-107. [DOI] [PubMed] [Google Scholar]
- 11.Davies, K., L. Bowden, P. Smith, W. Dean, D. Hill, H. Furuumi, H. Sasaki, B. Cattanach, and W. Reik. 2002. Disruption of mesodermal enhancers for Igf2 in the minute mutant. Development 129:1657-1668. [DOI] [PubMed] [Google Scholar]
- 12.Dean, W., L. Bowden, A. Aitchison, J. Klose, T. Moore, J. J. Meneses, W. Reik, and R. Feil. 1998. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125:2273-2282. [DOI] [PubMed] [Google Scholar]
- 13.Elson, D. A., and M. S. Bartolomei. 1997. A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol. Cell. Biol. 17:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedoriw, A. M., P. Stein, P. Svoboda, R. M. Schultz, and M. S. Bartolomei. 2004. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303:238-240. [DOI] [PubMed] [Google Scholar]
- 15.Feil, R., J. Charlton, A. P. Bird, J. Walter, and W. Reik. 1994. Methylation analysis on individual chromosomes: improved protocol for bisulphite genomic sequencing. Nucleic Acids Res. 22:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 17.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 18.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 19.Kaffer, C. R., M. Srivastava, K. Y. Park, E. Ives, S. Hsieh, J. Batlle, A. Grinberg, S. P. Huang, and K. Pfeifer. 2000. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14:1908-1919. [PMC free article] [PubMed] [Google Scholar]
- 20.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. F. Qi, A. Wolffe, R. Ohlsson, and V. V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 21.Laird, P. W., A. Zijderveld, K. Linders, M. A. Rudnicki, R. Jaenisch, and A. Berns. 1991. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 19:4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 23.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34-39. [DOI] [PubMed] [Google Scholar]
- 24.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9:2079-2089. [DOI] [PubMed] [Google Scholar]
- 25.Li, E., C. Beard, A. C. Forster, T. H. Bestor, and R. Jaenisch. 1993. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harbor Symp. Quant. Biol. 58:297-305. [DOI] [PubMed] [Google Scholar]
- 26.Martienssen, R. A. 2003. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 35:213-214. [DOI] [PubMed] [Google Scholar]
- 27.Neumann, B., P. Kubicka, and D. P. Barlow. 1995. Characteristics of imprinted genes. Nat. Genet. 9:12-13. [DOI] [PubMed] [Google Scholar]
- 28.Okamura, K., Y. Hagiwara-Takeuchi, T. Li, T. H. Vu, M. Hirai, M. Hattori, Y. Sakaki, A. R. Hoffman, and T. Ito. 2000. Comparative genome analysis of the mouse imprinted gene impact and its nonimprinted human homolog IMPACT: toward the structural basis for species-specific imprinting. Genome Res. 10:1878-1889. [DOI] [PubMed] [Google Scholar]
- 29.Olek, A., J. Oswald, and J. Walter. 1996. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24:5064-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pant, V., P. Mariano, C. Kanduri, A. Mattsson, V. Lobanenkov, R. Heuchel, and R. Ohlsson. 2003. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearsall, R. S., C. Plass, M. A. Romano, M. D. Garrick, H. Shibata, Y. Hayashizaki, and W. A. Held. 1999. A direct repeat sequence at the Rasgrf1 locus and imprinted expression. Genomics 55:194-201. [DOI] [PubMed] [Google Scholar]
- 32.Pearsall, R. S., H. Shibata, A. Brozowska, K. Yoshino, K. Okuda, P. J. deJong, C. Plass, V. M. Chapman, Y. Hayashizaki, and W. A. Held. 1996. Absence of imprinting in U2AFBPL, a human homologue of the imprinted mouse gene U2afbp-rs. Biochem. Biophys. Res. Commun. 222:171-177. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer, K., P. A. Leighton, and S. M. Tilghman. 1996. The structural H19 gene is required for transgene imprinting. Proc. Natl. Acad. Sci. USA 93:13876-13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed, M. R., A. D. Riggs, and J. R. Mann. 2001. Deletion of a direct repeat element has no effect on Igf2 and H19 imprinting. Mamm. Genome 12:873-876. [DOI] [PubMed] [Google Scholar]
- 35.Reinhart, B. J., and D. P. Bartel. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297:1831. [DOI] [PubMed] [Google Scholar]
- 36.Schoenherr, C. J., J. M. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33:66-69. [DOI] [PubMed] [Google Scholar]
- 37.Sunahara, S., K. Nakamura, K. Nakao, Y. Gondo, Y. Nagata, and M. Katsuki. 2000. The oocyte-specific methylated region of the U2afbp-rs/U2af1-rs1 gene is dispensable for its imprinted methylation. Biochem. Biophys. Res. Commun. 268:590-595. [DOI] [PubMed] [Google Scholar]
- 38.Szabo, P., S. H. Tang, A. Rentsendorj, G. P. Pfeifer, and J. R. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10:607-610. [DOI] [PubMed] [Google Scholar]
- 39.Takai, D., F. A. Gonzales, Y. C. Tsai, M. J. Thayer, and P. A. Jones. 2001. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum. Mol. Genet. 10:2619-2626. [DOI] [PubMed] [Google Scholar]
- 40.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorvaldsen, J. L., M. R. W. Mann, O. Nwoko, K. L. Duran, and M. S. Bartolomei. 2002. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 22:2450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tremblay, K. D., K. L. Duran, and M. S. Bartolomei. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tremblay, K. D., J. R. Saam, R. S. Ingram, S. M. Tilghman, and M. S. Bartolomei. 1995. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9:407-413. [DOI] [PubMed] [Google Scholar]
- 44.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 45.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-340. [DOI] [PubMed] [Google Scholar]
- 46.Yoon, B. J., H. Herman, A. Sikora, L. T. Smith, C. Plass, and P. D. Soloway. 2002. Regulation of DNA methylation of Rasgrf1. Nat. Genet. 30:92-96. [DOI] [PMC free article] [PubMed] [Google Scholar]