Abstract
Objective
To design, develop and prototype clinical dashboards to integrate high-frequency health and wellness data streams using interactive and real-time data visualisation and analytics modalities.
Materials and methods
We developed a clinical dashboard development framework called electronic healthcare data visualization (EHDViz) toolkit for generating web-based, real-time clinical dashboards for visualising heterogeneous biomedical, healthcare and wellness data. The EHDViz is an extensible toolkit that uses R packages for data management, normalisation and producing high-quality visualisations over the web using R/Shiny web server architecture. We have developed use cases to illustrate utility of EHDViz in different scenarios of clinical and wellness setting as a visualisation aid for improving healthcare delivery.
Results
Using EHDViz, we prototyped clinical dashboards to demonstrate the contextual versatility of EHDViz toolkit. An outpatient cohort was used to visualise population health management tasks (n=14 221), and an inpatient cohort was used to visualise real-time acuity risk in a clinical unit (n=445), and a quantified-self example using wellness data from a fitness activity monitor worn by a single individual was also discussed (n-of-1). The back-end system retrieves relevant data from data source, populates the main panel of the application and integrates user-defined data features in real-time and renders output using modern web browsers. The visualisation elements can be customised using health features, disease names, procedure names or medical codes to populate the visualisations. The source code of EHDViz and various prototypes developed using EHDViz are available in the public domain at http://ehdviz.dudleylab.org.
Conclusions
Collaborative data visualisations, wellness trend predictions, risk estimation, proactive acuity status monitoring and knowledge of complex disease indicators are essential components of implementing data-driven precision medicine. As an open-source visualisation framework capable of integrating health assessment, EHDViz aims to be a valuable toolkit for rapid design, development and implementation of scalable clinical data visualisation dashboards.
Keywords: early warning systems, biomedical informatics, data visuzalization, clinical decision systems, clinical dashboard
Strengths and limitations of this study.
Developing scalable and sustainable healthcare information technology (healthIT) solutions for data management, visual analytics and predictive modeling are critical to improving the quality of affordable healthcare delivery.
We developed electronic healthcare data visualization (EHDViz) as a cost-effective, open-source, extensible toolkit for rapid design, development and implementation of clinical dashboards to address the need to improve data visualisation in different aspects of healthcare delivery, including population health management, patient engagement and simulation-based learning.
A limitation of current version of EHDViz is that developers need skills in R and web development; also, extensive data cleaning and quality control steps are a priority before importing large quantity data for visualisation.
While EHDViz is designed as vendor agnostic framework, importing data from external systems such as sensor devices and fitness monitors needs authorisations for access to the data and technical support from the device manufacturers or data integration services.
To understand the benefits and limitations of the user experience, EHDViz and other open-source or commercial solutions with similar capabilities must be compared.
Introduction
The subclinical features and symptoms vary for individual patients as diseases progress and can be affected by lifestyle and medical interventions. These variations deviate greatly even among people with similar demographics, clinical profiles, family history and disease burdens. The patient-specific intervention a physician incorporates into a treatment plan relies heavily on the course of the illness. Electronic health record (EHR) software is widely used to capture longitudinal data and record vital signs, medications, laboratory values, diagnostic reports, fluid inputs/outputs, mental states, patient transfers and other health status parameters. However, EHR software often presents data with tabular views or static text formats, which does not reveal the underlying trends in a patient's disease progression nor the similarities among patient trends within a given department. EHRs have limited capabilities to integrate biomedical, clinical and patient-generated data integrated, and physicians often have to use multiple tools to gather patient status from heterogeneous databases to get a complete health assessment.1 Decades of research have shown that graphical summaries of patient information provide faster and more accurate medical diagnoses, thus improving the healthcare quality.2–4 Reproducible studies demonstrate that practicing clinicians are unlikely to adopt any information retrieval task that takes longer than 30 s.5 6 As patients are becoming more empowered through an increase in patient-generated data, physicians are now being challenged to comprehensively visualise increasingly complex patient histories and associated data streams in a short span of time in the clinical setting. There is an unmet need in the continuum of healthcare delivery to develop better ways of visualising and interpreting EHR data, on which physicians can base critical treatment decisions. Electronic health data (EHD) includes the acquisition of physiological values, diagnostic reports (radiology reports), laboratory values, pathology reports (biopsy report), physician consults and clinically actionable genetic information.7–9 In recent years, this is further supplemented by information that patients provide directly, such as blood pressure and food logs, and by continuous physiologic data from wearable devices from patient portals10 or mobile phones.3 Some institutions have implemented remote monitoring of patients using implanted devices, such as the implantable cardioverter defibrillator, as well as augmented clinical management using data streams from health monitoring devices,11 leading to improved outcomes,12 13 cost savings12 14 and earlier identification of device malfunction.15 A subset of modern clinical trials are also incorporating remote monitoring devices, including ones capable of collecting physiological data and cloud computing electrophysiological data.16 As an example, several ongoing clinical trials make use of Apple ResearchKit for evaluating patients with asthma,17 cardiovascular disease, diabetes and Parkinson's disease (see http://researchkit.org/), and some efforts implement mHealth-based solutions to engage patients and visualise data (AppCore: https://github.com/researchkit/AppCore). This trend is growing and generating an influx of data that patients or physicians typically do not handle in the clinical setting. Better tools are now required to integrate such data streams and provide detailed summary of a patient for improving patient engagement. See Khader et al for an extensive discussion on real-time data streams in healthcare.
The expanding number of data streams that can integrate with EHR contributes to the increase in the volume of big data in healthcare, which lays the foundation for paradigm shifts in modern medicine. Furthermore, acceleration in massive data influx is expected with the wide adoption and maturity of Internet of Healthcare Things (IoHT; see https://en.wikipedia.org/wiki/Internet_of_Things), where health sensors, fitness monitors and implantables will be able to upload physiological data to patient-authorised and secure systems, which can provide a data-rich portrait of a patient at the point of care. Recent reviews on the application of big data to healthcare identify actionability and decentralised data18 as key challenges to fulfil the potential of big data in healthcare.19 20 The EHD of a patient can be aggregated and normalised using data from personal logs, health monitoring or fitness devices and medical devices (eg, continuous positive airway pressure pumps for sleep apnoea). To get a comprehensive picture of wellness or illness state of patients, the data from hospital administration and operations data can also be aggregated. Information retrieval systems that integrate multiple data streams have been developed or suggested for particular applications, including antibiotic clinical decision systems,3 recording events during surgical operations,21 diabetic patient data collection programs,22 text mining, concept extraction from clinical documentation using natural language processing19 and wearable devices using software application programming interfacing (API) services (eg, Human API; see: https://www.humanapi.co/) that allow communication between health monitoring devices and provider databases using a secure, programmatic data access protocol. As of yet, there is no system that permits flexible retrieval and interactive visualisation of EHR, medical and patient-generated data streams and provides tools for real-time visualisation.
Clinical data visualisation
Visual descriptions of the health status of patients in clinical settings have been a challenging problem since the introduction of computer programs for care management. At that time, the principal limitations were resource availability, including appropriate graphical engines for rendering, and specialised hardware to visualise patient status using computer programs.2 Decades later, visualisation of clinical information and communication of wellness trajectories or disease risk trajectories of a patient using visual cues remains an emerging challenge in the current era of data-driven medicine.23 24 Visualisation tools can track the biochemical variation and physiological status of patients, as well as quantify biomarkers. Such visualisation aids were originally part of particular medical devices designed to monitor one or more specific physiological variables, such as heart rate and pulse rate, both within and external to EHRs. Efficient tools, algorithms and risk prediction models are now required for visual communication of clinical information to manage the high volume of biomedical and healthcare data in the hospital setting. Integrating such visualisation tools with predictive models and risk estimation tools could support accelerated patient stratification for improved care.
Visualising healthcare data using clinical dashboards
Clinical dashboards are tools that can visually capture the cross-sectional view of a variety of quality metrics, including patient statuses, progress in cohort aggregations, patient safety and healthcare delivery measures, performance improvement for care providers and aid in understanding the key features of the overall patient satisfaction and improved outcomes. Clinical dashboards are often developed using commercial or custom-built tools internally developed by hospitals or health systems; thus, little to no interoperability with tools is available for statistical analysis, machine learning or integration with predictive modelling that can aid in tasks including acuity prediction and readmission evaluation. Designing visual tools to graphically explain risk scores and predictive models would help to accelerate patient risk stratification for improved care. While there are a variety of technical challenges for integrated visualisation of multiple clinical visualisation tools, the interoperability of EHR applications and data feeds from medical devices remains a significant challenge. Vendor standards also hinder the integration of diverse data elements into a common platform. Data feeds often need extensive quality control, normalisation or other preprocessing procedures before the utilisation in risk scoring engines. Visualisation also plays a crucial role in shared decision-making (SDM),25 26 where a care provider and patient participate in a discussion regarding therapeutic strategies or clinical care delivery pathways that are supported by a variety of tools including those with visualisation. SDM is currently used in the treatment and management of cardiovascular diseases,27 28 diabetes29–32 and osteoporosis.33 Irrespective of the medical specialty, visualisation improves the ability to understand trends in a patient's health and the effects of interventions over time.
Visualisation and forecast of risk estimations in clinical setting
Adverse events during hospitalisation such as hospital-acquired infections (HAIs) or hospital-acquired conditions (HACs) including falls, cardiac arrests and unanticipated intensive care unit (ICU) transfers, and death are frequently preceded by several useful and predictive features that can be used for accelerate triaging to improve the care delivery. For example, slow and progressive physiological decompensation was identified in cardiac arrests (79%), unexpected ICU transfers (55%) and death during hospitalisation (54%)34 in a retrospective study that compared cohorts from different countries. Failure to recognise and respond to signs of deterioration includes infrequent or incomplete vital sign assessments, poor design of vital sign charts and reduced accuracy of ‘track-and-trigger’ systems.35 Several single parameter and multiparameter risk scoring methods have been proposed to implement a ‘track-and-trigger’ method of alerting for patients in clinical wards within 24 h of an adverse event for accelerated clinical intervention aids. The most established methods are based on vital signs and neurologic status, including Modified Early Warning Score (MEWS), Standardized Early Warning System (SEWS) and National Early Warning Score (NEWS) that differ on the inclusion of oxygen saturation and supplemental oxygen and the weight of different features. When assessed retrospectively, these vital-based systems have an area under receiver operator curves (AUROCs) of 0.76–0.83 for cardiac arrests, 0.73–0.77 for ICU transfers and 0.87–0.88 for mortality, effective for triggering follow-up evaluation.36 Implementations of these warning systems have required that staff perform rounds and fill out paper sheets37 or electronically enter38 the vital signs. Reports of real-time EHR information retrieval-based implementations of early warning systems have had some success in reducing adverse events in randomised control trials39 40 and crossover trials,41 although the risk models have restrictions due to the limited physiological feature space. Algorithms developed using the entire set of discrete health characteristics in the clinical data warehouse have incorporated significantly predictive laboratory values, physician orders and medications. When assessed retrospectively, these features consistently outperform the vital-constrained approaches.42–44
In this paper, we propose the design, development and implementation of an extensible clinical dashboard development framework by leveraging open-source technologies. The EHD visualization (EHDViz) framework is an interactive and extensible framework implemented using a modern statistical computing language. Biomedical informatics scientists and solution architects can use EHDViz to develop clinical decision aids to empower patients. EHDViz provides an EHR-agnostic visualisation framework that can be implemented in real time to assist healthcare providers in identifying patients with decompensating physiology via a visual aid. Thus, healthcare delivery management teams, healthcare executives and medical professionals can use the dashboards developed using EHDViz to retrieve, integrate and explore diverse healthcare data streams to assess patient health trends in a clinical unit, hospital or health system.
Methods
Description of EHDViz framework
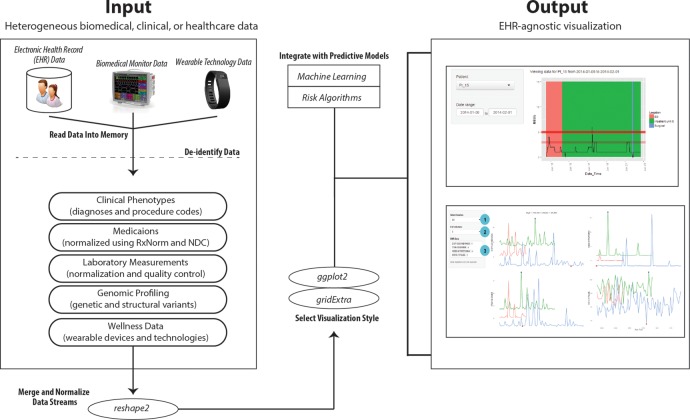
EHDViz is a software framework designed to interactively generate web-based healthcare data visualisation using various R packages (R language; R V.3.0.2; 2013-09-25). We provide an infographic of the client–server architecture of EHDViz in figure 1. We compiled various packages to organise a unified software framework for data input/output operations, data management of healthcare data, data cleaning and normalisation from diverse sources, generation of plots and statistical analysis. Data cleaning and quality control steps including the removal of outliers were performed using reshape2 package (https://cran.r-project.org/web/packages/reshape2/index.html). EHDViz uses the packages ggplot2 (http://ggplot2.org) and gridExtra (https://cran.r-project.org/web/packages/gridExtra/index.html) for developing plots. Native R plots can be generated and visualised using PDF viewers, generic image viewers and web browsers, but base R offers limited options for visualising real-time data streams. We developed a custom algorithm to combine individual R plots and visualise as a continuous, real-time data stream. We implemented the web server implementation using R/Shiny to deploy the plots created as part of EHDViz framework. We used the Shiny server architecture (https://github.com/rstudio/shiny-server) because it can be implemented over multiple desktop and server environments and can be distributed as suitable software modules. Data from wearable devices are compiled using the device-specific API for Fitbit. Wearable-specific APIs offer a secure way to collect and aggregate data generated by personal fitness monitoring devices. The package fitbitScraper (https://cran.r-project.org/web/packages/fitbitScraper/index.html) was used to extract the data from the wearable device.
Figure 1.
Client–server architecture of EHDViz. EHDViz, electronic healthcare data visualization.
Data handling in EHDViz
Various biomedical and healthcare data types, including disease and procedure indexes, clinical dictionaries and ontologies, namely the International Statistical Classification of Diseases and Related Health Problems (ICD-9: http://www.who.int/classifications/icd/en/) codes, are indexed in the current implementation to define specific disease terms pertaining to patients as part of diagnoses. Patients undergoing specific clinical procedures can also be retrieved and aggregated using Current Procedural Terminology (CPT: http://www.ama-assn.org/ama/pub/physician-resources/solutions-managing-your-practice/coding-billing-insurance/cpt/about-cpt.page) codes or Systematized Nomenclature of Medicine—Clinical Terms (SNOMED-CT) codes. EHDViz can also parse and normalise medication data using National Drug Codes (NDCs) and RxNorm and use medication data as part of the data aggregation methods in EHDViz (NDC: http://www.fda.gov/Drugs/InformationOnDrugs/ucm142438.htm; RxNorm: https://www.nlm.nih.gov/research/umls/rxnorm/). EHDViz can also handle data from operational and administrative datasets generated as part of healthcare delivery, including patient transfer data (ie, from the emergency department to surgery to ward and discharge), to query or aggregate patient cohorts in an adaptive fashion and to precisely visualise their health trends.
Input and output specifications of EHDViz
EHDViz can handle data in tab-delimited file format (.tsv) or comma-delimited file format (.csv). Data can also be extracted from various other formats and database using native R packages. For example, EHDViz can extract data from Excel files (xlsx: https://cran.r-project.org/web/packages/xlsx/index.html) or relational database systems that conform to Open Database Connectivity (ODBC) standards (RODBC: https://cran.r-project.org/web/packages/RODBC/index.html), Java Database Connectivity (JDBC) (RJDBC: https://cran.r-project.org/web/packages/RJDBC/index.html), MySQL (RMySQL: https://cran.r-project.org/web/packages/RMySQL/index.html) or modern, NoSQL database systems such as MongoDB (rmongodb: https://cran.r-project.org/web/packages/rmongodb/index.html). Various examples of input formats and sample files are provided at the URL: http://ehdviz.dudleylab.org/help.html#introduction. The data gathered from sources including EHRs, flat files, data warehouse or database connections will be used as the input for EHDViz dashboards and customised visualisation as output. The diverse set of data from various sources after parsing, quality control and normalisation can be integrated into the visualisation templates of an individual project. The output of EHDViz is in the format of customised visualisation dashboards rendered using a standard, modern web browser that supports HTML5 and responsive web development standards. The current version of EHDViz dashboards was successfully tested using modern browsers (Chrome Browser, Mozilla Firefox and Safari) on Windows, Linux or Mac operating systems.
Clinical dashboards developed using EHDViz
To evaluate the technical challenges in developing and deploying a real-time biomedical, clinical and patient-generated data visualisation dashboard, we created multiple prototype web applications using R language in the back end and the R/Shiny web server architecture in the front end as outlined above. Prototype dashboards are developed using three different datasets: (1) data from a single patient (n-of-1) with data streams not captured in a clinical setting demonstrate quantified-self visualisation, (2) simulated cohort of inpatients (n=445) and (3) simulated cohort of outpatients (n=14 221). The data simulation was performed using a deidentified EHR compiled at Icahn School of Medicine at Mount Sinai (ISMMS), a hospital of the Mount Sinai Health System in New York City. Data from fitness monitoring devices were aggregated using an API capable of secure retrieval of data from the fitness monitoring device of a user, and a custom web service function was designed to pull and integrate user-defined data features in real time. Dashboards discussed in this manuscript are implemented on a web server with Nginx (http://nginx.org/) on a secure, cloud-based virtual private server running on Ubuntu. The web interface is implemented using HTML, CSS and JavaScript, and visualisation dashboards are rendered using R/Shiny architecture.
Results
Availability: the source code of EHDViz and various clinical dashboard implementations are available from the URL: http://ehdviz.dudleylab.org/.
Clinical dashboards developed using EHDViz
Collaborative data visualisations, wellness trend predictors, risk estimation algorithms, proactive acuity status monitoring in a clinical setting and complex disease indicators are essential components of implementing data-driven precision medicine. In the following section, we discuss various dashboards developed using EHDViz. Briefly, we parsed the source data and removed the outliers as part of the data cleaning step. A custom web service function was designed to pull and integrate user-defined data features in real time from simulations of the clinical cohorts and fitness monitors using normalised data. The final dashboards were designed to show specific visualisations.
Dashboard 1: Visualising time series health data (quantified self)
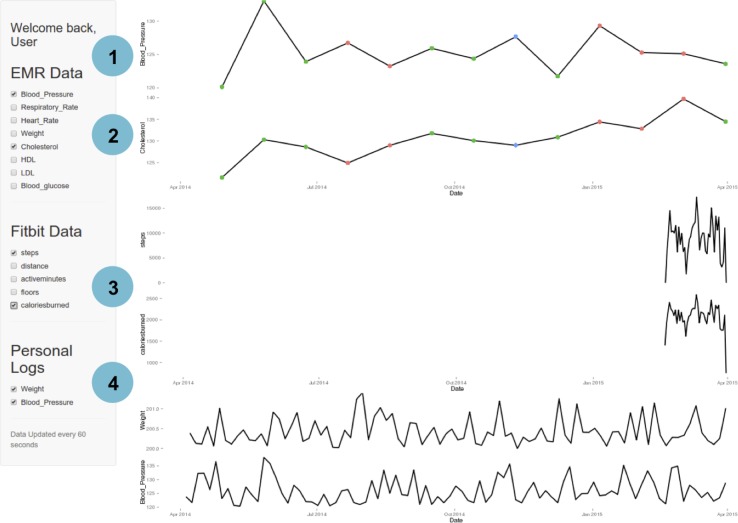
The quantified-self movement involves an increasing interest in individuals and patient communities in tracking many types of biometric data to gain insight into their health.45 Increasingly, patients are able to access and control their clinically collected health data.46 47 Our first demonstration addresses the challenge in quantified-self area of integrating and visualising time series health data from multiple data sources. The example in figure 2 demonstrates the integration of an individual patient's EHD sources. For this example, the patient has three primary sources of health data: (1) clinical data from outpatient visits, (2) continuous activity data from a wearable device (Fitbit, San Francisco, California, USA) and (3) a self-recorded blood pressure log. The clinical data from ambulatory visits were simulated by randomly sampling aggregated physiologic and lab values from 14 221 patients in an ISMMS outpatient cohort. The continuous activity data were scraped from one of the author's (MAB) wearable devices using the API at an interval of 15 min. The blood pressure log is simulated as weekly measurements from normal distributions N(13 015) and N(8510).The user interface features a main panel with ‘sparklines’ for each health feature and a sidebar with widgets for the user to select the health features of interest. In this example, a checkbox is provided to group patients for each data source: (1) EHR, (2) data from fitness monitoring device and (3) personal log. The user can select any combination of health features to be displayed. The main panel displays a stack of sparklines with selected health features sorted according to values selected in the sidebar. Minimums and maximums are highlighted with red and blue dots, respectively. In this application, the data source that updates most frequently was from the wearable device collected at 15 min intervals; the application was programmed to autorefresh every 15 min to retrieve new data.
Figure 2.
A quantified-self, healthcare data visualisation dashboard developed using EHDViz. Different features of the dashboard are highlighted as (1) user management, (2) dynamic selection, (3) integration with data streams and (4) integration with manual data input. EHDViz, electronic healthcare data visualization.
Dashboard 2: Visual analytics of data streams in clinical setting
Next, we demonstrate the retrieval of continuous data contained in a collection of patient's EHRs during an inpatient stay, where data will be much more dynamic than in the previous outpatient example. This implementation was tested with a simulated cohort of 445 inpatients with clinical labs recorded throughout their encounter and with simulated data (figures 3A–D and 4).
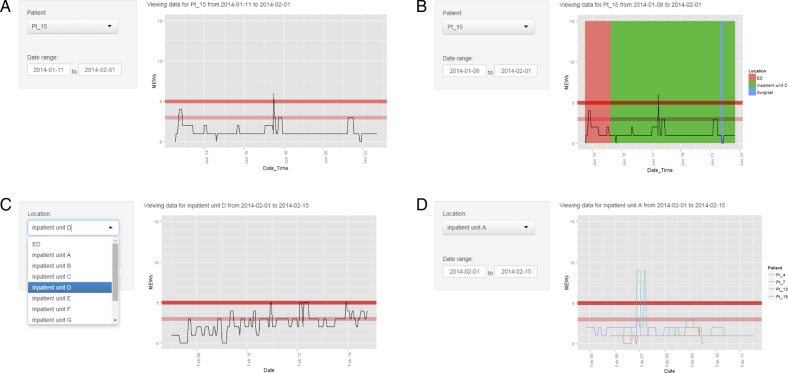
Figure 3.
Different scenarios of implementing a visual aid for MEWS using EHDViz framework. (A) Visualisation of a single patient; (B) visualisation of a single patient layered on patient admission, discharge and transfer data; (C) visualisation of trends of MEWS in different inpatient units; (D) visualisation of multiple patients in a same unit. EHDViz, electronic healthcare data visualization; MEWS, Modified Early Warning Score.
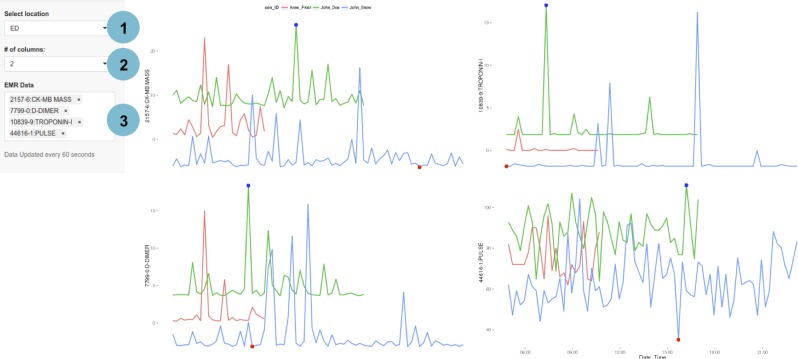
Figure 4.
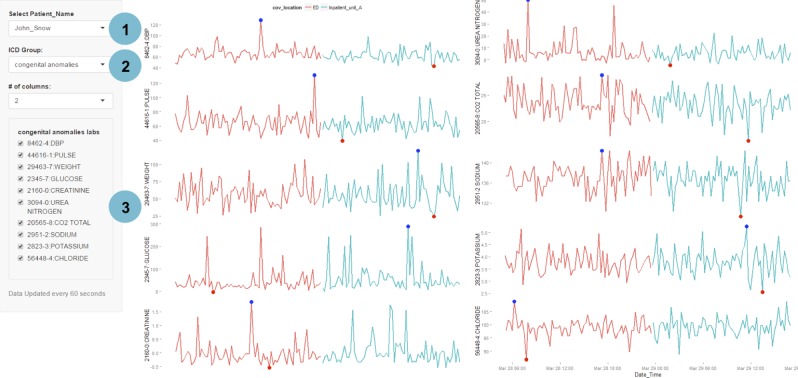
A customised, clinical evaluation dashboard developed using EHDViz that illustrates data in emergency department. Features of this dashboard include selection of specific clinical units using a drop-down menu, controlling for the layout and selecting patients that are tested for specific biomarkers. Different features of the dashboard are highlighted as (1) selection of individuals, (2) options to control visual layouts and (3) integration with ICD-9 codes. EHDViz, electronic healthcare data visualization; ICD-9, International Classification of Diseases, Ninth Revision.
User of this particular dashboard can use the sidebar to select a patient and the date range of interest. The relevant information is then retrieved from the EHR or data warehouse throughout the encounter (figure 3A–D). Within a single hospital visit, a patient could go through different hospital units including the emergency department, ICUs, inpatient units, surgical suite or ambulatory wards depending on the clinical status of the patient. In this example, patient transfers including admission, transfers and discharge were colour coded by location to intuitively show the dynamic trends in the health status (figure 3B). For the simulated data, we randomly retrieved data from the EHR for an age-matched and gender-matched cohort with 14 221 patients to populate each of the 375 continuous health features contained in the EHR.
For each of the 7000 unique diagnoses, we pooled corresponding patient data and found the most frequently measured health features for each ICD-9 class. The simulated patient dashboard (figure 4) allows the user to select a patient and an ICD-9 class from the drop-down menus in the side panel, which then populates the main panel with the most common health features measured for that ICD-9 class. The list of health features corresponding to the selected ICD-9 class is additionally displayed as a checkbox group in the side panel, so the user can further refine the displayed feature set. This enables the user to rapidly retrieve and assess trends in the most relevant biomarkers. We also provide a demonstration at http://ehdviz.dudleylab.org/providers/full that allows a keyword-based search and multiselection of all 375 health features to make customised dashboards. Real-time displays were also designed from the simulated data, demonstrated at http://ehdviz.dudleylab.org/providers/real-time.
Dashboard 3: High-velocity patient acuity status monitoring and data visualisation in the clinical setting
The examples in figures 3C and D and 5 demonstrate the use of EHDViz for developing visual aids for patient safety and cohort analysis. These dashboards provide risk estimation visualisation for users to track all patients simultaneously in a unit, which facilitates the identification of atypical and destabilising features to trigger interventions. Patient vital signs were retrieved from the EHR warehouse from 445 inpatients and processed to calculate the MEWS. Figure 3C and D shows the dashboard for monitoring these patients’ MEWS and shows the clinical stability trends. The user can select the clinical unit of interest with the drop-down menu, and sparklines with MEWS are displayed for each patient in the unit with alert-triggering thresholds displayed for reference. When there are multiple patients in the unit, MEWS are coloured by patient (figure 3D). Data for online demonstrations were simulated as discussed in scenario 2 and the ‘location’ and ‘patient’ covariates were switched from a data-colouring covariate to a user-filtering covariate and vice versa for use in a cohort application. As shown in figure 5, a user can select the clinical unit of interest and text search different clinical parameters, and the main panel will display the values of these features for all the patients in the selected unit, coloured by patient. This design allows rapid evaluation of various clinical features or predictors. Multiple values relevant to clinical manifestations of patient population can be compiled and new scores (eg, MEWS) can be computed for a population of patients. Demonstrations of ICD-9-class-based feature selection are provided at the URL: http://ehdviz.dudleylab.org/visualizations/Population_Management_ICD9/, and a real-time monitoring dashboard implemented using EHDViz is provided at the URL: http://ehdviz.dudleylab.org/visualizations/Population_Management_RealTime/.
Figure 5.
A population health management visualisation dashboard implemented using EHDViz. Different features of the dashboard are highlighted as (1) visualisation of data from floor using admission, discharge transfer data, (2) dynamic control of visualisation and (3) real-time user interaction. EHDViz, electronic healthcare data visualization.
Discussion
The treatment pathway for a patient depends on a number of factors that can be collected from different sources including patient-generated data, medications, vital signs, diagnoses and responses to therapies or other interventions. Physicians can collect data from the EHRs, patient health records, patient portals, electronic patient diaries, fitness trackers and the patient's recollections of medical history. In most presentations, however, these data overwhelm physicians instead of guiding them to informed decision-making. Real-time clinical monitoring and automated alerting provide better tools to improve patient safety, clinical outcomes and quality of healthcare delivery. Tools are currently available to monitor patient acuity, infectious diseases and adverse events. Specifically, there are customised tools that target specific needs of the clinical unit including operating rooms or ICUs. Developing a unified visualisation tool that can provide an overview of a patient by integrating different healthcare, biomedical or clinical data streams remains an open challenge. EHDViz, an open-source data visualisation framework capable of real-time data visualisation, can be used to address many of these issues. EHDViz aims to unify heterogeneous biomedical and healthcare data integration through R language, a popular and preferred programming language for scientific computing, predictive analytics and machine learning. R language is typically used for desktop or client cluster-based visualisation models. Here, we have improvised an R visualisation package designed to generate static plots and rendered it as a real-time data visualisation engine. Real-time displays can also be implemented and deployed over the web browsers using other programming languages including Python and JavaScript, and future releases of EHDViz could extend to these languages. Close integration with R also enables visual analytics and predictive modelling using the large library of R packages to run seamlessly within EHDViz. Users can customise the different levels of implementation of EHDViz dashboards for disease-specific, division-specific or institutional-specific applications. EHDViz offers features to integrate risk prediction algorithms for patient stratification with data mining algorithms to use underlying data repositories to refine the user experience and automatically retrieve the most relevant data for a selected context. Integrating various risk assessment algorithms with the traditional clinical dashboard style interface offers a powerful toolkit for clinicians. EHDViz could aid in designing dashboard development projects that combine visualisation, analytics and predictive modelling in healthcare and wellcare.
Application of EHDViz in simulation-based medical education
Simulation-based learning is at the core of the pedagogical principles of modern medicine. Medical students, residents and physicians extensively use EHR at the bedside during care delivery. EHDViz is an EHR and vendor-agnostic dashboard development toolkit that users can leverage as a teaching aid capable of generating custom EHR instances and visualisations. Simulated EHR systems can be designed based on single-use cases to evaluate an individual patient or number of patients that a resident is managing on a floor or unit.
Comparison with related healthcare data visualisation applications
Multiple visualisation tools are currently available for effective integration of actionable information in the workflow of clinical care pathways. A systematic review of data visualisation tools assessed multiple clinical data visualisation tools: tools such as EventFlow,48 LifeLines,49 LifeLines2,50 VISualization of Time-Oriented RecordS (VISITORS)51 and Dynamics Icon (DICON)52 were listed as tools capable of clinical data visualisation and dashboard development. Deng and Denecke53 used a tag cloud from radiology reports, pathology reports and surgical reports to summarise unstructured patient data. Data visualisation tools, such as HARVEST,24 offer web-based infrastructure for integrating, discovering and reporting data but are restricted to the data captured in a data warehouse. The design philosophy of EHDViz is to provide a tool that can integrate and visualise data from different sources in addition to data warehouses. LifeLines and LifeLines2 offer options to align, rank and summarise temporal visualisations. LifeFlow,54 a tool based on LifeLines and LifeLines2, is capable of visualising care-related events, including patient transfers. The focus of LifeFlow is temporal clinical event visualisation and implemented in Java and is deployed as stand-alone software. Thus, integration of different healthcare delivery or operational data is a challenge for LifeFlow. EHDViz, on the other hand, offers various options for customised visualisation and integration with a large library of predictive or statistical learning algorithms available as part of R language. CrowdED55 is another visualisation aid that is specific to the specific clinical locations; the tool can be used for data visualisation in the emergency departments but offers very limited extensibility. An objective comparison of user experiences, usability parameters and utilities by implementing various applications in same healthcare or clinical setting would provide quantitative estimates of the preference of data sources and user interface. Several of the existing healthcare data visualisation tools, however, are designed to address a single task and lack extensibility. EHDViz addresses this important challenge by leveraging widely used, scalable technologies to create clinical data visualisation dashboards to aid care providers.
Conclusions
Owing to the implementation of the Affordable Care Act (http://www.hhs.gov/healthcare/about-the-law/index.html) and the emerging trend of hospitals to rebuilding healthcare operations as affordable care organization (ACO), there is a growing need for health information technology (healthIT) solutions to be more agile and sustainable across different levels of hospitals and health systems. The need for delivering high-quality care by leveraging biomedical and healthcare data calls for the appropriation of healthITs capable of handling and managing healthcare big data. Open-source technologies offer a complementary option for healthIT developers to design, develop and deploy cost-effective clinical dashboards with no cost for the software licence and reuse. Adoption of these technologies (automated phenotyping, visual analytics and predictive modeling) may thus reduce overall healthcare spending. We developed EHDViz to integrate data from diverse sources including biomedical and healthcare data visualisation for integrated health assessment. Further, EHDViz could also play a significant role as a toolkit to emulate EHR environment to improve simulation-based learning. Hospitals and healthcare systems are emerging as learning health systems, and as such, data capture, smart clinical dashboards and adaptive visual analytics could play an integral role in managing the patient population. We envisage that design and development of real-time patient status assessment tools coupled with risk estimation using heterogeneous data could enhance the quality of healthcare delivery and improve patient outcomes.
Acknowledgments
The authors would like to thank the following members of Mount Sinai Health System—Hospital Big Data initiative: Dr Adel Bassily-Marcus, Dr Bruce Darrow, Dr Alan Moskowitz, Dr Annetine Gelijns, Dr Patricia Kovatch and Kash Patel. KS and JTD would like to thank Drs Carol Horowitz and Janice Gabrilove for useful discussion about the applications of EHDViz in the areas of patient engagement and simulation-based learning. The authors would also like to thank Harris Center for Precision Wellness (http://precisionwellness.org/) and Icahn Institute for Genomics and Multiscale Biology (http://icahn.mssm.edu/research/genomics) for infrastructure support.
Footnotes
Twitter: Follow Marcus A Badgeley at @MABadge, Khader Shameer at @kshameer, Benjamin Glicksberg at @BenGlicksberg, Patrick McCormick at @patrickmdnet, and Joel T Dudley at @jdudley
Contributors: MAB, KS, BSG and MST contributed to the data integration, software package implementation, clinical dashboards and web server development. KS, MAL, PJM, AK, DLR and JTD formulated visualisation strategy and designed illustrative examples. DLR and JTD contributed to the overall planning of the project, the development of an extensible software package for clinical dashboard development and the manuscript. All authors have contributed to the writing and compilation of the final manuscript. All authors approved their contributions and the final draft of the manuscript.
Funding: This work was supported by a grant from the National Institutes of Health, National Center for Advancing Translational Sciences (NCATS), Clinical and Translational Science Awards (UL1TR001433-01) to KS and JTD.
Competing interests: AK has received consulting fees from Mars, Inc. JTD has received consulting fees or honoraria from Janssen Pharmaceutica, GSK, AstraZeneca and Hoffmann-La Roche and holds equity in NuMedii, Inc, Ayasdi, Inc and Ontomics, Inc. No writing assistance was used in the production of this manuscript.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Source code and sample data are available on the companion website: http://ehdviz.dudleylab.org
References
- 1.Shameer K, Badgeley MA, Miotto R et al. . Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams. Brief Bioinform 2016;pii:bbv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torsvik T, Lillebo B, Mikkelsen G. Presentation of clinical laboratory results: an experimental comparison of four visualization techniques. J Am Med Inform Assoc 2013;20:325–31. 10.1136/amiajnl-2012-001147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powsner SM, Tufte ER. Graphical summary of patient status. Lancet 1994;344:386–9. http://www.ncbi.nlm.nih.gov/pubmed/7914312 (accessed 12 Sep 2014). [DOI] [PubMed] [Google Scholar]
- 4.Forsman J, Anani N, Eghdam A et al. . Integrated information visualization to support decision making for use of antibiotics in intensive care: design and usability evaluation. Inform Health Soc Care 2013;38:330–53. 10.3109/17538157.2013.812649 [DOI] [PubMed] [Google Scholar]
- 5.Merry P. Healthcare information. Slow to learn. Health Serv J 1997;107:28–9. http://www.ncbi.nlm.nih.gov/pubmed/10169554 (accessed 19 Mar 2015). [PubMed] [Google Scholar]
- 6.Sackett DL. Finding and applying evidence during clinical rounds: the “evidence cart”. JAMA 1998;280:1336–8. 10.1001/jama.280.15.1336 [DOI] [PubMed] [Google Scholar]
- 7.Kullo IJ, Jouni H, Olson JE et al. . Design of a randomized controlled trial of disclosing genomic risk of coronary heart disease: the Myocardial Infarction Genes (MI-GENES) study. BMC Med Genomics 2015;8:51 10.1186/s12920-015-0122-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012;13:395–405. 10.1038/nrg3208 [DOI] [PubMed] [Google Scholar]
- 9.Royston G, Hagar C, Long LA et al. . Mobile health-care information for all: a global challenge. Lancet Glob Health 2015;3:e356–7. 10.1016/S2214-109X(15)00054-6 [DOI] [PubMed] [Google Scholar]
- 10.Kruse CS, Bolton K, Freriks G. The effect of patient portals on quality outcomes and its implications to meaningful use: a systematic review. J Med Internet Res 2015;17:e44 10.2196/jmir.3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van der Velde ET, Foeken H, Witteman TA et al. . Integration of data from remote monitoring systems and programmers into the hospital electronic health record system based on international standards. Neth Heart J 2012;20:66–70. 10.1007/s12471-011-0234-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Khatib SM, Piccini JP, Knight D et al. . Remote monitoring of implantable cardioverter defibrillators versus quarterly device interrogations in clinic: results from a randomized pilot clinical trial. J Cardiovasc Electrophysiol 2010;21:545–50. 10.1111/j.1540-8167.2009.01659.x [DOI] [PubMed] [Google Scholar]
- 13.Raatikainen MJP, Uusimaa P, van Ginneken MME et al. . Remote monitoring of implantable cardioverter defibrillator patients: a safe, time-saving, and cost-effective means for follow-up. Europace 2008;10:1145–51. 10.1093/europace/eun203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bikou O, Licka M, Kathoefer S et al. . Cost savings and safety of ICD remote control by telephone: a prospective, observational study. J Telemed Telecare 2010;16:403–8. 10.1258/jtt.2010.090810 [DOI] [PubMed] [Google Scholar]
- 15.Guédon-Moreau L, Chevalier P, Marquié C et al. . Contributions of remote monitoring to the follow-up of implantable cardioverter-defibrillator leads under advisory. Eur Heart J 2010;31:2246–52. 10.1093/eurheartj/ehq203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahoo SS, Jayapandian C, Garg G et al. . Heart beats in the cloud: distributed analysis of electrophysiological “Big Data” using cloud computing for epilepsy clinical research. J Am Med Inform Assoc 2014;21:263–71. 10.1136/amiajnl-2013-002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asthma Mobile Health Study. http://apps.icahn.mssm.edu/asthma/ (accessed 1 Jan 2015).
- 18.Schneeweiss S. Learning from big health care data. N Engl J Med 2014;370:2161–3. 10.1056/NEJMp1401111 [DOI] [PubMed] [Google Scholar]
- 19.Dhar V. Big data and predictive analytics in health care. 2014;2:113–6. 10.1089/big.2014.1525 [DOI] [PubMed] [Google Scholar]
- 20.Thornton J. What you need to know to make the most of big data in biology. Lancet 2015;385(Suppl 1):S5–6. 10.1016/S0140-6736(15)60321-X [DOI] [PubMed] [Google Scholar]
- 21.Agarwal S, Joshi A, Finin T et al. . A pervasive computing system for the operating room of the future. Mob Networks Appl 2007;12:215–28. 10.1007/s11036-007-0010-8 [DOI] [Google Scholar]
- 22.Benhamou PY. Improving diabetes management with electronic health records and patients' health records. Diabetes Metab 2011;37(Suppl 4):S53–6. 10.1016/S1262-3636(11)70966-1 [DOI] [PubMed] [Google Scholar]
- 23.Farri O, Rahman A, Monsen KA et al. . Impact of a prototype visualization tool for new information in EHR clinical documents. Appl Clin Inform 2012;3:404–18. 10.4338/ACI-2012-05-RA-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poh C-L, Kitney RI, Shrestha RBK. Addressing the future of clinical information systems—Web-based multilayer visualization. IEEE Trans Inf Technol Biomed 2007;11:127–40. 10.1109/TITB.2006.875680 [DOI] [PubMed] [Google Scholar]
- 25.Pennington JW, Ruth B, Italia MJ et al. . Harvest: an open platform for developing web-based biomedical data discovery and reporting applications. J Am Med Inform Assoc 2014;21:379–83. 10.1136/amiajnl-2013-001825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montori VM, Breslin M, Maleska M et al. . Creating a conversation: Insights from the development of a decision aid. PLoS Med 2007;4:1303–7. 10.1371/journal.pmed.0040233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coylewright M, Blumenthal RS, Post W. Placing COURAGE in context: review of the recent literature on managing stable coronary artery disease. Mayo Clin Proc 2008;83:799–805. 10.4065/83.7.799 [DOI] [PubMed] [Google Scholar]
- 28.Nannenga MR, Montori VM, Weymiller AJ et al. . A treatment decision aid May increase patient trust in the diabetes specialist. The Statin Choice randomized trial. Health Expect 2009;12: 38–44. 10.1111/j.1369-7625.2008.00521.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah ND, Mullan RJ, Breslin M et al. . Translating comparative effectiveness into practice: the case of diabetes medications. Med Care. 2010;48(6 Suppl):S153–8. 10.1097/MLR.0b013e3181d5956c [DOI] [PubMed] [Google Scholar]
- 30.Mullan RJ, Montori VM, Shah ND et al. . The diabetes mellitus medication choice decision aid: a randomized trial. Arch Intern Med 2009;169:1560–8. 10.1001/archinternmed.2009.293 [DOI] [PubMed] [Google Scholar]
- 31.Breslin M, Mullan RJ, Montori VM. The design of a decision aid about diabetes medications for use during the consultation with patients with type 2 diabetes. Patient Educ Couns 2008;73:465–72. 10.1016/j.pec.2008.07.024 [DOI] [PubMed] [Google Scholar]
- 32.LeBlanc A, Ruud KL, Branda ME et al. . The impact of decision aids to enhance shared decision making for diabetes (the DAD study): protocol of a cluster randomized trial. BMC Health Serv Res 2012;12:130 10.1186/1472-6963-12-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montori VM, Shah ND, Pencille LJ et al. . Use of a decision aid to improve treatment decisions in osteoporosis: the osteoporosis choice randomized trial. Am J Med 2011;124:549–56. 10.1016/j.amjmed.2011.01.013 [DOI] [PubMed] [Google Scholar]
- 34.Kause J, Smith G, Prytherch D et al. . A comparison of antecedents to cardiac arrests, deaths and emergency intensive care admissions in Australia and New Zealand, and the United Kingdom—the ACADEMIA study. Resuscitation. 2004;62:275–82. 10.1016/j.resuscitation.2004.05.016 [DOI] [PubMed] [Google Scholar]
- 35.Deakin CD, Nolan JP, Soar J et al. . European Resuscitation Council Guidelines for Resuscitation 2010 Section 4. Adult advanced life support. Resuscitation 2010;81:1305–52. 10.1016/j.resuscitation.2010.08.017 [DOI] [PubMed] [Google Scholar]
- 36.Churpek MM, Yuen TC, Edelson DP. Risk stratification of hospitalized patients on the wards. Chest 2013;143:1758–65. 10.1378/chest.12-1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subbe CP. Validation of a modified Early Warning Score in medical admissions. QJM 2001;94:521–6. 10.1093/qjmed/94.10.521 [DOI] [PubMed] [Google Scholar]
- 38.Prytherch DR, Smith GB, Schmidt PE et al. . ViEWS—Towards a national early warning score for detecting adult inpatient deterioration. Resuscitation 2010;81:932–7. 10.1016/j.resuscitation.2010.04.014 [DOI] [PubMed] [Google Scholar]
- 39.Kollef MH, Chen Y, Heard K et al. . A randomized trial of real-time automated clinical deterioration alerts sent to a rapid response team. J Hosp Med 2014;9:424–9. 10.1002/jhm.2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey TC, Chen Y, Mao Y et al. . A trial of a real-time alert for clinical deterioration in patients hospitalized on general medical wards. J Hosp Med 2013;8:236–42. 10.1002/jhm.2009 [DOI] [PubMed] [Google Scholar]
- 41.Evans RS, Kuttler KG, Simpson KJ et al. . Automated detection of physiologic deterioration in hospitalized patients. J Am Med Informatics Assoc 2015;22:350–60. 10.1136/amiajnl-2014-002816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Churpek MM, Yuen TC, Winslow C et al. . Multicenter development and validation of a risk stratification tool for ward patients. Am J Respir Crit Care Med 2014;190:649–55. 10.1164/rccm.201406-1022OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alvarez CA, Clark CA, Zhang S et al. . Predicting out of intensive care unit cardiopulmonary arrest or death using electronic medical record data. BMC Med Inform Decis Mak 2013;13:28 10.1186/1472-6947-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escobar GJ, LaGuardia JC, Turk BJ et al. . Early detection of impending physiologic deterioration among patients who are not in intensive care: development of predictive models using data from an automated electronic medical record. J Hosp Med 2012;7:388–95. 10.1002/jhm.1929 [DOI] [PubMed] [Google Scholar]
- 45.Swan M. The quantified self: fundamental disruption in big data science and biological discovery. Big Data 2012;1 http://online.liebertpub.com/doi/pdfplus/10.1089/big.2012.0002 [DOI] [PubMed] [Google Scholar]
- 46.Serrato CA, Retecki S, Schmidt DE. MyChart-A New Mode of Care Delivery: 2005 Personal Health Link Research Report. Perm J 2007;11:14–20. 10.7812/TPP/07-002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinbrook R. Personally controlled online health data—the next big thing in medical care? N Engl J Med 2008;358: 1653–6. 10.1056/NEJMp0801736 [DOI] [PubMed] [Google Scholar]
- 48.Monroe M, Rongjian L, Hanseung L et al. . Temporal Event Sequence Simplification. IEEE Trans Vis Comput Graph 2013;19:2227–36. 10.1109/TVCG.2013.200 [DOI] [PubMed] [Google Scholar]
- 49.Plaisant C, Mushlin R, Snyder A et al. . LifeLines: using visualization to enhance navigation and analysis of patient records. Proc AMIA Symp 1998:76–80. [PMC free article] [PubMed] [Google Scholar]
- 50.Wang TD, Plaisant C, Shneiderman B et al. . Temporal summaries: supporting temporal categorical searching, aggregation and comparison. IEEE Trans Vis Comput Graph 2009;15:1049–56. 10.1109/TVCG.2009.187 [DOI] [PubMed] [Google Scholar]
- 51.Klimov D, Shahar Y. A framework for intelligent visualization of multiple time-oriented medical records. AMIA Annu Symp Proc 2005:405–9. [PMC free article] [PubMed] [Google Scholar]
- 52.Gotz D, Sun J, Cao N et al. . Visual cluster analysis in support of clinical decision intelligence. AMIA Annu Symp Proc 2011;2011:481–90. [PMC free article] [PubMed] [Google Scholar]
- 53.Deng Y, Denecke K. Visualizing unstructured patient data for assessing diagnostic and therapeutic history. Stud Health Technol Inform. 2014;205:1158–62. 10.3233/978-1-61499-432-9-1158 [DOI] [PubMed] [Google Scholar]
- 54.Wongsuphasawat K, JAG G, #243 et al. . LifeFlow: visualizing an overview of event sequences. Proceedings of the SIGCHI Conference on Human Factors in Computing Systems Vancouver, BC, Canada: ACM, 2011:1747–56. [Google Scholar]
- 55.Greci LS, Parshalle CE, Calvitti A et al. . CrowdED: crowding metrics and data visualization in the emergency department. J Public Health Manag Pract 2011;17:E20–8. 10.1097/PHH.0b013e3181e8b0e9 [DOI] [PubMed] [Google Scholar]