Abstract
Introduction:
Telemicrobiology is a growing component of clinical microbiology informatics. However, few studies have been performed to assess the diagnostic utility of telemicroscopy systems in evaluating infectious agents.
Objective:
Evaluate multiple contemporary digital pathology platforms for use in diagnostic telemicrobiology.
Materials and Methods:
A mix of thirty cases that included viral, bacterial, fungal, and parasitological findings were evaluated by four experts using ×40 whole slide imaging (WSI) scans, ×83 oil-immersion WSI scans, ×100 oil-immersion WSI scans, digital photomicrographs, and glass slides.
Results:
The ×83 WSI, ×100 WSI, and photomicrograph interpretations were not significantly different in quality and accuracy when compared to glass slide interpretations. The ×40 WSI interpretations were of lower quality and were more likely to be incorrect when compared to glass slide interpretations.
Conclusions:
In this study, high magnification, oil-immersion digital pathology platforms are better suited to support telemicrobiology applications and yield interpretations on par with glass slide evaluations.
Keywords: Digital pathology, infectious diseases, microbiology, telemicroscopy, telemicrobiology, whole slide imaging
INTRODUCTION
Telepathology is a growing component of the practice of pathology and laboratory medicine, which includes telemicrobiology.[1,2,3] Telemicrobiology is an important and growing component of clinical microbiology informatics.[3,4] Telemicrobiology has been incorporated into routine use by some microbiology core laboratories that supports satellite laboratories. Telemicrobiology has also been used for seeking expert consultation, such as the consultation services established by the Centers for Disease Control and Prevention's (CDC) Division of Parasitic Diseases and Malaria's service for diagnostic assistance (DPDx).[3] However, to the best of our knowledge, no published studies have evaluated the diagnostic utility of multiple contemporary digital pathology systems for use in telemicrobiology. In clinical practice, detecting and identifying microscopic organisms using glass slides is often performed using a ×100 oil-immersion objective lens. However, commercial whole slide imaging (WSI) systems used in digital pathology typically scan at lower magnifications and without oil immersion such as ×20, ×40, or sometimes ×60. The diagnostic utility of lower magnification WSI for the identification of microorganisms has been questioned.[5,6]
We evaluated different WSI platforms using ×40 dry scanning, ×83 oil-immersion, and ×100 oil-immersion to determine their diagnostic utility specifically for telemicrobiology. The accuracy of these WSI digital slides was compared to glass slides and manually captured ×100 oil-immersion photomicrographs, which have been more extensively used in telemicrobiology practice.[1,2,3,7,8]
MATERIALS AND METHODS
Specimen Selection
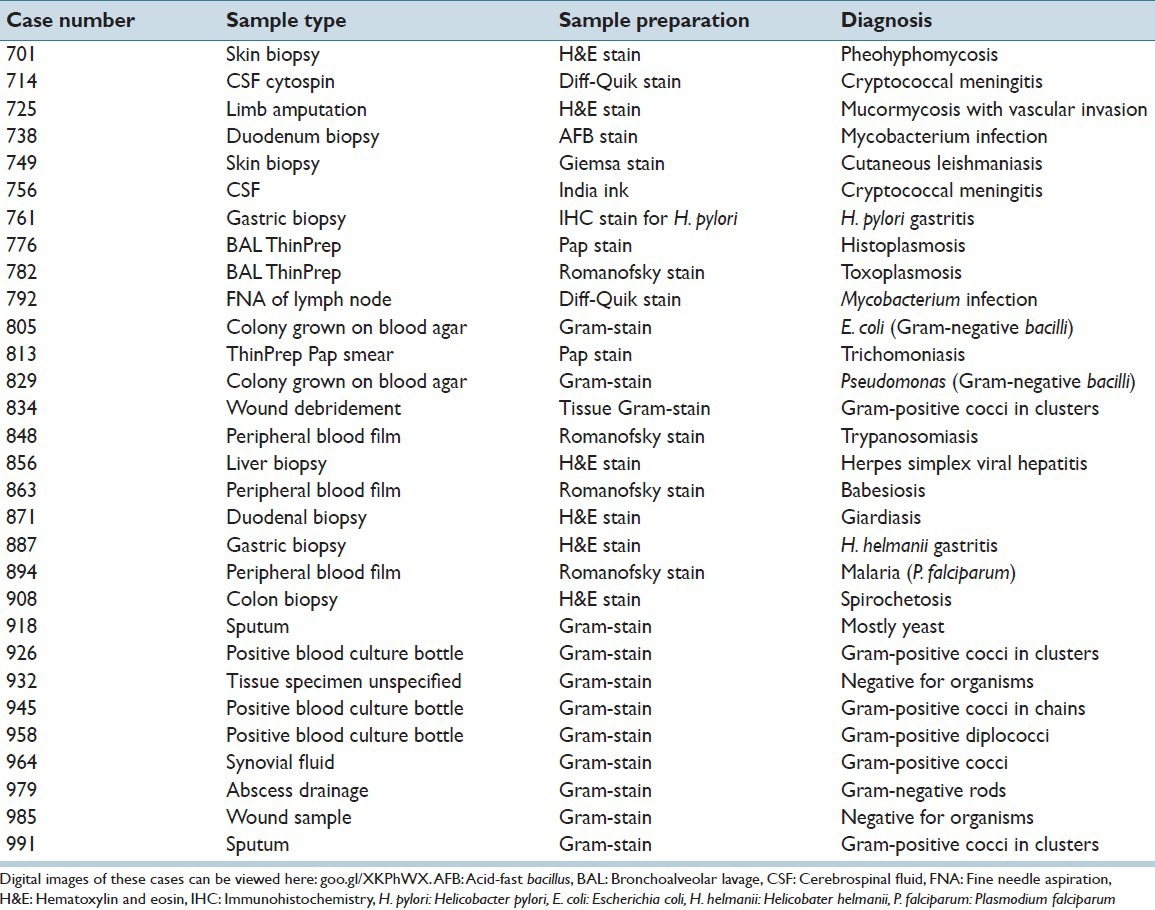
Two authors (DR, LP) reviewed previously characterized glass slides containing microbiology-relevant specimens, which correlated with known diagnoses. These authors selected a variety of specimen types, specimen preparations, and organism types in an attempt to broadly test the digital pathology systems’ telemicrobiology capabilities. In total, 30 cases [Table 1] with one glass slide per case were selected. Glass slides were 1.0 mm thick and had permanently mounted #1 or #1.5 glass coverslips. Diagnostically relevant regions on each slide were circled (approximately 5–10 mm in diameter) using a dotting marker pen, and digital images of these circled regions were generated using various imaging modalities. Slides were de-identified before being sent for scanning and review.
Table 1.
Description of specimens used in this study and their correct diagnoses
Digital Image Acquisition
Static images
Photomicrographs of diagnostically-relevant fields on the glass slides were captured using an Olympus BX46 light microscope (Center Valley, Pennsylvania, USA) with a mounted Olympus DP73 digital camera. Two jpeg images of each case were captured: One using the ×10 objective lens and one using the ×100 objective lens. The ×10 lens used was UPlanApo ×10/0.40 infinity/0.17, and the ×100 oil-immersion lens was UPlanSApo ×100/1.40 infinity/0.17/FN26.5. Images were captured using cellSens Standard 1.11 software (Center Valley, Pennsylvania, USA) (Olympus) with the following settings: Automatic exposure, 4800 × 3600 (pixel shift) resolution, and natural camera contrast. No postprocessing software was used on the digital images except to convert the jpegs to ScanScope virtual slide (.svs) files for online evaluation. No substantial loss in image quality was identified due to the conversion. For the ×100 photos each jpeg file was 1.4 ± 0.6 Mb in size, and each case took about 4.5 min to locate a diagnostically appropriate field and photograph.
Whole Slide Images
Prior to scanning, glass slides were cleaned. Pen markings indicating the region of interest on the glass slide to be scanned were not removed, albeit that such markings may sometimes cause digital image artifacts.[9] No artifacts were identified in the digital slides used for this study.
Slides were scanned at ×40 using a single z-plane via an Aperio XT whole slide scanner (Leica Biosystems, USA). Scanned images were evaluated for quality and to ensure that they were in focus. Digital files in (.svs) format were stored on an image server for remote evaluation using the WebScope viewer from Aperio. Each. svs file was 817 ± 242 Mb in size. Acquisition time for each slide averaged 1 min.
Slides were scanned at ×83 with oil-immersion using a single z-plane via an Aperio SC-O whole slide scanner (Leica Biosystems, USA). The svs files were uploaded to the cloud and then downloaded to the image server. Each svs file was 455 ± 297 Mb in size. Acquisition time for each slide was about 1 min; and total time spent setting up, scanning, and reviewing each scan for quality was about 8 min per slide.
Slides were scanned at ×100 with oil-immersion using a Zeiss Axio Imager M1 with an AxioCam MRc camera (Carl Zeiss; Gottingen, Germany); a Zeiss Alpha Plan-Apochromat ×100, 1.46 NA objective lens (Carl Zeiss; Gottingen, Germany); and AxioVision Rel 4.8 software (Carl Zeiss; Munich, Germany). The camera's white balance and exposure were manually adjusted for each slide, and shade correction was employed to minimize illumination variation between fields. Multiple z-planes were acquired for each field and merged into an extended depth of field image (EDF). The final single-plane composite image was exported as a jpeg with a compression quality factor of 100 before being converted to a svs file. Each file was 966 ± 137 Mb in size. Image acquisition averaged about 45 min per slide, including roughly 30 min for capture and then 10–15 min for processing (i.e., stitching, EDF merging, etc.).
Image Sharing
All de-identified digital image files were securely stored on IBM SONAS (IBM, Armonk, New York) storage where they could be remotely viewed by the evaluators.
Evaluator Selection and Training
Four evaluators with varied daily exposure to microbiology and an interest in digital pathology were chosen to participate in the study. Their backgrounds were as follows: cytopathologist, gastrointestinal pathologist, clinical microbiologist, and general pathologist with microbiology expertise. Two of the evaluators were experienced using digital pathology, and two evaluators were novices in digital pathology. All of the evaluators underwent training before the study, so they were proficient in navigating the digital images.
Slide Review
The four evaluators each evaluated the 30 cases using all five modalities: ×40 WSI, ×83 WSI, ×100 WSI, ×10 and ×100 digital photomicrographs, and glass slides. For this study, the photomicrograph modality includes both ×10 and ×100 static images, and this set were both available for evaluation. Each case was evaluated for the type of infection present (i.e., no pathogen, virus, bacterium, fungus, or parasite), and a free text interpretation could be entered into the data capture sheet to further describe microscopic findings (e.g., Giardia, Helicobacter, Gram-positive cocci in clusters, etc.). Evaluators also assessed the confidence in their interpretations and identified technical limitations for each case using an Excel (Microsoft; Redmond, CA, USA) data collection sheet. The electronic data collection sheet automatically populated case information (e.g., stain used and specimen type) based on the case number selected by the reviewer. The sheet also fostered standardized reporting and facilitated data compilation. A minimum 2 weeks washout period was employed between evaluations of the different imaging modalities. The evaluators were all from the same institution, but located at different hospitals. They used different monitors to review these images including a 24-inch HP ZR24w, 21.5-inch HP ProDisplay P221, and 22-inch Samsung SyncMaster 2243BWX display.
Data Analysis
The diagnostic interpretation for each case was compared to the expected diagnosis [Table 1], and the evaluators’ interpretations were converted into quality scores. A quality score of 4 indicated a correct interpretation with correct microorganism identification (e.g., “Cryptococcus sp.” or “Plasmodium falciparum”). A quality score of three indicated one of the following: A correct diagnosis with some degree of uncertainty, a differential diagnosis that included the correct diagnosis, or a diagnosis that lacked specific microorganism details (e.g., “Yeast, possibly Cryptococcus sp.” or “malaria”). A quality score of two indicated a correct diagnosis that was vague or lacked microorganism specifics (e.g., “Yeast” or “parasites in erythrocytes”). A quality score of 1 indicated the correct type of infection (e.g., fungus) but incorrect microorganism specifics (e.g., Cryptococcus interpreted as Candida or P. falciparum interpreted as Babesia sp.). A quality score of zero included false positives, false negatives, and grossly misidentified organisms (e.g., yeast interpreted as bacteria). Quality scores of 0 or 1 were both considered incorrect interpretations. Quality scores of 2, 3, or 4 were all considered correct interpretations; however, higher scores (i.e. 4 > 3 > 2) were considered better interpretations. The quality of intraevaluator interpretations was compared using this scoring system. A “best interpretation” score classification is evaluator and case specific. Each evaluator's best interpretation for each case was identified by comparing the interpretation scores for each of the five imaging modalities. In the five interpretations for each evaluator for each case, the highest correct score (i.e. 2, 3, or 4) achieved by an evaluator was deemed the “best interpretation.” Multiple imaging modalities could be classified as a “best interpretation” for an evaluator's case if each interpretation achieved the evaluator's maximum score for the case. Cases in which the evaluator failed to achieve a correct interpretation using all five modalities, and cases in which the evaluator deferred interpretation were not considered in the analyses.
Statistical analyses were performed using two-tailed Student's t-tests in Microsoft Excel 2010 (Redmond, Washington, USA). A P value of 0.05 was chosen as a cut-off, and values below this cut-off were defined as statistically significant.
RESULTS
Diagnostic Accuracy
Glass slide interpretations were better than ×40 WSI interpretations (P = 0.04) [Figure 1] in that the intraobserver quality scores were greater for glass slide interpretations than for ×40 WSI interpretations. The ×83 WSI interpretations, photomicrograph interpretations, and ×100 WSI interpretations trended toward better interpretations than ×40 WSI (P values were not significant). Correct interpretations were achieved using all modalities, and statistically significant differences between platforms were not identified. However, glass slide interpretations, photomicrograph interpretations, and ×83 WSI interpretations trended toward achieving significantly more frequent correct interpretations than ×40 WSI (P values were not significant). Incorrect interpretations were obtained more frequently using ×40 WSI than using glass slides, photomicrographs, or ×83 WSI (P < 0.01, P = 0.01, and P = 0.03, respectively). The modality that yielded the best interpretations [Figure 2], the highest rate of correct interpretations, and the highest rate of incorrect interpretations [Figure 3] varied between evaluators.
Figure 1.
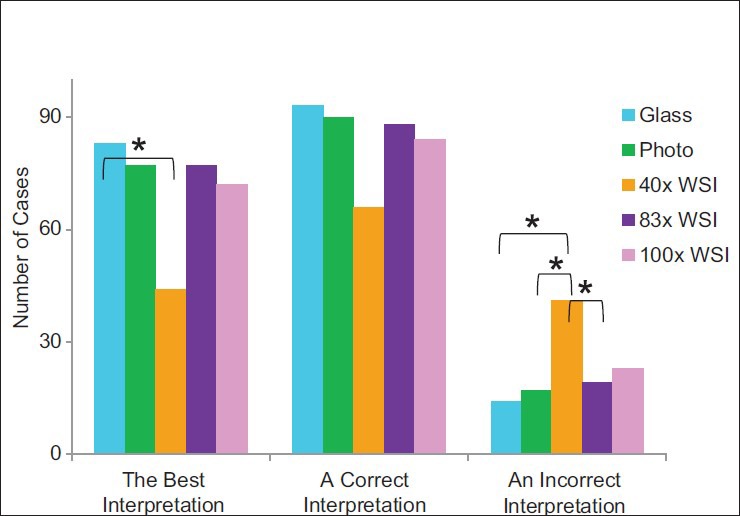
Comparison of interpretation quality by imaging modality. Asterisks designate groups with P < 0.05. The photo modality includes both ×10 and ×100 photomicrographs. The best interpretation is the greatest correct quality score (i.e., 2, 3, or 4) achieved by an evaluator for a case across all modalities. An incorrect interpretation includes quality scores of 0 and 1
Figure 2.
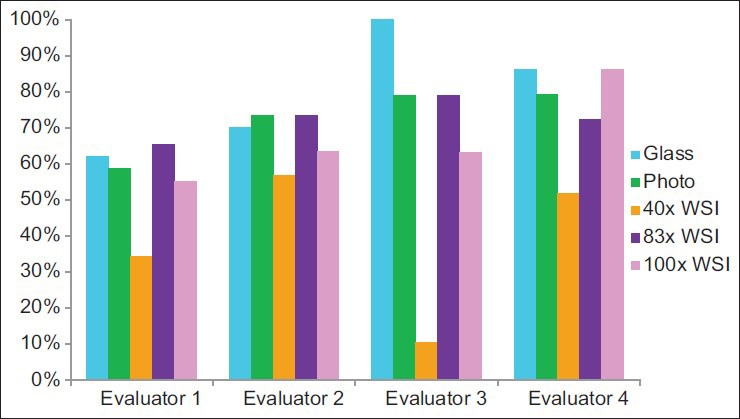
Frequency of achieving the best interpretation
Figure 3.
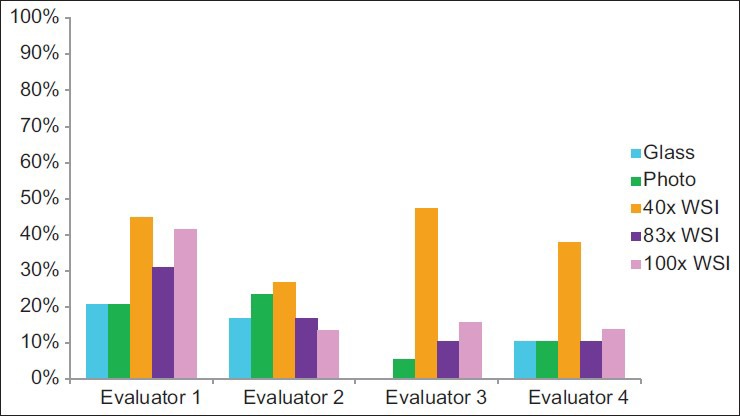
Frequency of achieving an incorrect interpretation
Perceived Limitations
In 13 instances, cases were not considered in the analyses. Eight of these cases were due to one evaluator's self-reported lack of expertise with specimens’ sources and/or stains. In five instances, cases were not considered because the evaluator did not achieve a correct diagnosis using any of the five modalities. In total, 137 (91%) evaluations (of a potential 150 case evaluations) were considered in the analyses. When considering only the three evaluators that did not defer the interpretation of any of the cases, these three individuals perceived resolution of the digital image to be inadequate more often with the ×40 WSI and ×100 WSI than in the photomicrographs [Figure 4]. However, the ×83 WSI results were almost as good as those for the photomicrographs. They also perceived image focus to be inadequate most often with the ×40 WSI cases and least often with the ×83 WSI cases [Figure 5].
Figure 4.
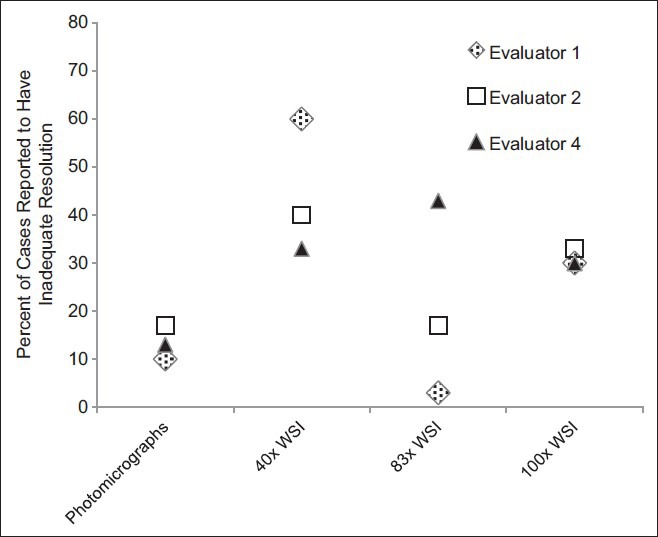
Resolution perceived as a limitation. Three of the four evaluators evaluated the resolution adequacy for every case using all digital modalities, and a summary of their interpretations is depicted
Figure 5.
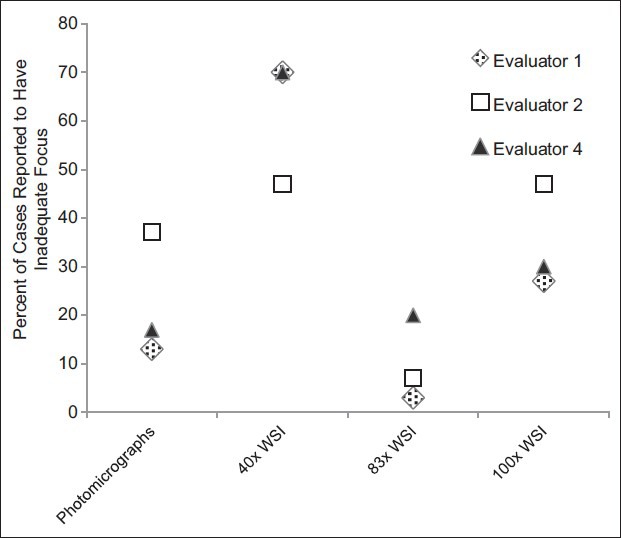
Focus perceived as a limitation. Three of the four evaluators evaluated the focus adequacy for every case using all digital modalities, and a summary of their interpretations is depicted
Technical Issues
Only minor technical challenges were encountered, and these occurred mainly with image acquisition. Scanning of one ×40 WSI case failed because the scanner could not automatically find the correct focal plane [Figure 6]. This required the operator to manually intervene and tune the focus before image acquisition. The ×100 WSI process involved custom development. The system used to generate ×100 WSI files posed difficulty correctly stitching tiles containing large amounts of feature-poor space [Figure 7]. In addition, because the ×100 WSI system collected multiple z-planes of each field that were later stitched together to include the sharpest (most in focus) frames, this required working with upward of 75 Gb of data at times, which resulted in software instability. In one instance, this instability caused the system to crash and corrupted the solid state drive resulting in loss of previously captured scans. When viewing converted static photomicrographs, the magnification slider using the Aperio image viewer appeared to be mislabeled [Figure 8], and one of the evaluators found this to be confusing.
Figure 6.
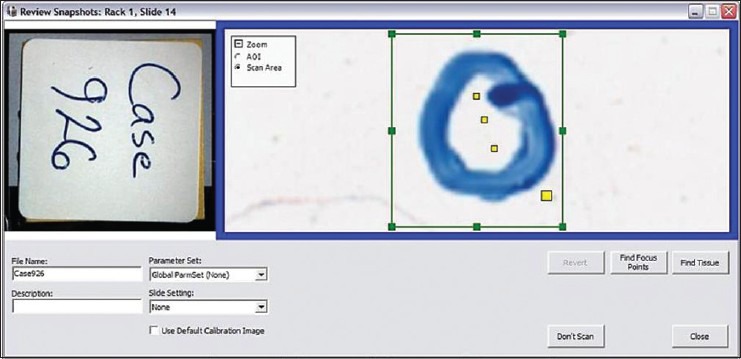
Out of focus scan. One case with Gram-positive cocci failed to automatically focus correctly when performing a ×40 whole slide imaging scan
Figure 7.
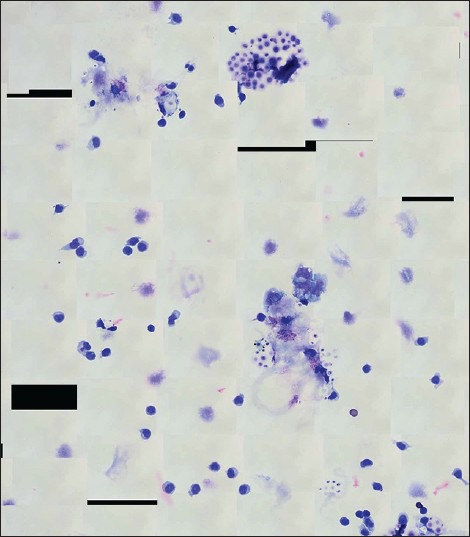
Custom developed ×100 whole slide image overview for a case with cerebrospinal fluid Cryptococcus. Note the obvious stitching of feature-poor areas (i.e. white space), which resulted in black polygon artifacts in the ×100 whole slide image scans
Figure 8.
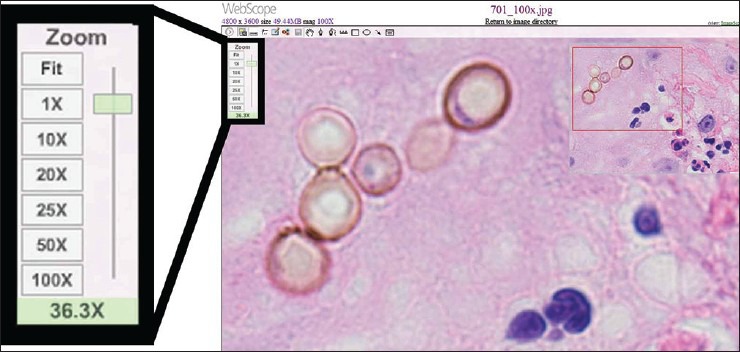
Photomicrograph using a whole slide image viewer. The photomicrographs were captured as jpegs and converted to ScanScope virtual slide files so that they could be viewed online with WebScope (Aperio; Vista, California, USA) software. Note that the magnification of the dematiaceous fungi depicted here is underestimated by the zoom indicator on the left sliding bar. Integrating components of multiple digital pathology systems can present unanticipated interoperability challenges, such as this
DISCUSSION
The results of this study are concordant with previous studies,[1,2,10] which have demonstrated that diagnoses made from static digital photomicrographs (snapshots) are on par with glass slide interpretations. In telemicrobiology, two main variables are commonly limiting factors in making an accurate and precise diagnosis in difficult cases. One variable is simply capturing images that adequately encompass the diagnostically relevant area(s), and the second variable is obtaining an adequate resolution for the interpreter to resolve the salient features of the microorganism(s) and/or the pathological features. Photomicrographs can be used successfully to perform telemicrobiology if the photographer captures diagnostically relevant images that are in focus, but this caveat is its major limitation in a clinical application.[3] It has been noted that the photographer may not capture adequate images to diagnose all of the infecting organisms (e.g. stool that is positive for multiple parasites or blood containing multiple Plasmodium spp.) or may select only images that support the photographer's suspected diagnosis. Capturing a sufficient amount of unbiased and diagnostically relevant area is most easily accomplished by capturing a large area of the slide and/or many representative areas, which WSI is designed to do. Unfortunately for telemicrobiology, many of today's WSI systems capture images with a resolution that is too low for interpreters to achieve a high-quality microbiology diagnosis with high confidence. Capturing images with adequate resolution is typically accomplished with image acquisition at a higher magnification. However, optical resolution is more important than digital magnification (i.e., zooming). For WSI, image resolution depends on a combination of the objective lens (both magnification and numerical aperture) used, the digital camera's sensor inside the instrument, and the computer monitor display used to view the final digital image. It is plausible that the different displays used in our study may have contributed to varying appearances of microorganisms when viewed by the participants. An important advantage of WSI over photomicrographs is that the individual generating the WSI scan does not need to have any microbiology acumen in order to consistently capture diagnostic images. While some studies have reported that some infections (e.g., bacterial vaginosis, fungi, Trichomonas vaginalis) can be identified with WSI,[11] other investigators have shown that microscopic organisms cannot be easily visualized when using WSI with ×20 scans.[5] The current study demonstrates that high magnification and oil-immersion WSI, which can effectively address both of the common limiting factors, has a better diagnostic performance with telemicrobiology than ×40 WSI. Utilization of oil immersion WSI is relatively new to the field of digital pathology, and one study has shown similar great potential for this technology for the interpretation of hematopathology slides.[12] In the current study, we used two oil-immersion WSI systems: An in-house developed ×100 WSI platform and the Aperio ×83 WSI system. The ×100 WSI system would need more development before being used clinically, but the ×83 WSI is currently available commercially. Since it is important to speciate microorganisms such as malaria, for identifying blood parasites laboratories may want to rely on instruments such as CellaVision that perform more standardized digital imaging with oil-immersion on peripheral blood smears.
The field of digital pathology would benefit from additional studies evaluating additional imaging modalities for telemicrobiology. The use of multiple z-planes (dynamic WSI) was not employed in this study. It is likely that ×40 WSI using multiple z-planes would provide diagnostic capabilities superior to single plane ×40 WSI and possibly on par with glass slide interpretations.[6] Some researchers have suggested that in order to reproduce clear focusing of microorganisms such as Helicobacter pylori with virtual microscopy, it may be necessary to construct three-dimensional (3D) virtual slides composed of WSIs with different focuses.[6] However, at present, the use of virtual 3D slides is not practical for routine clinical practice. It is likely that that the greater the burden of Helicobacter organisms present the easier it will be to identify them. However, even though the cases we selected for this study had ample microorganisms they still could not be detected with low-resolution images. In addition, remotely controlled live video high-dry telemicroscopy that is capable of displaying multiple z-planes (e.g., Zeiss or formerly Trestle, Sakura VisionTek M6), static/video hybrid microscopy (e.g., ViewsIQ Panoptiq),[13] and live video human-to-human telemicroscopy would all be potentially useful alternate platforms for telemicrobiology that warrant further study. The CDC Division of Parasitic Diseases and Malaria's telemicrobiology service (called DPDx) has extensive experience successfully using photomicrographs in its telemicrobiology service and is beginning to explore the use of live video human-to-human telemicroscopy for handling teleconsults.[7]
In the present “proof-of-concept” study, there were some limitations related to testing the various imaging modalities by means of subjective human interpretations. The evaluators were sometimes asked to interpret specimens outside their routine scope of work. In an attempt to avoid any effect this may have had on the quality of their interpretations, cases where this was an issue were deferred by the interpreter and not considered in the analyses. It is possible that the number of deferred cases would have been reduced if all of the evaluators routinely encountered the spectrum of cases covered in the study set as part of their daily practice. The microbiology expertise of the individuals in this limited study was not revealed in order to keep their identity anonymous. In some instances, minor intraevaluator interpretation variability was noted. For example, an evaluator that reported “probably mycobacteria” 1 month later reported simply “mycobacteria” for the same case. The former evaluation is less confident and was thus scored as a 3 instead of a 4. The scoring criteria were not communicated in detail to the evaluators before beginning the study, which may have helped to minimize variances in their choice of words. This study also has the potential for confounding due to recall bias. Although the recommended 2-week washout period was used, it is possible that recall bias may have influenced some interpretations.[14] The field of clinical microbiology and infectious diseases pathology is broad, and this study only covered some representative examples of microbial pathology and morphology. Hopefully, this study will serve as a resource for those seeking to implement clinical telemicrobiology or perform more in-depth studies about telemicrobiology.
In summary, single-plane ×40 WSI was inferior to glass slide interpretation for the purpose of telemicrobiology. However, evaluators achieved interpretations similar to glass slide interpretations when using high magnification digital pathology systems including ×83 WSI with oil-immersion, ×100 WSI with oil-immersion, and ×100 static digital photomicrographs. More extensive evaluations of suitable imaging modalities and applications in microbiology need to be performed to validate digital imaging systems for their use in microbiology.
Financial Support and Sponsorship
Nil.
Conflicts of Interest
Dr. Liron Pantanowitz is a consultant for Omnyx, LLC, Digital Pathology. Dr. Mohamed Salama serves on the medical advisory board to Leica.
Acknowledgments
The authors thank Dr. Grant Bullock for generously offering the use of his microscope and camera for this study.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2016/7/1/10/177687
REFERENCES
- 1.McLaughlin WJ, Schifman RB, Ryan KJ, Manriquez GM, Bhattacharyya AK, Dunn BE, et al. Telemicrobiology: Feasibility study. Telemed J. 1998;4:11–7. doi: 10.1089/tmj.1.1998.4.11. [DOI] [PubMed] [Google Scholar]
- 2.Scheid P, Lam DM, Thömmes A, Zöller L. Telemicrobiology: A novel telemedicine capability for mission support in the field of infectious medicine. Telemed J E Health. 2007;13:108–17. doi: 10.1089/tmj.2007.0043. [DOI] [PubMed] [Google Scholar]
- 3.Rhoads DD, Mathison BA, Bishop HS, da Silva AJ, Pantanowitz L. Review of telemicrobiology. Am J Clin Pathol. 2015 doi: 10.5858/arpa.2015-0116-RA. Available form: http://www.archivesofpathology.org/doi/pdf/10.5858/arpa.2015-0116-RA . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhoads DD, Sintchenko V, Rauch CA, Pantanowitz L. Clinical microbiology informatics. Clin Microbiol Rev. 2014;27:1025–47. doi: 10.1128/CMR.00049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell WS, Lele SM, West WW, Lazenby AJ, Smith LM, Hinrichs SH. Concordance between whole-slide imaging and light microscopy for routine surgical pathology. Hum Pathol. 2012;43:1739–44. doi: 10.1016/j.humpath.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 6.Kalinski T, Zwönitzer R, Sel S, Evert M, Guenther T, Hofmann H, et al. Virtual 3D microscopy using multiplane whole slide images in diagnostic pathology. Am J Clin Pathol. 2008;130:259–64. doi: 10.1309/QAM22Y85QCV5JM47. [DOI] [PubMed] [Google Scholar]
- 7.Mathison B. Pathology Informatics Summit. Pittsburgh, PA: 2015. May 7, Telediagnosis in the Identification of Parasites of Public Health Concern - The DPDx Perspective. Oral Presentation. [Google Scholar]
- 8.Mathison BA. Atlanta, GA: 2014. Aug 24-27, What Can the CDC Do for You? Telediagnosis and Molecular Technologies for Diagnosing Parasitic Diseases. 114th ASM General Meeting. [Google Scholar]
- 9.Collins RA, Parwani AV, Khalbuss WE, Duboy J, Pantanowitz L. Effect of pen marking on glass slides for whole slide image scanning in abstracts: Pathology informatics 2012. J Pathol Inform. 2012;3:S3. [Google Scholar]
- 10.Mathison BA, Bishop H, Johnston S, Xayavong M, Arguin P, da Silva AJ. Atlanta, GA: 2010. Nov 3-7, Trends in Malaria Diagnosis: Combining the Use of Telediagnosis, Microscopy and PCR in the Identification of Plasmodium Spp. 59th American Society of Tropical Medicine and Hygiene Annual Meeting. [Google Scholar]
- 11.Tawfik O, Davis M, Dillon S, Tawfik L, Diaz FJ, Fan F. Whole slide imaging of pap cell block preparations versus liquid-based thin-layer cervical cytology: A comparative study evaluating the detection of organisms and nonneoplastic findings. Acta Cytol. 2014;58:388–97. doi: 10.1159/000365046. [DOI] [PubMed] [Google Scholar]
- 12.Chen ZW, Kohan J, Perkins SL, Hussong JW, Salama ME. Web-based oil immersion whole slide imaging increases efficiency and clinical team satisfaction in hematopathology tumor board. J Pathol Inform. 2014;5:41. doi: 10.4103/2153-3539.143336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhoads DD, Ahmed I, Pantanowitz L. Feasibility of using the panoptiq imaging system for telemicrobiology in abstract: Pathology informatics 2015. J Pathol Inform. 2015;6:S51–2. [Google Scholar]
- 14.Pantanowitz L, Sinard JH, Henricks WH, Fatheree LA, Carter AB, Contis L, et al. Validating whole slide imaging for diagnostic purposes in pathology: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Am J Clin Pathol. 2013;137:1710–22. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]


