Abstract
The ETS transcription factor complex GABP consists of the GABPα protein, containing an ETS DNA binding domain, and an unrelated GABPβ protein, containing a transactivation domain and nuclear localization signal. GABP has been shown in vitro to regulate the expression of nuclear genes involved in mitochondrial respiration and neuromuscular signaling. We investigated the in vivo function of GABP by generating a null mutation in the murine Gabpα gene. Embryos homozygous for the null Gabpα allele die prior to implantation, consistent with the broad expression of Gabpα throughout embryogenesis and in embryonic stem cells. Gabpα+/− mice demonstrated no detectable phenotype and unaltered protein levels in the panel of tissues examined. This indicates that Gabpα protein levels are tightly regulated to protect cells from the effects of loss of Gabp complex function. These results show that Gabpα function is essential and is not compensated for by other ETS transcription factors in the mouse, and they are consistent with a specific requirement for Gabp expression for the maintenance of target genes involved in essential mitochondrial cellular functions during early cleavage events of the embryo.
The E26 transformation-specific (ETS) DNA binding domain was first discovered as part of a fusion protein product of the replication-defective E26 avian erythroblastosis virus that can lead to acute leukemia (4, 26, 27). Since then, ETS genes have been identified in species as diverse as Drosophila melanogaster (23), Caenorhabditis elegans (14), Xenopus laevis (25), and metazoa, but not plants, fungi, and protozoa (13). The number of genes containing the highly conserved DNA binding domain that defines the ETS family has been amplified during evolution, with 1 gene (Lin1) present in the earthworm and more than 30 in mammals (23), suggesting divergence of gene function.
GABPα is an ETS protein that, together with an unrelated GABPβ subunit, forms the functional GABP transcription factor complex, also known as nuclear respiratory factor 2 (NRF-2) or adenovirus E4 transcription factor 1 (E4TF-1). GABPα is the mammalian orthologue of the D. melanogaster protein D-Elg, which has been shown by mutation studies to be involved in egg chamber patterning and development during oogenesis (10). The mammalian GABP complex, together with an unrelated transcription factor, NRF-1, has been shown in vitro to regulate the expression of many genes necessary for cellular respiration in mitochondria (30). These include genes encoding cytochrome c oxidase subunits IV and Vb (39), VIA1 (41), VIIAL (31), VIIC (32), and XVII (35) and mitochondrial transcription factor A (MTFA) (38), the principal transcription factor in mitochondrial gene expression. In addition, GABP is a proposed regulator of ribosomal proteins L27A, L30, and S16 (11); the cell cycle and cell survival regulators retinoblastoma protein (33) and Fas ligand (24); neuromuscular junction proteins such as utrophin (18) and nicotinic acetylcholine receptor subunits δ and ɛ (9, 19); the hematopoietic protein thrombopoietin (17) and coagulation factor IX (5); interleukin 2 (2) and interleukin 16 (3); and long terminal repeats of human immunodeficiency virus (37) and mouse mammary tumor virus (1).
In order to establish the in vivo role of GABP, we have targeted the Gabpα gene in mice. The results establish that Gabpα is essential for early embryogenesis in mammals, and its loss of function results in a preimplantation lethal phenotype. Gabpα protein levels in tissues of Gabpα heterozygous mice are not significantly different from those in wild-type mice, consistent with a tight means of regulation of Gabpα expression. The early death of mice null for Gabpα protein expression is presumed to be due to a simultaneous decrease in the expression of MTFA and of other Gabp target genes, since a high rate of mitochondrial transcription is known to occur during cleavage of the embryo (22), and a similar peri-implantation lethal phenotype is observed in mouse embryos lacking the other key regulator of nucleus-encoded mitochondrial protein expression, NRF-1 (15).
MATERIALS AND METHODS
Determination of Gabpα gene structure.
Sequence in the 5′ region of Gabpα (including exons 1 to 5) was obtained by screening a λfixII murine 129/SvJ genomic DNA library (Stratagene) with an NcoI-BamHI restriction digestion product spanning bp 413 to 678 of the Gabpα cDNA (GenBank identity accession no. [gi] M74515). Sequence of Gabpα introns 5 and 6 was obtained by screening a set of 129/SvJ mouse bacterial artificial chromosome (BAC) filters (BACPAC) with a 977-bp SacII-SpeI genomic fragment within intron 3 (spanning bp 12695 to 13672 of gi:27960443). Genomic sequence spanning exons 7 to 10 was derived by PCR analysis of 129/SvJ embryonic stem (ES) cell DNA.
Intron-exon boundaries of mouse Gabpα were amplified by using the Elongase (Invitrogen) polymerase mix on a λ phage, ES cell, or BAC genomic DNA template. Primer positions, given in reference to gi:193382 (with annealing temperatures in parentheses), were as follows: for I1, bp 413 to 432 (5′) and 484 to 503 (3′) (60°C); for I2, bp 512 to 531 (5′) and 538 to 557 (3′) (55°C); for I3, bp 556 to 574 (5′) and 682 to 700 (3′) (55°C); for I4, bp 722 to 739 (5′) and 793 to 810 (3′) (49°C); for I5, bp 813 to 830 (5′) and 1084 to 1101 (3′) (53°C); for I6, bp 1016 to 1033 (5′) and 1210 to 1227 (3′) (58°C); for I7, bp 1219 to 1236 (5′) and 1364 to 1381 (3′) (52°C); for I8, bp 1330 to 1347 (5′) and 1447 to 1464 (3′) (56°C); and for I9, bp 1441 to 1460 (5′) and 1688 to 1707 (3′) (57°C). PCR products were gel purified by using QiaexII beads (QIAGEN), subcloned into the pGEM-T (Promega) vector, sequenced by using ABI PRISM BigDye Terminator chemistry (version 3.4.1), and analyzed on an ABI 377 DNA sequencer (Perkin-Elmer Applied Biosystems). Sequence analysis and alignment were performed using Sequencher (version 3) software.
Generation of a Gabpα targeting vector and gene-targeted mice.
The first coding exon of Gabpα (exon 2) was targeted via insertion of an IRES-lacZ-neomycin cassette (described in reference 7) into a NotI site immediately downstream of the start codon. The resulting construct, spanning the area from an XbaI site upstream of exon 1 to a HindIII site downstream of exon 2 (bp 3766 to 9610 of gi:27960443), was linearized and electroporated into 129/SvJ ES cells, as previously described (20). A targeting frequency of 7% was obtained. Correctly targeted ES cell clones were then injected into the blastocysts of C57BL/6 females to yield chimeric pups that were bred to wild-type 129/SvJ mice to generate Gabpα heterozygotes. Two correctly targeted ES cell lines were independently injected and generated chimeric pups which were bred to generate heterozygotes. Because there was no discernible difference between the lines, only one of these was used in this study. Mice were housed in windowless rooms with controlled temperature (22 ± 2°C) and a 12-h light and dark cycle (8:00 am to 8:00 pm). All experimentation involving mice was performed with approval of the appropriate ethics committee within Monash University, in accordance with Australian federal regulations.
Genotyping of targeted ES cells.
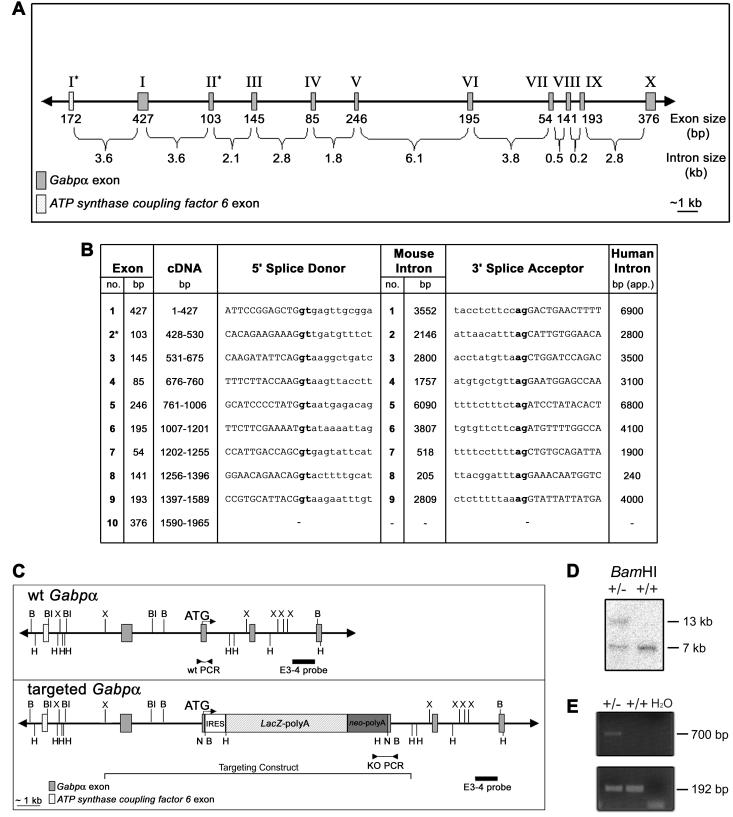
Genomic DNA was extracted from ES cells by lysis in a solution containing 200 mM NaCl, 100 mM Tris-HCl (pH 8.0), 5 mM EDTA (pH 8.0), 0.2% (wt/vol) sodium dodecyl sulfate, and 100 μg of proteinase K (Roche)/ml for 4 h at 55°C, followed by isopropanol precipitation. Targeting events were identified by Southern blot analysis as previously described (16), where 10 μg of genomic DNA was digested with BamHI and hybridized with a 977-bp SacII-SpeI fragment (E3-4) external to the targeting construct, within intron 3 (spanning bp 12695 to 13672 of gi:27960443), detecting a 7-kb wild-type band and a 13-kb targeted band.
The ATP synthase coupling factor 6 gene is adjacent to the 5′ end of Gabpα, running in the opposite direction (see gi:27960443; ATP synthase coupling factor 6 exon I is at positions 685 to 855, and Gabpα exon I is at positions 4474 to 4878). Genomic PCR spanning bp 3544 to 6970 and 3544 to 8627 of gi:27960443 was performed to examine for genomic disruption due to the targeting event.
Genotyping of embryos and adult mice.
Genomic DNA was extracted from mouse tail segments (taken at weaning, at 21 to 28 days after birth) or whole embryos by lysis in a solution containing 1% sodium dodecyl sulfate, 0.1 M NaCl, 0.1 M EDTA (pH 8.0), 0.05 M Tris-HCl (pH 8.0), and 0.5 mg of proteinase K (Roche)/ml at 55°C overnight, following by phenol-chloroform extraction and ethanol precipitation. PCR was performed with 250 ng of genomic DNA as a template to amplify a 192-bp wild-type product with oligonucleotides F3 (5′-CTTCCAGGACTGAACTTTTGAACG-3′) and B3 (5′-AAAAACAAGCACACTGGCCTACTC-3′) and a 700-bp targeted product with oligonucleotides F8 (5′-TCTCCTGTCATCTCACCTTGC-3′) and B3 by using Taq DNA polymerase (Promega) with 2 mM MgCl2 and 1 M betaine (Sigma) and annealing at 55°C.
Generation of a Gabpα-specific antibody.
A unique region of 108 amino acids, corresponding to bp 572 to 895 of the Gabpα cDNA (gi:193382), was identified by performing BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST) of the mouse Gabpα open reading frame (gi:6679899) against the nonredundant cDNA database. This sequence was excised by BstOI-BanI digestion and subcloned into the SmaI site of the pQE-31 His tag expression vector (QIAGEN). The recombinant Gabpα-His fusion protein was expressed in BL21 DE3(pLysS) cells (Stratagene) with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) induction for 1 h, and protein was eluted via TALON nickel resin (Clontech). Polyclonal antibodies specific to mouse Gabpα were then generated in two rabbits, each immunized with an initial injection of 0.8 mg of recombinant His-tagged Gabpα protein in Freund's complete adjuvant and three equivalent booster injections in Freund's incomplete adjuvant.
Immunodetection of Gabpα protein by Western blot analysis. Gabpα protein levels were determined by Western blotting using protein lysates from wild-type and Gabpα-heterozygous 129/SvJ-derived ES cells, subsequent to their growth without a fibroblast feeder cell layer for several days, and pooled tissue lysates from four male and four female 6- to 8-week-old Gabpα-heterozygous and wild-type mice. An optimized dilution of polyclonal rabbit antisera was used, followed by 0.15 μg of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (DAKO)/ml. β-Tubulin was detected by using 0.05 μg of a mouse anti-β-tubulin monoclonal antibody (Chemicon)/ml, followed by 0.3 μg of horseradish peroxidase-conjugated rabbit anti-mouse immunoglobulin G (DAKO)/ml. Quantities of Gabpα protein relative to those of β-tubulin were determined by measuring the intensities of protein bands upon Western blot analysis, performed on three independent occasions, by using MacBas (version 2.5) software.
Gabpα mRNA expression during embryogenesis.
A Gabpα fragment spanning bp 413 to 1884 of gi:193382 (the open reading frame spans the area from position 454 to 1815) was subcloned into pBluescript SK(+) and used to generate digoxigenin-labeled sense and antisense transcripts according to the manufacturer's instructions (Roche). Subsequent hybridization was performed as previously described (29). A sense control yielded no detectable signal. Furthermore, the probe is specific for Gabpα. Ristevski et al. previously examined the embryonic expression of Ets2 (29), which has the Ets domain sequence most closely related to that of Gabpα. There is no overlap in expression between these genes, confirming the specificity of the probe.
Nucleotide sequence accession numbers.
The Gabpα genomic fragments sequenced in this study have been submitted to GenBank (gi:27960443 to gi:27960445; accession no. AF346288 to AF346290).
RESULTS
Targeted disruption of the mouse Gabpα gene.
To aid in the generation of a Gabpα targeting construct, the structure and sequence of the Gabpα gene of the mouse strain 129/SvJ were determined by lambda phage and BAC library screening, followed by PCR using genomic DNA from λ phage clones, BAC clones, and ES cells (Fig. 1A). Gabpα spans ∼26 kb of genomic DNA and consists of 10 exons and 9 introns. As with human GABPα, the 5′ end of mouse Gabpα lies several kilobases from the gene encoding the mitochondrial protein ATP synthase coupling factor 6, which is transcribed in the reverse orientation to Gabpα (8). In the mouse, 3.6 kb of genomic DNA separates the first exons of these two genes.
FIG. 1.
Genomic structure and targeted disruption of mouse Gabpα. (A) Schematic of mouse Gabpα genomic structure. Asterisks indicate the first coding exons of Gabpα and the closely linked gene ATP synthase coupling factor 6. (B) Intron-exon boundary sequences of Gabpα and comparison of human (12) and mouse intron sizes. Coding sequences are capitalized, introns are lowercased, and the first coding exon is indicated by an asterisk. Splice sites (gt-ag) are boldfaced. (C) Wild-type (wt) and targeted mouse Gabpα alleles. An IRES-LacZ-neomycin cassette was inserted into exon 2, immediately downstream of the start codon, and the position of the targeting construct (from the XbaI to the HindIII site) is indicated. The positions of the intron-3 probe (external to the targeting construct) used for Southern blot analysis (E3-4) and of PCR primers are indicated. Restriction endonuclease sites are as follows: B, BamHI; Bl, BalI; H, HindIII; N, NotI; X, XbaI. (D) Southern blot analysis of ES cell clones by BamHI digestion and hybridization with the E3-4 probe detecting 7-kb wild-type and 13-kb targeted alleles. (E) PCR screen of wild-type (+/+) and heterozygous (+/−) genomic DNA performed using a 5′ primer within intron 1 (F3) or the neomycin cassette (F8) and a common 3′ primer (B3) immediately downstream of exon 2 to generate 192-bp wild-type and 700-bp targeted PCR products.
The mouse Gabpα intron-exon boundaries were predicted by alignment of mouse (gi:193382) and human (gi:286026) cDNA sequences, because those of the human gene had been defined previously (12). Primers were designed to include 50 to 100 bp on either side of the predicted boundaries, and PCRs were performed using Elongase DNA polymerase (Invitrogen). Sequence analysis (with Sequencher software [version 3.0]) generated three genomic fragments (gaps within introns 5 and 6 due to their large size), the sequences of which have been submitted to GenBank (gi:27960443 to gi:27960445). Subsequent to this analysis, these three fragments have been aligned with the public genome-sequencing database (gi:20897082 bases 18725599 to 20709283) and found to be identical, yielding the complete 26,595-bp genomic sequence of Gabpα. As shown in Fig. 1B, the intron-exon boundaries of mouse Gabpα conform to the gt-ag splice rule, and the positions and relative sizes of introns are conserved between the mouse and human genes (see reference 12 for the structure of the human gene).
By using this sequence information, a Gabpα targeting cassette was generated. An IRES-lacZ-neomycin cassette was inserted downstream of the ATG, disrupting exon 2, the first coding exon (Fig. 1C). The targeting vector, spanning bp 3766 to 9610 of gi:27960443, was electroporated into 129/SvJ-derived ES cells, and Southern blot analysis revealed a targeted band of 13 kb relative to a wild-type band of 7 kb (Fig. 1D). PCR was performed to generate 700-bp targeted and 192-bp wild-type allele products (Fig. 1E). Given the close proximity of Gabpα to ATP synthase coupling factor 6 (the first exons are separated by 3.6 kb) (Fig. 1A), we examined the 5′ targeting end for any genomic disruption during homologous recombination. An external PCR primer and two internal primers gave rise to PCR products of the expected size in both wild-type and heterozygous targeted genomic DNA (data not shown), suggesting that genomic rearrangement and disruption of ATP synthase coupling factor 6 expression were unlikely.
Several positive ES clones were microinjected into C57BL/6 blastocysts to produce chimeras showing germ line transmission. The resulting Gabpα-heterozygous mice were found to be viable and fertile.
Analysis of Gabpα heterozygote matings.
Breeding of heterozygous Gabpα mice yielded no Gabpα−/− mice at weaning (Table 1), no Gabpα−/− embryos at embryonic day 7.5 (E7.5), E8.5, and E9.5, and no Gabpα−/− blastocysts from a total of 36 analyzed (9 would be expected in accordance with a Mendelian distribution) (blastocyst data not shown). Therefore, lack of Gabpα protein expression results in preimplantation lethality in mice. Of the 274 weanling mice genotyped, Gabpα+/+ and Gabpα−/− genotypes were observed at the expected ratio. This indicates that Gabpα is essential very early in embryogenesis and that monoallelic expression from the wild-type allele in Gabpα+/− cells produces normal levels of Gabpα and is thus sufficient for survival and development.
TABLE 1.
Genotype analysis derived from Gabpα heterozygous intercross matings
| Agea | Observed (expected) no. with the following Gabpα genotype:
|
χ2 | Probabilityb | ||
|---|---|---|---|---|---|
| Wild type | Heterozygous | Knockout | |||
| Weaning (n = 201) | 78 (67) | 123 (134) | 0 | 2.7 | 0.2-0.3 |
| E9.5 (n = 47) | 18 (15.7) | 29 (31.3) | 0 | 0.52 | 0.7-0.9 |
| E8.5 (n = 53) | 25 (17.7) | 28 (35.3) | 0 | 4.56 | 0.1-0.2 |
| E7.5 (n = 23) | 12 (7.7) | 11 (15.3) | 0 | 3.66 | 0.1-0.2 |
n, total number examined at each age.
Based on 2 degrees of freedom.
Gabpα expression during embryogenesis.
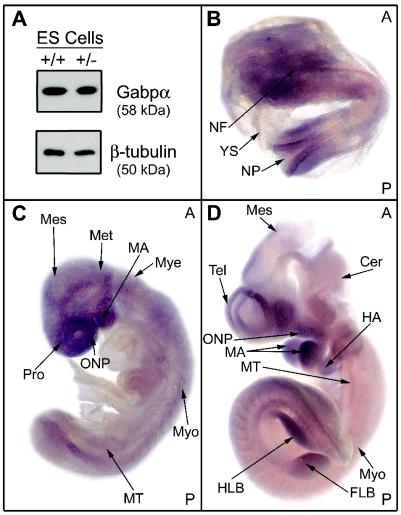
Expression of Gabpα protein was examined by Western blotting using a rabbit polyclonal antibody raised against a unique His-tagged 108-amino-acid region of Gabpα. This antiserum did not cross-react with the closely related proteins Ets-1 and Ets-2 (similarly His tagged). The relatively high level of Gabpα expression in ES cells (Fig. 2A) suggests an important role for Gabp in early embryogenesis. No obvious reduction in Gabpα protein expression was observed in heterozygous ES cells.
FIG. 2.
Gabpα expression during embryogenesis. (A) Western blot analysis of Gabpα and β-tubulin protein levels in wild-type (+/+) and Gabpα heterozygous (+/−) ES cells. (B through D) Whole-mount mRNA in situ hybridization analysis of Gabpα was performed at E8.0 (B), E9.0 (C), and E10 (D). Arrows indicate areas of relatively abundant expression. A and P indicate the anterior and posterior sides of the embryo in each panel. Tissues are as follows: NF, neural fold; N, neural pore; YS, yolk sac; Mes, mesencephalon; Met, metencephalon; MA, mandibular arch; Mye, myelencephalon; Pro, prosencephalon; ONP, oronasopharyngeal region; Myo, myotome; MT, mesonephric tubule; Cer, cerebellum; Tel, telencephalon; HA, hyoid arch; HLB, hindlimb bud; FLB, forelimb bud.
The distribution of Gabpα mRNA expression throughout the developing embryo was determined by whole-mount mRNA in situ hybridization. As shown in Fig. 2, Gabpα is expressed broadly at E8.0 (B), E9.0 (C), and E10 (D). Gabpα is expressed in the developing myotome, consistent with the role of Gabp in the regulation of neuromuscular genes. Other areas of high Gabpα expression include the neural fold at E8, as well as the mesencephalon and cerebellum from E10. In addition, Gabpα expression is detected at all developmental stages in the fore- and hindlimb buds, mandibular and hyoid arches, and nasopharyngeal ectoderm and mesoderm. Therefore, Gabpα is broadly expressed in the developing mouse embryo from an early stage.
Phenotypic analysis of Gabpα-heterozygous mice.
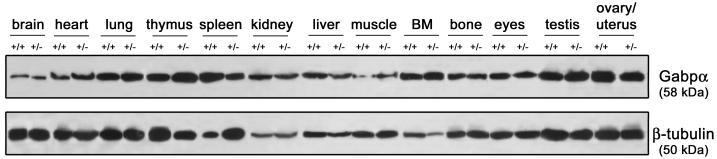
We examined Gabpα+/− mice for histopathology, growth rate, immunophenotype, X-ray tomography, and gait and grip strength in comparison to those of their wild-type littermates (data not shown), and found no abnormalities. We examined wild-type and Gabpα-heterozygous mouse tissue extracts by Western blot analysis and found that Gabpα protein levels were similar in heterozygous mice and their wild-type littermates (Fig. 3). This indicates that Gabpα protein levels are tightly regulated, resulting in constant levels of Gabpα protein despite a single gene copy number, and is consistent with the absence of any phenotype in Gabpα+/− mice. In order to establish that no C-terminal truncations of Gabpα were generated from the targeted allele, we examined these tissue extracts with a monoclonal antibody generated against the C terminus (a gift from Steve Burden). No additional bands were observed by using this antibody, suggesting that intact protein is made at normal levels from the untargeted allele (data not shown).
FIG. 3.
Protein expression in adult Gabpα-heterozygous mice. Shown are results of Western blot analysis of Gabpα protein in lysates from Gabpα wild-type (+/+) and heterozygous (+/−) mouse tissues, pooled from four animals of each sex for each genotype at the age of 6 to 8 weeks. The β-tubulin protein was used as a loading control. Muscle was sampled from the quadriceps. BM, bone marrow.
DISCUSSION
GABPα is a gene of fundamental importance to cell function and development, as demonstrated by the early lethality of mice carrying a homozygous deletion in the Gabpα gene. This is consistent with the conservation of the human GABPα sequence with that of the mouse (8), rat (8) and puffer fish (36), implying maintenance of protein function throughout evolution. Furthermore, steady-state Gabpα protein levels are maintained in heterozygous mice despite the presence of only a single Gabpα allele. We show that Gabpα protein is expressed in ES cells and Gabpα mRNA is detectable by whole-mount in situ hybridization from as early as E8.0, consistent with previous reports that transcripts are widely expressed in late embryogenesis (28), and in the adult mouse (6, 21).
The embryonic lethality of Gabpα-null mice is not surprising, because interestingly, mouse models of knockout of ETS factors such as Fli-1 (34), Tel (40), and Ets-2 (42) also display embryonic lethal phenotypes. These findings highlight the fact that although the ETS transcription factor family is large and consists of many highly homologous proteins, ETS factors are not functionally redundant; each plays a unique role in the embryo during mammalian development and in adult tissues.
Gabp (also known as Nrf-2) and Nrf-1 are two unrelated transcription factors that act to regulate many genes encoding protein products that function in the mitochondrial respiratory chain. Interestingly, Nrf-1-deficient mice exhibit peri-implantation lethality (15) while Nrf1-heterozygous mice, like Gabpα heterozygotes, are phenotypically normal. Nrf-1−/− blastocysts have reduced amounts of mitochondrial DNA and show a decrease in rhodamine 123 staining of mitochondria relative to that for wild-type blastocysts, suggesting that Nrf-1 is required for mitochondrial maintenance in vivo (15). A high rate of mitochondrial transcription is known to occur during cleavage of the embryo and may require the new synthesis and transport of nucleus-encoded components (22). Therefore, the early embryonic lethality of mice lacking either Nrf-1 or Gabpα may be due to decreased expression of nucleus-encoded genes of mitochondrial function that are transcriptionally regulated by these two factors, such as MTFA. Complete loss of MTFA protein expression in the mouse results in depletion of mitochondrial DNA and death prior to E10.5 (22). Therefore, the earlier death of mice null for Gabpα protein expression may be due to decreased expression of MTFA and other Gabp target genes. The role of Gabpα in the development and function of specific tissues will require the establishment of conditional deletion of this gene.
Acknowledgments
We thank Tony Ward, Vivien Vasic, and the animal house staff for technical assistance.
REFERENCES
- 1.Aurrekoetxea-Hernández, K., and E. Buetti. 2000. Synergistic action of GA-binding protein and glucocorticoid receptor in transcription from the mouse mammary tumor virus promoter. J. Virol. 74:4988-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avots, A., A. Hoffmeyer, E. Flory, A. Cimanis, U. R. Rapp, and E. Serfling. 1997. GABP factors bind to a distal interleukin 2 (IL-2) enhancer and contribute to c-Raf-mediated increase in IL-2 induction. Mol. Cell. Biol. 17:4381-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannert, N., A. Avots, M. Baier, E. Serfling, and R. Kurth. 1999. GA-binding protein factors, in concert with the coactivator CREB binding protein/p300, control the induction of the interleukin 16 promoter in T lymphocytes. Proc. Natl. Acad. Sci. USA 96:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bister, K., M. Nunn, C. Moscovici, B. Perbal, M. A. Baluda, and P. H. Duesberg. 1982. Acute leukemia viruses E26 and avian myeloblastosis virus have related transformation-specific RNA sequences but different genetic structures, gene products, and oncogenic properties. Proc. Natl. Acad. Sci. USA 79:3677-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boccia, L. M., D. Lillicrap, K. Newcombe, and C. R. Mueller. 1996. Binding of the Ets factor GA-binding protein to an upstream site in the factor IX promoter is a critical event in transactivation. Mol. Cell. Biol. 16:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, T. A., and S. L. McKnight. 1992. Specificities of protein-protein and protein-DNA interaction of GABPα and two newly defined ets-related proteins. Genes Dev. 6:2502-2512. [DOI] [PubMed] [Google Scholar]
- 7.Castrillo, A., D. J. Pennington, F. Otto, P. J. Parker, M. J. Owen, and L. Boscá. 2001. Protein kinase Cɛ is required for macrophage activation and defense against bacterial infection. J. Exp. Med. 194:1231-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinenov, Y., C. Coombs, and M. E. Martin. 2000. Isolation of a bi-directional promoter directing expression of the mouse GABPα and ATP synthase coupling factor 6 genes. Gene 261:311-320. [DOI] [PubMed] [Google Scholar]
- 9.Duclert, A., N. Savatier, L. Schaeffer, and J.-P. Changeux. 1996. Identification of an element crucial for the sub-synaptic expression of the acetylcholine receptor ɛ-subunit gene. J. Biol. Chem. 271:17433-17438. [DOI] [PubMed] [Google Scholar]
- 10.Gajewski, K. M., and R. A. Schulz. 1995. Requirement of the ETS domain transcription factor D-ELG for egg chamber patterning and development during Drosophila oogenesis. Oncogene 11:1033-1040. [PubMed] [Google Scholar]
- 11.Genuario, R. R., and R. P. Perry. 1996. The GA-binding protein can serve as both an activator and repressor of ribosomal protein gene transcription. J. Biol. Chem. 271:4388-4395. [DOI] [PubMed] [Google Scholar]
- 12.Goto, M., T. Shimizu, J.-I. Sawada, C. Sawa, H. Watanabe, H. Ichikawa, M. Ohira, M. Ohki, and H. Handa. 1995. Assignment of the E4TF1-60 gene to human chromosome 21q21.2-q21.3. Gene 166:337-338. [DOI] [PubMed] [Google Scholar]
- 13.Graves, B. J., and J. M. Petersen. 1998. Specificity within the ets family of transcription factors. Adv. Cancer Res. 75:1-55. [DOI] [PubMed] [Google Scholar]
- 14.Hart, A. H., R. Reventar, and A. Bernstein. 2000. Genetic analysis of ETS genes in C. elegans. Oncogene 19:6400-6408. [DOI] [PubMed] [Google Scholar]
- 15.Huo, L., and R. C. Scarpulla. 2001. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol. Cell. Biol. 21:644-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang, S. Y., P. J. Hertzog, K. A. Holland, S. H. Sumarsono, M. J. Tymms, J. A. Hamilton, G. Whitty, I. Bertoncello, and I. Kola. 1995. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc. Natl. Acad. Sci. USA 92:11284-11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamura, T., H. Handa, N. Hamasaki, and S. Kitajima. 1997. Characterization of the human thrombopoietin gene promoter. A possible role of an Ets transcription factor, E4TF1/GABP. J. Biol. Chem. 272:11361-11368. [DOI] [PubMed] [Google Scholar]
- 18.Khurana, T. S., A. G. Rosmarin, J. Shang, T. O. B. Krag, S. Das, and S. Gammeltoft. 1999. Activation of utrophin promoter by heregulin via the ets-related transcription factor complex GA-binding protein α/β. Mol. Biol. Cell 10:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koike, S., L. Schaeffer, and J.-P. Changeux. 1995. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor δ-subunit gene. Proc. Natl. Acad. Sci. USA 92:10624-10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahoud, M. H., S. Ristevski, D. J. Venter, L. S. Jermiin, I. Bertoncello, S. Zavarsek, S. Hasthorpe, J. Drago, D. de Kretser, P. J. Hertzog, and I. Kola. 2001. Gene targeting of Desrt, a novel ARID class DNA-binding protein, causes growth retardation and abnormal development of reproductive organs. Genome Res. 11:1327-1334. [DOI] [PubMed] [Google Scholar]
- 21.LaMarco, K., C. C. Thompson, B. P. Byers, E. M. Walton, and S. L. McKnight. 1991. Identification of Ets- and Notch-related subunits in GA binding protein. Science 253:789-792. [DOI] [PubMed] [Google Scholar]
- 22.Larsson, N.-G., J. Wang, H. Wilhelmsson, A. Oldfors, P. Rustin, M. Lewandoski, G. S. Barsh, and D. A. Clayton. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231-236. [DOI] [PubMed] [Google Scholar]
- 23.Laudet, V., C. Hänni, D. Stéhelin, and M. Duterque-Coquillaud. 1999. Molecular phylogeny of the ETS gene family. Oncogene 18:1351-1359. [DOI] [PubMed] [Google Scholar]
- 24.Li, X. R., A. S.-F. Chong, J. Wu, K. A. Roebuck, A. Kumar, J. E. Parrillo, U. R. Rapp, R. P. Kimberly, J. W. Williams, and X. Xu. 1999. Transcriptional regulation of Fas gene expression by GA-binding protein and AP-1 in T cell antigen receptor.CD3 complex-stimulated T cells. J. Biol. Chem. 274:35203-35210. [DOI] [PubMed] [Google Scholar]
- 25.Marchioni, M., S. Morabito, A. L. Salvati, E. Beccari, and F. Carnevali. 1993. XrpFI, an amphibian transcription factor composed of multiple polypeptides immunologically related to the GA-binding protein α and β subunits, is differentially expressed during Xenopus laevis development. Mol. Cell. Biol. 13:6479-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn, M., H. Weiher, P. Bullock, and P. Duesberg. 1984. Avian erythroblastosis virus E26: nucleotide sequence of the tripartite onc gene and of the LTR, and analysis of the cellular prototype of the viral ets sequence. Virology 139:330-339. [DOI] [PubMed] [Google Scholar]
- 27.Nunn, M. F., P. H. Seeburg, C. Moscovici, and P. H. Duesberg. 1983. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature 306:391-395. [DOI] [PubMed] [Google Scholar]
- 28.Reymond, A., V. Marigo, M. B. Yaylaoglu, A. Leoni, C. Ucla, N. Scamuffa, C. Caccioppoli, E. T. Dermitzakis, R. Lyle, S. Banfi, G. Eichele, S. E. Antonarakis, and A. Ballabio. 2002. Human chromosome 21 gene expression atlas in the mouse. Nature 420:582-586. [DOI] [PubMed] [Google Scholar]
- 29.Ristevski, S., P. P. L. Tam, P. J. Hertzog, and I. Kola. 2002. Ets2 is expressed during morphogenesis of the somite and limb in the mouse embryo. Mech. Dev. 116:165-168. [DOI] [PubMed] [Google Scholar]
- 30.Scarpulla, R. C. 2002. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta 1576:1-14. [DOI] [PubMed] [Google Scholar]
- 31.Seelan, R. S., L. Gopalakrishnan, R. C. Scarpulla, and L. I. Grossman. 1996. Cytochrome c oxidase subunit VIIa liver isoform. J. Biol. Chem. 271:2112-2120. [DOI] [PubMed] [Google Scholar]
- 32.Seelan, R. S., and L. I. Grossman. 1997. Structural organization and promoter analysis of the bovine cytochrome c oxidase subunit VIIc gene. J. Biol. Chem. 272:10175-10181. [DOI] [PubMed] [Google Scholar]
- 33.Sowa, Y., Y. Shiio, T. Fujita, T. Matsumoto, Y. Okuyama, D. Kato, J.-I. Inoue, J.-I. Sawada, M. Goto, H. Watanabe, H. Handa, and T. Sakai. 1997. Retinoblastoma binding factor 1 site in the core promoter region of the human RB gene is activated by hGABP/E4TF1. Cancer Res. 57:3145-3148. [PubMed] [Google Scholar]
- 34.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, Y., K. Kako, H. Arai, T. Ohishi, Y. Inada, A. Takehara, A. Fukamizu, and E. Munekata. 2002. Characterization and identification of promoter elements in the mouse COX17 gene. Biochim. Biophys. Acta 1574:359-364. [DOI] [PubMed] [Google Scholar]
- 36.Tassone, F., L. Villard, K. Clancy, and K. Gardiner. 1999. Structures, sequence characteristics, and synteny relationships of the transcription factor E4TF1, the splicing factor U2AF35 and the cystathionine beta synthetase genes from Fugu rubripes. Gene 226:211-223. [DOI] [PubMed] [Google Scholar]
- 37.Verhoef, K., R. W. Sanders, V. Fontaine, S. Kitajima, and B. Berkhout. 1999. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-κB enhancer element into a GABP binding site. J. Virol. 73:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virbasius, J. V., and R. C. Scarpulla. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA 91:1309-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Virbasius, J. V., C.-M. A. Virbasius, and R. C. Scarpulla. 1993. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 7:380-392. [DOI] [PubMed] [Google Scholar]
- 40.Wang, L. C., F. Kuo, Y. Fujiwara, D. G. Gilliland, T. R. Golub, and S. H. Orkin. 1997. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 16:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong-Riley, M., A. Guo, N. J. Bachman, and M. I. Lomax. 2000. Human COX6A1 gene: promoter analysis, cDNA isolation and expression in the monkey brain. Gene 247:63-75. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, H., M. L. Flannery, S. Kupriyanov, J. Pearce, S. R. McKercher, G. W. Henkel, R. A. Maki, Z. Werb, and R. G. Oshima. 1998. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 12:1315-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]