Abstract
BACKGROUND
Postoperative atrial fibrillation (AF) is a potentially life-threatening complication after coronary artery bypass graft (CABG) surgery. Genetic predisposition may predict risk for developing postoperative AF.
METHODS
Study subjects underwent CABG surgery with cardiopulmonary bypass at Duke University Medical Center. In a discovery cohort of 877 individuals from the Perioperative Genetics and Safety Outcomes Study (PEGASUS), we performed a genome-wide association study (GWAS) using a logistic regression model with a covariate adjustment for AF risk index. Single-nucleotide polymorphisms (SNPs) that met a P<5×10−5 were further tested using a replication dataset of 304 individuals from the CATHeterization GENetics (CATHGEN) biorepository, followed by meta-analysis. Potential pathways related to postoperative AF were identified through gene enrichment analysis using the top GWAS SNPs (P<10−4).
RESULTS
Nineteen SNPs met the a priori defined discovery threshold for replication, but only 3 met nominal significance (P<0.05) in the CATHGEN group, with only one – rs10504554, in the intronic region in lymphocyte antigen 96 (LY96) – showing the same direction of the effect for postoperative AF (odds ratio [OR]=0.48; 95% CI: 0.34–0.68, P=2.9×10−5 vs OR=0.55; 95% CI: 0.31–0.99, P=0.046), and strong overall association by meta-analysis (meta-P=4.0×10−6). Gene enrichment analysis highlighted the role of LY96 in pathways of biologic relevance to activation and modulation of innate immune responses. Our analysis also showed potential association between LY96 and nuclear factor NF-kappa-B interaction, and postoperative AF through their relevance to inflammatory signaling pathways.
CONCLUSIONS
In patients undergoing CABG surgery, we found genetic polymorphisms in LY96 associated with decreased risk for postoperative AF.
Keywords: genome-wide association study, atrial fibrillation, coronary artery bypass graft, lymphocyte antigen 96
INTRODUCTION
Postoperative atrial fibrillation (AF) is one of the most common complications following coronary artery bypass graft surgery (CABG), occurring in 25% to 40% of patients.1 It is associated with an increased risk for adverse neurologic events, congestive heart failure, myocardial infarction, and perioperative mortality as well as prolonged hospital length of stay and increased hospital costs.1–3 Indeed, the impact of postoperative AF on hospital resources is substantial with extra costs exceeding $10,000 per patient, which translates into more than $2 billion each year in the United States alone.4
Several patient-specific and procedure-related risk factors associated with increased risk for postoperative AF have been described in cardiac surgery patients. These include advanced age, a history of atrial fibrillation, chronic obstructive pulmonary disease, valve surgery, and discontinuation of beta-blocker or angiotensin converting enzyme inhibitor therapy.1, 5 The identification of these risk factors led to the development and validation of several comprehensive risk indices that are used to predict postoperative AF and possible preventive strategies; 1, 5–7 however, their reliability and hence, generalizability remain limited due to significant differences in the risk factors that were selected and validated for each index. Indeed, advanced age is the only consistent independent risk factor for postoperative AF.6 This suggests that the etiology of postoperative AF after cardiac surgery is multifactorial, and that genetic variations may play a significant role.8–10
Genetic predisposition in AF has been suggested by a few candidate gene and genome-wide association studies (GWAS). These variants, or single-nucleotide polymorphisms (SNPs), reported to date for AF have mostly been located in genes that modulate intrinsic cardiac pacemaker activity, signal transduction, and cardiopulmonary development.11, 12 However, most of these studies were performed in subjects selected from large-scale, community-based cohort studies, and compared subjects with incident or prevalent AF vs subjects without AF. In the subsequent association analyses, the investigators adjusted only for a limited number of subject-specific characteristics such as age and gender. 11, 12 In contrast, only a few candidate gene studies have attempted to identify and/or validate genetic variants that are independently associated with new-onset postoperative AF in cardiac surgery patients.13, 14 Therefore, we conducted the first genome-wide association study to identify common genetic variants associated with new-onset postoperative AF in the setting of CABG surgery.
MATERIALS AND METHODS
The study design and data reporting followed the recommendations of “Strengthening the Reporting of Genetic Association Studies” (STREGA).11 In the present study, 2 independent cohorts of patients who underwent CABG at the Duke Heart Center at Duke University Medical Center, Durham, North Carolina, were used for intial common variant discovery by GWAS, and replication analysis of top candidate SNPs. Each of the parent studies in our investigation was approved by the Institutional Review Board at Duke University Medical Center, and all subjects provided written informed consent.
The discovery cohort for GWAS was comprised of patients from the Perioperative Genetics and Safety Outcomes Study (PEGASUS), a longitudinal study that enrolled 1,004 patients who underwent isolated non-emergent CABG surgery with cardiopulmonary bypass (CPB) between 1997 and 2006.12 For patients who had more than one cardiac surgery during that period, only data from the first surgery were included. Of the original 1,004 patients, 877 patients with self-reported European ancestry and complete phenotypic and genotypic data met our inclusion criteria for the discovery cohort.
The replication cohort for our study was selected from 475 patients in the CATHeterization GENetics (CATHGEN) study13 who underwent cardiac catheterization between 2001 and 2010 to evaluate ischemic heart disease, and who subsequently underwent CABG surgery with CPB between 2006 and 2010. Of these, 304 patients without valve surgery met our inclusion criteria.
Intraoperative anesthetic, perfusion, and cardioprotective management was standardized. General anesthesia was maintained with a combination of fentanyl and isoflurane. Perfusion support consisted of nonpulsatile CPB (30°C– 32°C), crystalloid prime, pump flow rates >2.4 L/min/m2, cold blood cardioplegia, α-stat blood gas management, activated clotting times >450 seconds maintained with heparin, ε-aminocaproic acid infusion administered routinely, and serial hematocrits maintained at >0.18.
Data Collection and End-point Definition
Patient demographics, preoperative and procedural factors, and perioperative medication use are components1 of the postoperative AF Risk Index (Supplemental Table 1), were recorded and collated using the Duke Information System for Cardiovascular Care, an integral part of the Duke Databank for Cardiovascular Disease. The postoperative AF Risk Index is a predictor of postoperative AF in cardiac surgery patients. Diagnosis of new-onset postoperative AF was based on postoperative electrocardiogram or rhythm strip, or at least 2 of the following forms of documentation: progress notes, nursing notes, discharge summary, or change in medication.
Genotyping and Quality Controls
Genotyping platforms and quality controls (QCs) for genotype and sample exclusion have been described previously.15 Briefly, 1004 samples from the PEGASUS cohort (discovery samples) were genotyped using the Illumina Human610-Quad BeadChip.Various QC criteria were applied to ensure the genotype quality. Markers were excluded if the GenCall (http://support.illumina.com/downloads/gencall_software.html) score was ≤ 0.15, the genotype call frequency was < 98%, or gender errors were detected At the sample level, we checked cryptic relatedness and duplications. For a pair of samples with identity by descent (IBD) > 0.1875 (between 2nd and 3rd degree relative), one sample was excluded. At this stage, the QC’ed genotype dataset at this stage consisted of 960 subjects with 561,091 markers. However, 83 subjects had missing data on postoperative AF status and therefore were excluded from subsequent analysis. Thus, the final discovery dataset (PEGASUS cohort) consisted of 877 patients, all of European descent who had both genotype and phenotype data available.
Population structure was investigated using the EigenSoft program16 to generate 15 principal components (PCs). However, as expected, we did not find any PCs associated with AF since only patients with self-reported European ancestry were included in our dataset. Therefore, no PCs were included in the final association analysis model.
All CATHGEN samples were genotyped using the Illumina OMNI 1-Quad BeadChip, and subject to the same marker and sample QC criteria as described above. We selected a subset of CATHGEN samples for the replication dataset based on the availability of postoperative AF data. Since different genotype platforms were used for the discovery and replication samples, we used imputed markers to maximize the shared SNPs between the 2 datasets, and to fine map the regions of interest. The IMPUTE217 program was used for imputing untyped SNPs using the post-QCed PEGASUS genotype dataset (960 samples with 561,091 markers) and phased haplotypes from the 1000 genome CEU reference panel. The best-guess imputed genotype for any untyped SNP per sample was chosen as the genotype with the highest imputation probability (imputation score). If the highest imputation score for an imputed SNP of a sample was < 90%, a missing imputed genotype was assigned.
Statistical Analysis
Descriptive statistics of clinical variables are presented as frequency and percentage for categorical variables, and mean ± SD for continuous variables. Univariable logistic regression analysis was performed to test the differences in demographic, and clinical and procedural characteristics between patients with and without postoperative AF. P values (P) were derived from 2-sided Wald tests. Analyses of clinical variables were conducted using SAS Version 9.2 (SAS Institute Inc, Cary, NC).
All association analyses below were performed using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) for all genotyped and imputed markers. At the data analysis stage, we included additional QC criteria by excluding markers that were significantly deviated from Hardy-Weinberg Equilibrium (HWE) (P < 10−6) or had a minor allele frequency (MAF) < 2%. For each of the SNPs, allelic associations with postoperative AF were assessed using logistic regression analyses adjusted for the postoperative AF Risk Index. These association tests, including those for imputed genotypes, were performed assuming an additive inheritance model (homozygote major allele vs heterozygote vs homozygote minor allele). Markers were considered to be significant genome-wide if P < 5 × 10−8, which is the most commonly accepted significant threshold for GWAS. In addition to this stringent criterion, a relaxed significance threshold of P < 5 × 10−5 was applied when choosing SNPs for replication in the CATHGEN cohort.18 The same logistic regression model adjusted for the postoperative AF Risk Index was applied in the replication dataset.
To assess the overall effect of candidate SNPs, we then conducted a meta-analysis using the weighted Z-score meta-analysis as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/metal). The final candidate gene(s) were prioritized based on 1) meeting nominal significance in the CATHGEN cohort, 2) showing the direction of their effect for postoperative AF in the discovery dataset, and 3) reaching statistical significance in the meta-analysis. For the top candidate gene(s) or region(s), we increased marker density by using all imputed markers within the gene or region in the discovery dataset to examine the association pattern within the region. Within a gene or region of interest, we also assessed whether other SNPs showed independent association from the top SNP initially identified by performing a conditional association analysis that included the top SNP in the logistic regression model. Pairwise linkage disequilibrium (LD) between markers within the final candidate gene(s) was computed and quantified as r2 × 100, using Haploview (http://www.broad.mit.edu/mpg/haploview/).
Pathway analysis was performed based on the Ingenuity Pathways Knowledge Base, a pathway database created by Ingenuity Systems Inc (http://www.ingenuity.com/products/ipa). In brief, this database consists of gene regulatory and signaling pathways that are manually curated from the literature for the database. The database uses the Ingenuity Pathways Analysis (IPA®) software to view and analyze pathway data, and for a given gene set, the software automatically generates the pathways that are related to those genes (http://www.ingenuity.com/wp-content/themes/ingenuity-qiagen/pdf/ipa/ipa_datasheet.pdf). First, top SNPs with P < 0.0001, based on the results of the discovery cohort, were selected and uploaded to IPA so that they could be mapped to their respective molecules. Next, the pathway analysis was performed based on these molecules and the results were compared to the reference set of molecules for each pathway. Using a right-tailed Fisher exact test, a pathway was considered statistically significant (P < 0.05) if the number of molecules in our dataset was significantly larger than the expected number of molecules by chance based on the reference set of molecules within that pathway. Finally, the convergence between the results of the enrichment analysis and GWAS results was examined to identify pathways that are significant and potentially relevant to postoperative AF.
We further investigated potential gene-gene interaction between candidate gene(s) identified from our association study, and other genes of interest within the same pathway. We selected the most significant SNP within each gene of interest as the representative marker for the gene. The SNP-SNP interaction term was tested for its association with postoperative AF using the logistic regression model adjusted for individual SNP effect and the AF Risk Index.
Finally, we also examined SNPs within previously identified genes or loci for postoperative AF in our dataset. The same logistic regression model adjusted for the AF Risk Index was performed for these analyses.
Sources of Funding
This work was supported, in part, by National Institutes of Health grants HL075273 and HL092071 (to Dr. Podgoreanu); AG09663, HL054316, and HL069081 (to Dr. Newman); HL096978, HL108280, and HL109971 (to Dr. Mathew); HL095987 (to Dr. Shah); and HL101621 (to Dr. Kraus); and by American Heart Association grants 9970128N (to Dr. Newman), 9951185U (to Dr. Mathew), and 0120492U (to Dr. Podgoreanu). CATHGEN provided logistics support. The authors are solely responsible for the design and conduct of this study, all study analyses, the drafting and editing of the paper and its final contents.
RESULTS
Our study consisted of 877 patients in the discovery dataset and 304 patients in the replication dataset. Demographics and clinical characteristics of the patients in these 2 datasets, stratified according to the documented presence or absence of postoperative AF, are shown in Table 1. The mean age was 63.3 ± 10.2 years in the discovery dataset and 60.7 ± 10.7 years in the replication dataset. Both datasets had higher proportions of male patients (656 [74.8%] in the discovery dataset and 198 [65.1%] in the replication dataset). The median [IQR] postoperative AF risk scores were similar between the 2 datasets (12 [6–22] vs 11 [2–18]). Of the 877 patients in the discovery cohort, 257 (29.3%) developed postoperative AF. These subjects had a significantly higher median postoperative AF risk score compared to controls without postoperative AF (22 [12–31] vs 11 [3–17]; OR = 1.10; 95% CI: 1.08–1.11; P < 0.0001). Similarly, patients in the replication dataset with postoperative AF had a significantly higher median postoperative AF risk score compared to patients without postoperative AF (20 [11–30] vs 6 [0–13]; OR = 1.11; 95% CI: 1.08–1.14; P < 0.0001).
Table 1.
Demographic, Clinical and Procedural Characteristics of the Study Populations based on Postoperative Atrial Fibrillation Risk Index
| DISCOVERY DATASET (N=877) | REPLICATION DATASET (N=304) | |||||||
|---|---|---|---|---|---|---|---|---|
| Predictor | No PoAF (n=620) |
Yes PoAF (n=257) |
OR (95%CI) | P-value* | No PoAF (n=220) |
Yes PoAF (n=84) |
OR (95%CI) | P-value* |
| Age, y | 61.58±9.86 | 67.51±9.61 | 1.07 (1.05,1.08) | 2.55×10−14 | 59.35±10.67 | 64.24±9.93 | 1.05 (1.02,1.07) | 0.0005 |
| Medical History | ||||||||
| Atrial Fibrillation | 0 | 0 | 0 | 0 | ||||
| Chronic obstructive pulmonary disease | 45 (7.26) | 34 (13.23) | 1.95 (1.22,3.12) | 0.0055 | 15 (6.82) | 6 (7.14) | 1.05 (0.39,2.81) | 0.9197 |
| Concurrent valve surgery | 0 | 0 | 0 | 0 | ||||
| Withdrawal of Postoperative Treatment | ||||||||
| Beta-blocker | 45 (7.26) | 85 (33.07) | 6.31 (4.23,9.42) | 1.54×10−19 | 13 (5.91) | 35 (41.67) | 11.37 (5.60, 23.10) | 1.77×10−11 |
| ACE inhibitor | 232 (37.42) | 128 (49.81) | 1.66 (1.24,2.23) | 0.0007 | 103 (46.82) | 48 (57.14) | 1.51 (0.91, 2.51) | 0.1084 |
| Beta-blocker Treatment | ||||||||
| Preoperative and Postoperative | 456 (73.55) | 113 (43.97) | 0.28 (0.21,0.38) | 3.59×10−16 | 180 (81.82) | 33 (39.29) | 0.14 (0.08, 0.25) | 8.09×10−12 |
| Postoperative | 547 (88.23) | 141 (54.86) | 0.16 (0.11,0.23) | 7.78×10−25 | 204 (92.73) | 42 (50) | 0.08 (0.04,0.15) | 6.12×10−14 |
| Preoperative and Postoperative ACE | 79 (12.74) | 21 (8.17) | 0.61 (0.37,1.01) | 0.0547 | 43 (19.55) | 5 (5.95) | 0.26 (0.10, 0.68) | 0.0062 |
| Inhibitor Treatment | ||||||||
| Preoperative and Postoperative Statin | 191 (30.81) | 60 (23.35) | 0.68 (0.49,0.96) | 0.0266 | 111 (50.45) | 21 (25) | 0.33 (0.19, 0.57) | 9.32×10−5 |
| Treatment | ||||||||
| Postoperative Treatment | ||||||||
| Potassium Supplementation | 544 (87.74) | 170 (66.15) | 0.27 (0.19,0.39) | 5.36×10−13 | 193 (87.73) | 56 (66.67) | 0.28 (0.15, 0.51) | 3.87×10−5 |
| NSAIDs | 129 (20.81) | 23 (8.95) | 0.37 (0.23,0.60) | 4.15×10−5 | 58 (26.36) | 8 (9.52) | 0.29 (0.13, 0.65) | 0.0023 |
Continuous variables are presented as means ± standard deviation, or median [interquartile range], and categorical variables as percent frequencies. OR (95%CI), univariate odds ratio (95% confidence interval); ACE, angiotensin converting enzyme; NSAIDs, non-steroidal anti-inflammatory drugs
Comparisons were made using logistic regression to test the differences in demographic, and clinical and procedural characteristics between cases and controls, where P-values were derived from the Wald tests. Patients in the discovery and replication datasets are separated by actual documented presence or absence of postoperative AF.
A total of 561,091 genotyped markers were available for analysis after our initial QCs. Of these, we excluded 13 SNPs due to deviation from HWE, and an additional 36,103 SNPs due to an MAF < 0.02. The remaining 524,975 markers were analyzed in the 877 subjects in the discovery cohort for any association with postoperative AF. The GWAS results in the discovery cohort are depicted using a Manhattan plot (Supplemental Figure 1A) and a quantile-quantile (QQ) plot, which showed good adherence to null expectations (Supplemental Figure 1B). None of the SNPs reached genome-wide significance. However, 19 SNPs met the a priori defined discovery threshold of P < 5×10−5, and were subsequently analyzed in the replication cohort (Table 2).
Table 2.
Logistic Regression Analysis of Genetic Predictors of Postoperative Atrial Fibrillation in the Study Populations
| DISCOVERY DATASET |
REPLICATION DATASET |
Combined |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
MAF |
|||||||||||||
| Chr | SNP | BP | Gene symbol* | Minor allele |
Major allele |
No PoAF |
Yes PoAF |
OR (95%CI) | P‡ | No PoAF |
Yes PoAF |
OR (95%CI) | P‡ | Meta-P |
| 2 | rs880726# | 236343827 | SH3BP4 I AGAP1 |
A | C | 0.15 | 0.23 | 1.98 (1.47–2.66) |
5.9×10−6 | 0.19 | 0.17 | 1.15 (0.67–1.97) |
0.62 | 2.5×10−5 |
| 3 | rs17711446# | 55534264 | LOC100289467 I ERC2 |
A | G | 0.11 | 0.18 | 2.12 (1.51–2.97) |
1.2×10−5 | 0.13 | 0.16 | 1.46 (0.79–2.69) |
0.22 | 1.0×10−5 |
| 4 | rs4425335# | 169618128 | PALLD | G | A | 0.22 | 0.31 | 1.75 (1.34–2.28) |
4.2×10−5 | 0.29 | 0.23 | 1.04 (0.64–1.69) |
0.88 | 0.0002 |
| 6 | rs6455 | 32006896 | CYP21A2* | G | C | 0.015 | 0.047 | 5.61 (2.75–11.42) |
2.1×10−6 | 0.025 | 0.029 | 1.96 (0.51–7.58) |
0.33 | 3.2×10−6 |
| 6 | rs9349540# | 50364629 | DEFB112 I TFAP2D |
G | A | 0.35 | 0.25 | 0.58 (0.45–0.75) |
4.6×10−5 | 0.31 | 0.31 | 0.96 (0.60–1.54) |
0.86 | 0.0003 |
| 8 | rs10504554 | 74919713 | LY96 | G | A | 0.18 | 0.12 | 0.48 (0.34–0.68) |
2.9×10−5 | 0.18 | 0.17 | 0.55 (0.31–0.99) |
0.046 | 4.0×10−6 |
| 9 | rs7870498 | 77146468 | RORB | G | A | 0.49 | 0.38 | 0.59 (0.46–0.74) |
1.0×10−5 | 0.49 | 0.48 | 0.85 (0.57–1.27) |
0.42 | 2.5×10−5 |
| 9 | rs1323369# | 77148835 | RORB | G | A | 0.49 | 0.38 | 0.59 (0.47–0.75) |
1.4×10−5 | 0.5 | 0.48 | 0.82 (0.54–1.23) |
0.33 | 2.2×10−5 |
| 10 | rs11015781# | 18994583 | ARL5B I LOC100130846 |
A | C | 0.3 | 0.4 | 1.84 (1.43–2.37) |
2.6×10−6 | 0.32 | 0.34 | 1.2 (0.76–1.91) |
0.44 | 7.1×10−6 |
| 10 | rs2210723# | 110233497 | LOC100128304 I LOC645318 |
G | A | 0.062 | 0.11 | 2.41 (1.60–3.63) |
2.6×10−5 | 0.08 | 0.077 | 0.92 (0.44–1.93) |
0.82 | 0.0004 |
| 10 | rs12242854# | 110259916 | LOC100128304 I LOC645318 |
A | G | 0.062 | 0.11 | 2.41 (1.60–3.63) |
2.8×10−5 | 0.08 | 0.077 | 0.91 (0.44–1.92) |
0.82 | 0.0004 |
| 12 | rs4760332 | 58222672 | CTDSP2 | A | C | 0.28 | 0.38 | 1.7 (1.34–2.16) |
1.4×10−5 | 0.31 | 0.31 | 0.78 (0.48–1.22) |
0.26 | 0.0008 |
| 12 | rs10783850 | 58229377 | CTDSP2 | G | A | 0.28 | 0.38 | 1.65 (1.30–2.09) |
4.7×10−5 | 0.31 | 0.31 | 0.75 (0.47–1.19) |
0.22 | 0.002 |
| 14 | rs2241121 | 89826113 | FOXN3 | A | G | 0.21 | 0.13 | 0.49 (0.35–0.68) |
2.1×10−5 | 0.22 | 0.16 | 0.67 (0.38–1.17) |
0.16 | 1.1×10−5 |
| 14 | rs1612612# | 93497111 | ITPK1 I C14orf85 |
A | G | 0.24 | 0.33 | 1.73 (1.33–2.25) |
4.1×10−5 | 0.26 | 0.21 | 0.58 (0.34–0.98) |
0.04 | 0.006 |
| 17 | rs1399293 | 63254424 | RGS9 I AXIN2 | G | A | 0.12 | 0.06 | 0.35 (0.23–0.55) |
4.9×10−5 | 0.071 | 0.13 | 1.74 (0.92–3.31) |
0.09 | 0.0055 |
| 17 | rs16961956# | 63259951 | RGS9 I AXIN2 | A | G | 0.11 | 0.06 | 0.36 (0.23–0.57) |
1.3×10−5 | 0.071 | 0.12 | 1.61 (0.84–3.07) |
0.15 | 0.006 |
| 17 | rs17763241 | 63257153 | RGS9 I AXIN2 | A | G | 0.11 | 0.06 | 0.38 (0.24–0.59) |
2.6×10−5 | 0.71 | 0.12 | 1.61 (0.84–3.08) |
0.15 | 0.009 |
| 20 | rs6099823 | 56469998 | PMEPA1 I LOC100129869 |
A | G | 0.07 | 0.13 | 2.21 (1.51–3.22) |
4.1×10−6 | 0.12 | 0.054 | 0.38 (0.16–0.88) |
0.02 | 0.005 |
Chr, chromosome; MAF, minor allele frequency in cases and controls; OR, multivariate odds ratio; PoAF, postoperative atrial fibrillation; SNP, single-nucleotide polymorphism
Intergenic region is expressed with “|” between two franking genes; SNP rs6455 in CYP21A2 is a coding SNP.
Adjusted for postoperative atrial fibrillation risk index as a continuous variable.
Indicates imputed markers in the CATHGEN cohort
Of the 19 SNPs analyzed in the replication cohort (Table 2), 10 SNPs were based on the imputed genotype, due to the difference between the 2 BeadChips used in these 2 datasets. We identified 3 SNPs with nominal significance (P < 0.05) in the replication dataset. However, only the minor allele of rs10504554, in the intronic region of the lymphocyte antigen 96 gene (LY96, allele G), had the same direction of the effect indicating a decreased risk for postoperative AF in both datasets (discovery dataset: OR = 0.48; 95% CI: 0.34–0.68; P = 2.9×10−5, and replication dataset: OR = 0.55; 95% CI: 0.31–0.99; P = 0.046). The meta-analysis of both datasets by METAL showed that this SNP remained significantly associated with postoperative AF (meta-P = 4.0×10−6) (Table 2).
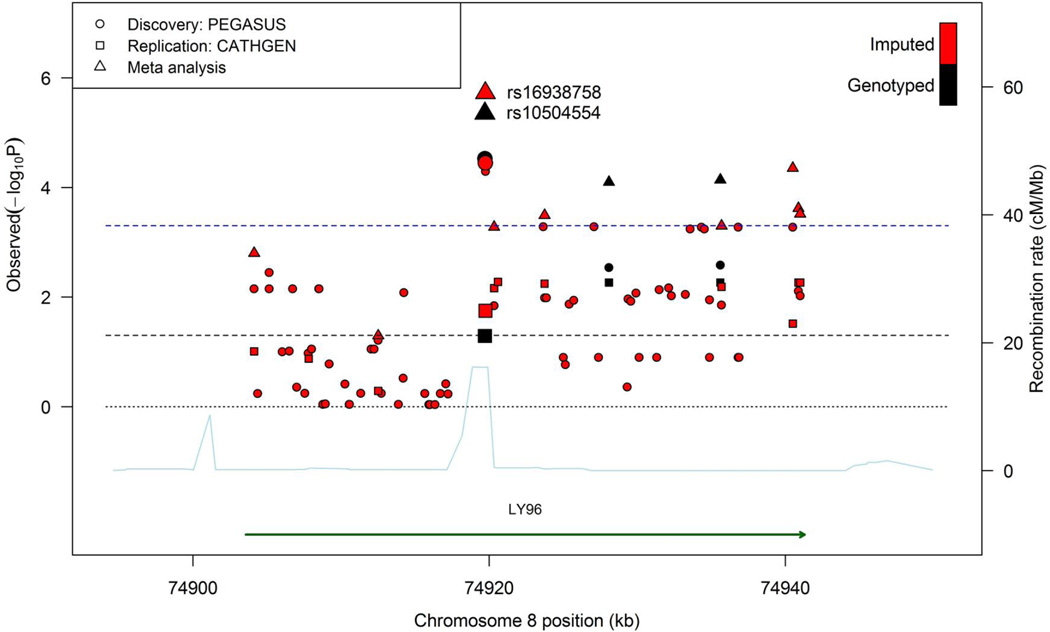
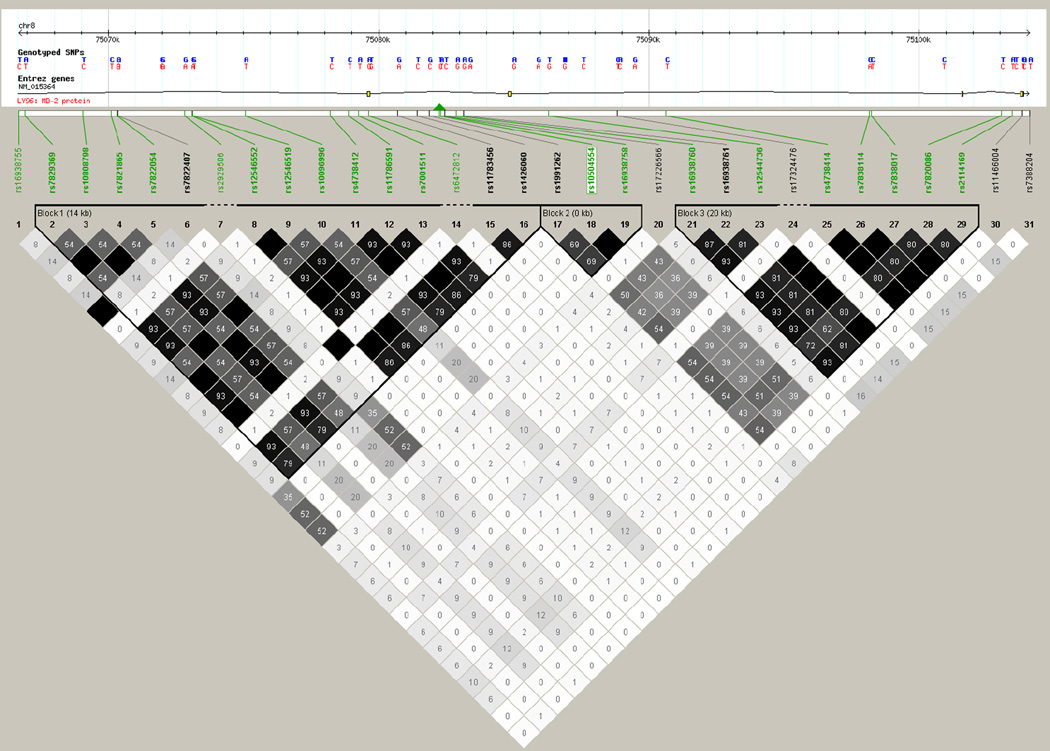
We increased the marker density in LY96 using a total of 71 imputed markers (MAF ≥ 0.02) to identify the SNP that was most strongly associated with postoperative AF, and potentially close to the causal variant in this gene. The average distance between markers is 526.71 bp (SD, 677.21 bp). Figure 1 depicts all genotyped and imputed markers investigated in this gene. The imputed SNP rs16938758 showed the most significant association with postoperative AF from the meta-analysis, and rs10504554 remained the most significant marker among the genotyped and imputed markers (Supplemental Table 2). Furthermore, when we adjusted for the effect of rs10504554 via the conditional association analysis, none of the SNPs, including rs16938758 (P=0.7) in LY96 gene region showed significant association with postoperative AF while rs10504554 remained significantly associated with postoperative AF. This was due to the strong LD between rs10504554 and rs16938958 (r2=0.97), as also indicated by Figure 2 where rs10504554 and rs16938758 are in the same LD block.
Figure 1.
Association plot of the LY96 region with high-density coverage by genotyped and imputed markers in the discovery dataset. Two adjacent SNPs, rs16938758 (imputed) and rs10504554 (genotyped), showed consistent association with postoperative AF in both discovery (PEGASUS: Perioperative Genetics and Safety Outcome Study) and replication datasets (CATHGEN: catheterization genetics).
Figure 2.
Haploview plot of linkage disequilibrium for single-nucleotide polymorphisms (SNPs) in LY96 gene based on the discovery dataset. Each diamond represents the correlation (r2) between each pair of SNPs with darker shades representing stronger linkage disequilibrium. True haplotype blocks in the population are marked with black lines in the correlation plot. The green triangle indicates the location of the SNP we identified in the LY96 gene, based on the UCSC Genome Browser (http://genome.ucsc.edu). SNPs highlighted in bold are present in LD blocks. Uploaded markers are highlighted in green, and markers downloaded by Haploview are highlighted in black. The SNP, rs10504554 identified in our study from GWAS is highlighted in a white rectangle.
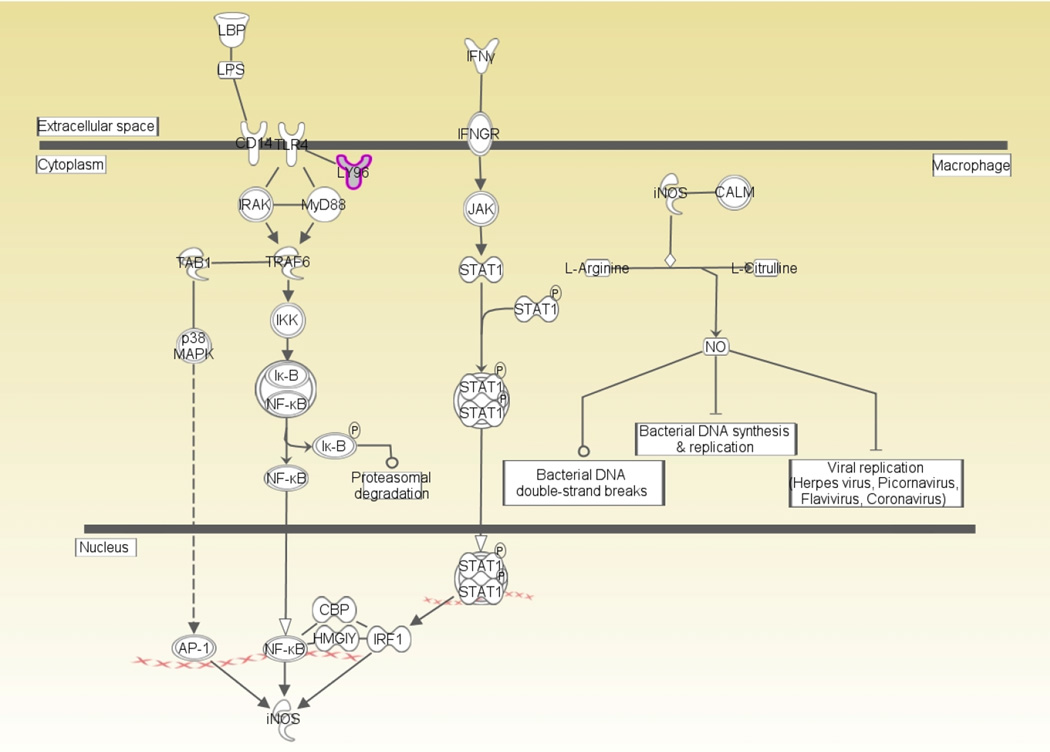
In parallel, 32 SNPs with P < 0.0001 in the discovery cohort were selected and uploaded into the IPA program for gene enrichment analysis. These SNPs were mapped to 12 unique genes, and the subsequent pathway analysis identified 19 nominally significant pathways (P < 0.05, Supplemental Table 3). We then examined the convergence between the results of the enrichment analysis and the results of the GWAS. This analysis highlighted the role of LY96 in several canonical pathways that are associated with activation and modulation of innate immune responses, and are potentially relevant to postoperative AF (Supplemental Figure 2 and Table 3). Of these pathways, the iNOS pathway (Figure 3), which is activated via toll-like receptor 4 (TLR-4)-LY96 receptor complex activation, has been previously identified as a relevant pathway to AF.19 Therefore, we analyzed the potential gene-gene interaction between LY96 SNP and each of the 4 functionally relevant candidate genes in the iNOS-related signaling pathway (Figure 3 and Supplemental Table 4) in terms of their association with postoperative AF. We found a nominally significant interaction between rs230530 in nuclear factor NF-kappa-B (NFκB1) and rs10504554 in LY96 that was associated with a decreased risk for postoperative AF (OR = 0.50; 95% CI: 0.29–0.86; P = 0.01), and that had the same level and direction of the effect as the rs10504554 alone (OR = 0.48; 95% CI: 0.34–0.68; P = 2.9 × 10−5).
Figure 3. The inducible nitric oxide synthase pathway.
AP-1, activator protein-1; CALM, calmodulin; CBP, cAMP response element-binding protein; CD14, cluster of differentiation; HMGIY, high-mobility group protein; Iκ-B, inhibitor of kappaB protein; IFNγ, interferon gamma; IFNGR, interferon-gamma receptor; IκB-NFκB, inhibitor of kappaB protein-NFkappaB; IKK, inhibitor-of-kappaB-protein kinase; iNOS, inducible nitric oxide synthase; IRAK, interleukin-1 receptor-associated kinase; IRF1, interferon regulatory factor-1; JAK, janus kinase; LBP, lipopolysaccharide binding protein; LPS, lipopolysaccharide; LY96, lymphocyte antigen 96; MyD88, myeloid differentiation primary response 88; NF-κB, nuclear factor-kappaB; NO, nitric oxide; p38MAPK, p38 mitogen-activated protein kinase; STAT1, signal transducer and activator of transcription 1; STAT1 dimer, signal transducer and activator of transcription 1 dimer; TAB1, transforming growth factor-beta activated kinase 1; TLR4, toll-like receptor 4; TRAF6, tumor necrosis factor receptor-associated factor 6.
Finally, we examined the relationship between SNPs identified in prior GWAS of AF and postoperative AF in our discovery dataset. Nominally significant associations were found between some of these genetic variants and incident postoperative AF (Supplemental Table 5), but these associations did not reach our pre-defined discovery threshold of P < 5 × 10−5 that was required to be selected for replication in the CATHGEN cohort. Further, we found no evidence for an association between incident postoperative AF in our covariate-adjusted analysis and the genetic variations at the 4q25 (PITX2) locus (Supplemental Table 6), which were previously found to be associated with AF in both ambulatory11, 12 and cardiac surgery cohorts.13
CONCLUSIONS
Using a genome-wide approach, we identified genetic variants in the LY96 gene that was associated with a reduced risk for postoperative AF in patients who underwent CABG with CPB. The encoded protein for the LY96 gene, LY96, is also known as myeloid differentiation protein 2 (MD-2), and is an essential co-receptor for TLR-4 signaling. Importantly, our findings suggest an independent association even after adjusting for clinical and procedural variables known to predict a risk for postoperative AF. Consistent with this finding, which was based on a combination of the genome-wide association results and a gene enrichment analysis, we also found several innate immune signaling pathways that were significant and potentially relevant to the development of postoperative AF. Within one of these pathways – the inducible nitric oxide synthase (iNOS) pathway – we observed a significant gene-gene interaction between LY96 and NFκB1. These findings add to mounting data that implicate a central role for inflammation in the development of postoperative AF, and suggest that LY96 may represent a novel target in a strategy to prevent postoperative AF after CABG surgery.
TLR4 is a prominent member of a family of pattern-recognition receptors that initiate the innate immune response to microbial pathogens.20 While TLR4 is expressed primarily by cells of myeloid lineage, it is also expressed in tissues without a recognized immune function, such as the heart and vasculature.21 In the heart, TLR4 is essential for LPS-induced LV dysfunction and for myocardial expression of tumor necrosis factor α (TNFα), interleukin (IL)-1β, and iNOS.22, 23 Furthermore, TLR4 plays a critical role in initiating the intense inflammatory response to myocardial ischemia-reperfusion, since TLR4-deficient murine strains are protected from myocardial infarction caused by ischemia-reperfusion injury.24
As noted above, the encoded protein for the LY96 gene is the TLR-4 co-receptor LY96, which is associated with the extracellular domain of TLR-4. This co-receptor is essential for TLR-4 pathway activation, whether by LPS,20 or through the binding of endogenous ligands such as heat-shock proteins (HSP) or fibrinogen.25 Indeed, ischemia-reperfusion injury in myocardial cells after CABG surgery with CPB induces the release of HSP70, an intracellular protein involved in protein folding and transport.26 Dybdahl et al21 discovered an early peak in inducible HSP70 in plasma after CABG surgery, which may originate from myocardial and coronary endothelial cells stressed by ischemia, or from blood cells damaged by the CPB machine. Their additional experiments also revealed that HSP70 is involved in activating the innate immune system by binding to the TLR-4 complex as an endogenous ligand. However, the downstream effect of TLR-4 complex activation and the resulting cytokine response to HSP70 could be inhibited by TLR-4 monoclonal antibodies.21
Few have studied the functional role and consequences of genetic variations in LY96. Nevertheless, recent evidence suggests that genetic variations in LY96 may have biological significance in the pathogenesis of sepsis and multiorgan dysfunction syndrome. For example, an SNP (rs11465996) within LY96 in the Chinese Han population has been associated with a higher sepsis morbidity rate and higher multiple organ dysfunction scores in patients with major trauma.27 This polymorphism was also associated with significantly higher TNFα production by peripheral blood leukocytes in response to LPS stimulation.27 Notably, this SNP was not represented on the Illumina Human610-Quad BeadChip used to genotype our discovery cohort. Using imputed genotype data for this SNP in the discovery cohort did not show a statistically significant association with postoperative AF (rs11775193: OR = 1.09; 95% CI: 0.85–1.40; P = 0.49; and rs2291217: OR = 1.06; 95% CI: 0.82–1.35; P = 0.67). Likewise, in a Caucasian patient undergoing surgery for cancer, Hanmann et al28 found that a rare coding mutation within the first exon in LY96 caused an amino acid exchange from threonine 35 to alanine, resulting in reduced LPS-induced signaling.
Together, these findings indicate a potential role for LY96 genetic variations in LPS responsiveness and disease susceptibility, and also highlight the potential importance of LY96 with its co-receptor TLR-4 in noninfectious disease processes such as activation of the innate immune system in response to myocardial ischemia-reperfusion injury.29 In fact, inhibition of the TLR-4-LY96 complex with eritoran, a synthetic analog of lipid A and a competitive antagonist of the LY96 receptor, terminates TLR-4-LY96-mediated signaling, due to proximal inhibition of innate immune responses.30 Furthermore, by inhibiting the TLR-4-LY96 complex, eritoran attenuates the inflammatory response to myocardial ischemia-reperfusion injury, as evidenced by decreased NFκB nuclear translocation, and decreased expression of inflammatory mediators.29
Consistent with the single-marker analysis above, we also identified several innate immune cellular pathways that were significant and potentially relevant to postoperative AF. Intriguingly, the most significant of these was the “hepatic fibrosis/hepatic stellate cell activation” pathway. The activation and signaling events of this pathway are similar to the fibroproliferative signaling pathways observed in the development and modulation of atrial fibrosis.12 Atrial fibrosis may result from a variety of cardiac pathological processes including senescence, cardiac dysfunction, mitral valve disease, and possibly myocardial ischemia.31 Indeed, Sinno et al32 proposed that coronary artery disease predisposes the atria to chronic ongoing ischemia, creating a substrate for atrial conduction slowing, and modulation of electrical remodeling processes leading to increased vulnerability to AF.12, 31
We also found that the iNOS signaling pathway, which can be activated via TLR-4-LY96 complex activation, is another potentially relevant pathway in the pathogenesis of AF.33 When we performed a gene-gene interaction analysis of the genes encoding the functional signaling molecules of this pathway, we observed a nominally significant interaction between LY96 and NFκB1, indicating that epistasis of the genes in this pathway can potentially contribute to changes in the response to activation of the TLR4-LY96 signaling and expression of NFκB that are potentially relevant to postoperative AF..
Injury caused by reperfusion after myocardial ischemia is associated with induction and activation of macrophages, neutrophils, endothelial cells, and cardiomyocytes, which release proinflammatory mediators such as cytokines and iNOS.34 In the atrial myocardium, expression of iNOS results in excess nitric oxide production, which in turn, leads to the generation of reactive oxygen species and oxyradical-mediated myocardial injury.33 The resulting oxidative damage to the atrial myocardium can contribute to atrial contractile dysfunction, which has been previously implicated in the pathogenesis of AF.35
In support of a role for local inflammatory responses in AF, a recent study of human atrial samples from patients with persistent AF who underwent cardiac surgery, revealed active adhesion and recruitment of macrophages across the endocardium in human fibrillating atria, indicating a potential role for immune cells in oxidative stress-mediated atrial pathogenesis and AF.36 Similarly, Han et al19 demonstrated that patients with persistent AF who underwent mitral valve replacement surgery, expressed higher levels of iNOS compared to patients with normal sinus rhythm. This finding was associated with protein nitration, cardiomyocyte apoptosis, and increased atrial and systemic inflammation. Indeed, selective inhibition of iNOS reduced myocyte apoptosis and myocardial damage in an animal ischemia-reperfusion model.37, 38 Although the time sequence and interrelationship between inflammation and AF perpetuation in the setting of cardiac surgery-related postoperative AF have not yet been explored, these preliminary observations support the notion that induction of iNOS and activation of subsequent inflammatory responses contribute to the substrates for AF.
The etiology of postoperative AF after cardiac surgery is multifactorial. Several potential genetic and nongenetic factors have been implicated. Among the previously implicated genetic factors, noncoding polymorphisms within the chromosome 4q25 region are associated with the development of postoperative AF in both ambulatory11, 12 and cardiac surgery cohorts.13 Although in our study, the direction of the effect size of the most frequently described SNP for PITX2 – rs2200733 – compared favorably to these previous studies, its magnitude was smaller (OR = 1.33 in our study; 1.71 in ambulatory patients;12 and 2.14 in cardiac surgery patients13). This discrepancy may be due to differences in sample size, variation in study design and patient population, or differences in allele frequencies. In our recent candidate gene study, we found an association between variants of G protein-coupled receptor kinase 5 (GRK5) gene polymorphisms and postoperative AF in the discovery and replication datasets from a subset of patients who exclusively received perioperative beta-blocker therapy.15 In the current larger dataset, this association was still present in our discovery dataset (Supplemental Table 5), but did not reach our discovery threshold of P < 5 × 10−5 that was required for replication in the CATHGEN cohort. This may be due to the fact that in the current study, we expanded our study cohorts to include patients with and without perioperative beta-blocker therapy.
This study has potential limitations. First, since the identified SNP is a tagging marker in the intronic region of the LY96, the observed association could be due to changes in regulation of gene expression, or linkage disequilibrium with the true unobserved causal SNP(s). Therefore, future research to identify casual SNP(s) and to decipher their relationship to expression or protein level of LY96 is important. Second, based on current sample size and an incidence of postoperative AF of 25%, allele frequency of 0.16 as our top SNP (rs10504554), and complete LD between SNP and causal variant, our power calculations show that our study has 83% power to detect a genotypic relative risk of 0.56, which is equivalent to an odds ratio of 0.45. Thus, although we used a relatively large population of cardiac surgery patients, this study is powered to detect only common variants with relatively large effect sizes. Since only variants with minor allele frequencies > 0.02 were assessed, the possibility of rare genetic variants that drive a pronounced clinical phenotype was not explored. Third, non-genetic factors associated with postoperative AF include a number of transient phenomena such as heightened parasympathetic tone, atrial stretch, electrolyte shifts, metabolic abnormalities, and pericarditis.39 Due to our study design, we were not able to explore the effect of these factors on postoperative AF. Finally, patients enrolled in our study were Caucasians, and therefore our findings cannot be generalized to other ethnic groups.
In conclusion, we present the results of a genome-wide association study in a cohort of patients at risk for postoperative AF after CABG surgery. Based on our integrated approach using postoperative AF as a well defined phenotype, and performing single-marker, pathway, and gene-gene interaction analyses, we identified a single-nucleotide polymorphism in the LY96 gene and its gene-gene interaction with NFκB1 in the iNOS-related signaling pathway. Not only does this finding add to the mounting evidence that innate immune responses play a central role in the development of postoperative AF, but it also provides novel targets for intervention to prevent this potentially devastating complication.
Supplementary Material
Acknowledgments
The authors thank Dr. Elizabeth R. Hauser, PhD (Professor of Biostatistics and Bioinformatics, Duke University Medical Center) for her help in reviewing the manuscript and providing constructive suggestions. Finally, we thank Kathy Gage (Research Development Associate, Department of Anesthesiology, Duke University Medical Center) for her editorial contributions to the manuscript.
Appendix: Duke Perioperative Genetics and Safety Outcomes (PEGASUS) Investigative Team Members
Allen AS, Davis RD, Fontes MJ, Funk B, Gaca JG, Ginsburg GS, Glower DD, Goldstein DB, Hall RL, Hauser E, Jones R, Kertai MD, Laskowitz DT, Li YJ, Lodge AJ, Mathew JP, Milano CA, Newman MF, Phillips-Bute B, Podgoreanu MV, Smith MP, Smith PK, Stafford-Smith M, Swaminathan M, Welsby IJ, White WD, and Willard HF.
Footnotes
Conflict of Interest Disclosure
William E. Kraus and Svati H. Shah report conflicts of interests that are not related to the content of the manuscript.
REFERENCES
- 1.Mathew JP, Fontes ML, Tudor IC, Ramsay J, Duke P, Mazer CD, et al. A multicenter risk index for atrial fibrillation after cardiac surgery. Jama. 2004;291(14):1720–1729. doi: 10.1001/jama.291.14.1720. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell LB. Incidence, timing and outcome of atrial tachyarrhythmias after cardiac surgery. In: Steinberg JS, editor. Atrial Fibrillation after Cardiac Surgery. US: Springer; 2000. pp. 37–50. [Google Scholar]
- 3.Borzak S, Tisdale JE, Amin NB, Goldberg AD, Frank D, Padhi ID, et al. Atrial fibrillation after bypass surgery: does the arrhythmia or the characteristics of the patients prolong hospital stay? Chest. 1998;113(6):1489–1491. doi: 10.1378/chest.113.6.1489. [DOI] [PubMed] [Google Scholar]
- 4.Echahidi N, Pibarot P, O’Hara G, Mathieu P. Mechanisms, prevention, and treatment of atrial fibrillation after cardiac surgery. J Am Coll Cardiol. 2008;51(8):793–801. doi: 10.1016/j.jacc.2007.10.043. [DOI] [PubMed] [Google Scholar]
- 5.Mathew JP, Collard CD, Fontes M, Miao Y, Mangano DT. Perioperative statin therapy decreases the risk of atrial fibrillation after cardiac surgery. Anesth Analg. 2009;108:SCA85. [Google Scholar]
- 6.Shen J, Lall S, Zheng V, Buckley P, Damiano RJ, Jr, Schuessler RB. The persistent problem of new-onset postoperative atrial fibrillation: a single-institution experience over two decades. J Thorac Cardiovasc Surg. 2011;141(2):559–570. doi: 10.1016/j.jtcvs.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariscalco G, Biancari F, Zanobini M, Cottini M, Piffaretti G, Saccocci M, et al. Bedside tool for predicting the risk of postoperative atrial fibrillation after cardiac surgery: the POAF score. J Am Heart Assoc. 2014;3(2):e000752. doi: 10.1161/JAHA.113.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldron NH, Cooter M, Klinger RY, Kertai MD, Podgoreanu MV, Stafford-Smith M, et al. Improved ability to predict postoperative atrial fibrillation with the McSPI AFRisk index versus the CHA2DS2Vasc or POAF scores. 37th Annual SCA Annual Meeting & Workshop; Washington, D. C.. 2015. p. 30. [Google Scholar]
- 9.Amar D, Shi W, Hogue CW, Jr, Zhang H, Passman RS, Thomas B, et al. Clinical prediction rule for atrial fibrillation after coronary artery bypass grafting. J Am Coll Cardiol. 2004;44(6):1248–1253. doi: 10.1016/j.jacc.2004.05.078. [DOI] [PubMed] [Google Scholar]
- 10.Magee MJ, Herbert MA, Dewey TM, Edgerton JR, Ryan WH, Prince S, et al. Atrial fibrillation after coronary artery bypass grafting surgery: development of a predictive risk algorithm. Ann Thorac Surg. 2007;83(5):1707–1712. doi: 10.1016/j.athoracsur.2006.12.032. discussion 1712. [DOI] [PubMed] [Google Scholar]
- 11.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44(6):670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darbar D, Roden DM. Genetic mechanisms of atrial fibrillation: impact on response to treatment. Nat Rev Cardiol. 2013;10(6):317–329. doi: 10.1038/nrcardio.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Body SC, Collard CD, Shernan SK, Fox AA, Liu KY, Ritchie MD, et al. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2(5):499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudino M, Andreotti F, Zamparelli R, Di Castelnuovo A, Nasso G, Burzotta F, et al. The −174G/C interleukin-6 polymorphism influences postoperative interleukin-6 levels and postoperative atrial fibrillation. Is atrial fibrillation an inflammatory complication? Circulation. 2003;108(Suppl 1):II195–II199. doi: 10.1161/01.cir.0000087441.48566.0d. [DOI] [PubMed] [Google Scholar]
- 15.Kertai MD, Li YW, Li YJ, Shah SH, Kraus WE, Fontes ML, et al. G protein-coupled receptor kinase 5 gene polymorphisms are associated with postoperative atrial fibrillation after coronary artery bypass grafting in patients receiving beta-blockers. Circ Cardiovasc Genet. 2014;7(5):625–633. doi: 10.1161/CIRCGENETICS.113.000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 18.Li YJ, Goh L, Khor CC, Fan Q, Yu M, Han S, et al. Genome-wide association studies reveal genetic variants in CTNND2 for high myopia in Singapore Chinese. Ophthalmology. 2011;118(2):368–375. doi: 10.1016/j.ophtha.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han W, Fu S, Wei N, Xie B, Li W, Yang S, et al. Nitric oxide overproduction derived from inducible nitric oxide synthase increases cardiomyocyte apoptosis in human atrial fibrillation. Int J Cardiol. 2008;130(2):165–173. doi: 10.1016/j.ijcard.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Oblak A, Jerala R. The molecular mechanism of species-specific recognition of lipopolysaccharides by the MD-2/TLR4 receptor complex. Mol Immunol. 2015;63(2):134–142. doi: 10.1016/j.molimm.2014.06.034. [DOI] [PubMed] [Google Scholar]
- 21.Dybdahl B, Wahba A, Lien E, Flo TH, Waage A, Qureshi N, et al. Inflammatory response after open heart surgery: release of heat-shock protein 70 and signaling through toll-like receptor-4. Circulation. 2002;105(6):685–690. doi: 10.1161/hc0602.103617. [DOI] [PubMed] [Google Scholar]
- 22.Pinsky DJ, Cai B, Yang X, Rodriguez C, Sciacca RR, Cannon PJ. The lethal effects of cytokine-induced nitric oxide on cardiac myocytes are blocked by nitric oxide synthase antagonism or transforming growth factor beta. J Clin Invest. 1995;95(2):677–685. doi: 10.1172/JCI117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishiyama S, Hiroe M, Nishikawa T, Abe S, Shimojo T, Ito H, et al. Nitric oxide contributes to the progression of myocardial damage in experimental autoimmune myocarditis in rats. Circulation. 1997;95(2):489–496. doi: 10.1161/01.cir.95.2.489. [DOI] [PubMed] [Google Scholar]
- 24.Oyama J, Blais C, Jr, Liu X, Pu M, Kobzik L, Kelly RA, et al. Reduced myocardial ischemia-reperfusion injury in toll-like receptor 4-deficient mice. Circulation. 2004;109(6):784–789. doi: 10.1161/01.CIR.0000112575.66565.84. [DOI] [PubMed] [Google Scholar]
- 25.Guo LH, Schluesener HJ. The innate immunity of the central nervous system in chronic pain: the role of Toll-like receptors. Cell Mol Life Sci. 2007;64(9):1128–1136. doi: 10.1007/s00018-007-6494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demidov ON, Tyrenko VV, Svistov AS, Komarova YY, Karpishenko AI, Margulis BA, et al. Heat shock proteins in cardiosurgery patients. Eur J Cardiothorac Surg. 1999;16(4):444–449. doi: 10.1016/s1010-7940(99)00291-2. [DOI] [PubMed] [Google Scholar]
- 27.Zeng L, Zhang AQ, Gu W, Zhou J, Zhang LY, Du DY, et al. Identification of haplotype tag SNPs within the whole myeloid differentiation 2 gene and their clinical relevance in patients with major trauma. Shock. 2012;37(4):366–372. doi: 10.1097/SHK.0b013e3182498c8f. [DOI] [PubMed] [Google Scholar]
- 28.Hamann L, Kumpf O, Muller M, Visintin A, Eckert J, Schlag PM, et al. A coding mutation within the first exon of the human MD-2 gene results in decreased lipopolysaccharide-induced signaling. Genes Immun. 2004;5(4):283–288. doi: 10.1038/sj.gene.6364068. [DOI] [PubMed] [Google Scholar]
- 29.Shimamoto A, Chong AJ, Yada M, Shomura S, Takayama H, Fleisig AJ, et al. Inhibition of Toll-like receptor 4 with eritoran attenuates myocardial ischemia-reperfusion injury. Circulation. 2006;114(1 Suppl):I270–I274. doi: 10.1161/CIRCULATIONAHA.105.000901. [DOI] [PubMed] [Google Scholar]
- 30.Opal SM, Laterre PF, Francois B, LaRosa SP, Angus DC, Mira JP, et al. Effect of eritoran, an antagonist of MD2-TLR4, on mortality in patients with severe sepsis: the ACCESS randomized trial. Jama. 2013;309(11):1154–1162. doi: 10.1001/jama.2013.2194. [DOI] [PubMed] [Google Scholar]
- 31.Burstein B, Nattel S. Atrial fibrosis: mechanisms and clinical relevance in atrial fibrillation. J Am Coll Cardiol. 2008;51(8):802–809. doi: 10.1016/j.jacc.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 32.Sinno H, Derakhchan K, Libersan D, Merhi Y, Leung TK, Nattel S. Atrial ischemia promotes atrial fibrillation in dogs. Circulation. 2003;107(14):1930–1936. doi: 10.1161/01.CIR.0000058743.15215.03. [DOI] [PubMed] [Google Scholar]
- 33.Bonilla IM, Sridhar A, Gyorke S, Cardounel AJ, Carnes CA. Nitric oxide synthases and atrial fibrillation. Front Physiol. 2012;3:105. doi: 10.3389/fphys.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wildhirt SM, Weismueller S, Schulze C, Conrad N, Kornberg A, Reichart B. Inducible nitric oxide synthase activation after ischemia/reperfusion contributes to myocardial dysfunction and extent of infarct size in rabbits: evidence for a late phase of nitric oxide-mediated reperfusion injury. Cardiovasc Res. 1999;43(3):698–711. doi: 10.1016/s0008-6363(99)00080-2. [DOI] [PubMed] [Google Scholar]
- 35.Mihm MJ, Yu F, Carnes CA, Reiser PJ, McCarthy PM, Van Wagoner DR, et al. Impaired myofibrillar energetics and oxidative injury during human atrial fibrillation. Circulation. 2001;104(2):174–180. doi: 10.1161/01.cir.104.2.174. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita T, Sekiguchi A, Iwasaki YK, Date T, Sagara K, Tanabe H, et al. Recruitment of immune cells across atrial endocardium in human atrial fibrillation. Circ J. 2010;74(2):262–270. doi: 10.1253/circj.cj-09-0644. [DOI] [PubMed] [Google Scholar]
- 37.Dohi K, Ohtaki H, Inn R, Ikeda Y, Shioda HS, Aruga T. Peroxynitrite and caspase-3 expression after ischemia/reperfusion in mouse cardiac arrest model. Acta Neurochir Suppl. 2003;86:87–91. doi: 10.1007/978-3-7091-0651-8_20. [DOI] [PubMed] [Google Scholar]
- 38.Ramasamy R, Hwang YC, Liu Y, Son NH, Ma N, Parkinson J, et al. Metabolic and functional protection by selective inhibition of nitric oxide synthase 2 during ischemia-reperfusion in isolated perfused hearts. Circulation. 2004;109(13):1668–1673. doi: 10.1161/01.CIR.0000124489.46660.2E. [DOI] [PubMed] [Google Scholar]
- 39.Rho RW. The management of atrial fibrillation after cardiac surgery. Heart. 2009;95(5):422–429. doi: 10.1136/hrt.2007.132795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.